Abstract
Background
Long noncoding RNAs (lncRNAs) play a key role in the development and progression of many cancer types, including lung cancer. The objective of this study is to examine the function and molecular mechanism of lncRNAs involved in non-small cell lung cancer (NSCLC).
Methods
First, 7 lung cancer-related differentially expressed LncRNAs were screened from 2 genomic profiling datasets. Of these lncRNAs, FOXF1 adjacent noncoding developmental regulatory RNA (FENDRR) was found to be the only one that was both significantly down-regulated in the patients with advanced pathology and negatively correlated with prognosis. Thus, lncRNA FENDRR was further studied in this project. Clinical correlation analysis was further conducted in the GSE30219 dataset and 73 paired lung cancer and noncancerous tissues stored in our lab; Subsequently, we evaluated FENDRR coding potential with the Phylogenetic Codon Substitution Frequencies (PhyloCSF), Coding-Potential Assessment Tool (CPAT), and Coding Potential Calculator (CPC) online analytical tool. The cell growth ability was measured by CCK8 assay and clonogenicity assay, the metastatic capacities were evaluated using Transwell migration and invasion assays. Mechanistically, we analyzed the correlation of FENDRR function in NSCLC with immune response by utilizing The Cancer Genome Atlas (TCGA) data.
Results
Results indicated a negative clinical correlation of FENDRR. Coding potential analysis showed FENDRR as a noncoding RNA. Elevated expression of FENDRR led to cell growth arrest, inhibition of proliferative ability, declined migration and invasion potential of NSCLC cells in vitro. Mechanistically, we discovered that FENDRR expression might be involved in aberrant immune response regulation.
Conclusions
Taken together, our results provide a greater understanding of lncRNA FENDRR as a tumor suppressor with respect to tumor-immune interactions in NSCLC.
Keywords: Long noncoding RNA (lncRNA), FOXF1 adjacent noncoding developmental regulatory RNA (FENDRR), tumor progression, immune regulation, non-small cell lung cancer (NSCLC)
Introduction
Lung cancer is emerging as the leading cause of cancer death among men and women worldwide (1), with small-cell lung cancer accounting for nearly 15% and non-small cell lung cancer (NSCLC) accounting for nearly 85% of all lung cancer patients (2). The last decade has witnessed significant growth in therapeutic options for lung cancer patients; however, the expected survival time of advanced NSCLC patients is still only a few months (3). Metastases account for most cancer-related deaths and remain as a dominant contributor to a lethal outcome (4,5). The complex mechanism underlying lung cancer progression still remains the least understood aspect. Therefore, the potential therapeutic targets involved in NSCLC urgently need to be explored.
Noncoding RNAs (ncRNAs) are known to encode a large proportion of mammalian genomes (6,7). One type of ncRNA, long noncoding RNAs (lncRNAs), are typically longer than 200 nucleotides in length and have been found to participate in a variety of cellular functions (8). LncRNAs, as classified by genome position (9), consist of many types of RNA, including sense, antisense, bidirectional, intergenic, and intronic transcripts types, which may account for the diversity of cellular functions of lncRNAs. LncRNAs have been demonstrated to play a role in diverse cancer types, including lung cancer (10). For example, one study found that BLACAT2 (long intergenic non-protein coding RNA 958, Linc00958) expression was positively correlated with LN metastasis in bladder cancer (11), while other research has observed that lncRNA LINK-A mediated drug resistance of AKT (protein kinase B) inhibitors by interacting with PtdIns-3,4,5-P3 to hyperactivate AKT (12). In further studies, several other lncRNAs have been identified to be capable of dysregulating lung cancer. For example, lncRNA SNHG1 (small nucleolar RNA host gene 1) could sponge miR-145-5p to facilitate tumor progression of NSCLC by improving MTDH (metadherin) expression (13); epithelial-mesenchymal transition (EMT), and cell migration were facilitated by lncRNA linc00460 in lung cancer cell lines (14), and lncRNA IGFBP4-1 (insulin like growth factor binding protein 4-1) accelerated lung cancer progression by reprogramming energy metabolism (15). It was also found that lncRNA AGAP2-AS1 (AGAP2 antisense RNA 1) could interact with LSD1 (LSD1 zinc finger family protein) and EZH2 (enhancer of zeste 2 polycomb repressive complex 2 subunit) to inhibit LATS2 (large tumor suppressor kinase 2) and KLF2 (KLF2-Kruppel like factor 2) expression in NSCLC cells (16). Meanwhile, EMT, stemness, and metastasis were shown to be promoted by the downregulation of lncRNA FOXF1-AS1 (also named FENDRR) in NSCLC cells (17). However, the underlying functions and molecular mechanisms of lncRNAs involved in NSCLC progression remain partially understood.
In this study, based on lncRNA screening by bioinformatics and clinical correlation analysis, lncRNA FOXF1 adjacent noncoding developmental regulatory RNA (FENDRR) was further identified to be significantly downregulated in NSCLC tumor tissues compared with normal tissues, and FENDRR expression was negatively related with distal metastasis and pathological stages. FENDRR was further evaluated in vitro to determine its biological role in NSCLC cell line, and it was found that elevated FENDRR expression led to dramatic cell growth arrest, and the inhibition of the migration and invasion potential of NSCLC cell H1299. Moreover, TCGA datasets were utilized to identify changes in the tumor immune microenvironment. Our overall data, for the first time, demonstrate that FENDRR can suppress tumor progression of NSCLC and has a significant correlation with immune response. These findings may equip us more with approaches in the understanding of function and mechanism of FENDRR in NSCLC.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2147).
Methods
Human tissue samples
The Department of Lung Cancer at Shanghai Chest Hospital, Shanghai Jiao Tong University kindly provided 73 paired fresh human NSCLC tissues and matched adjacent noncancerous tissues. All clinical specimens were immediately frozen in liquid nitrogen and then stored at the condition of −80 °C for subsequent study. All specimens used in this study were obtained based on the informed consent from all patients. The study was conducted in accordance with the Declaration of Helsinki. The study was approved by Ethics Committee of The Department of Lung Cancer at Shanghai Chest Hospital, Shanghai Jiao Tong University (No. XS2017-007) and written informed consent was obtained from all patients.
Cell culture
NSCLC cell lines, NCI-H1299 cell line (ATCC, Cat# CRL-5803, RRID:CVCL_0060), A549 cell line (ATCC, Cat# CCL-185, RRID:CVCL_0023), NCI-H460 cell line (ATCC, Cat# HTB-177, RRID:CVCL_0459), NCI-H358 cell line (ATCC, Cat# CRL-5807, RRID:CVCL_1559) were used in this study. HEK293T cell line (ATCC, Cat# CRL-3216, RRID:CVCL_0063) was used for packaging of lentivirus containing FENDRR. All cell lines were maintained in Dulbecco Modified Eagle’s Medium (DMEM) (HyClone, Cat# SH30243.01) or Roswell Park Memorial Institute Medium (RPMI)-1640 (HyClone, Cat# SH30809.01B) supplemented 10% fetal bovine serum (FBS) (Yeasen Biotech, Cat# 40130ES76), 1% penicillin-streptomycin (Sigma-Aldrich, Cat A5955), at 37 °C in a humidified air atmosphere containing 5% CO2.
Analysis of the coding potential
Codon substitution frequencies (CSF) analysis (PhyloCSF) from the University of California Santa Cruz (UCSC) was utilized to analyze the coding potential of FENDRR. Two other online tools, the Coding Potential Calculator (CPC) and the Coding-Potential Assessment Tool (CPAT), were also used for potential noncoding analysis.
RNA isolation and real-time polymerase chain reaction (RT-PCR)
Total RNA of tissue samples and cells were extracted with TRIzol reagent (Invitrogen, Cat# 15596026, CA, USA), PrimeScript RT Reagent Kit (TaKaRa, Cat# RR036A) was used to synthesize cDNA, SYBR Green Premix Ex Taq (TaKaRa, Cat# RR820A) was used to conduct RT-PCR, The primers sequences used are listed in Table 1. β-actin served as the internal control.
Table 1. Primers used in this study.
| Name | Forward-primer | Reverse-primer |
|---|---|---|
| FENDRR | AGCCTACTCGTCAAAAGCCC | GCCTAGATCCGAAGGCTGTC |
| β-actin | GTCATTCCAAATATGAGATGCGT | GCATTACATAATTTACACGAAAGCA |
Subcellular fractionation
PARISTM Kit (Ambion, Cat#AM1921) was used to conduct the experiment. After cells were collected, according to the protocol from the kit, 500 µL of cell fractionation buffer was added to the cell pellets and incubated for 10 minutes on ice. Then, the tube was flicked to loosen the pellets and centrifuged at 4 °C for 5 minutes at a low speed to keep the pellets intact. Next, the supernatant was put into an RNase-free tube to extract cytoplasmic RNA. The nuclear pellets were then washed once with the cell fractionation buffer, and cold cell disruption buffer was added to lyse the pellets. At last, cellular fractions were split into 2 parts: cytoplasmic and nuclear fractions.
Construction of lentiviral vector and cell lines with stable gene expression
Full-length FENDRR was commercially synthesized (GENEWIZ), and then was cloned and inserted into vector pWPXL (A kind gift from Dr. Didier Trono) to produce lentiviral vector pWPXL FENDRR. Then, all 3 plasmids, including pWPXP FENDRR, lentiviral vector packaging system pMDG, and PSPAX2, were co-transfected into HEK293T cells mediated by Lipofectamine 2000 (Invitrogen, Cat# 11668-019). HEK293T cell culture medium was changed into DMEM without antibiotics before cell transfection. Six to 8 hours after cell transfection, the culture medium was changed back to DMEM containing antibiotics. The supernatant containing lentivirus was then collected from the dish after 48 hours, stored at −80 °C, or immediately added to a 6-well plate with corresponding cell lines drop by drop in a 1:1 ratio (800 µL virus: 800 µL DMEM).
In vitro cell proliferation assays
For the colony formation assay, 500 cells were seeded into 6-well plates for approximately 2 weeks of culture until the cells formed colonies. Then, 100% methanol was added to the plates to fix the colonies for 30 minutes, and then stained for 10–20 minutes with 0.1% crystal violet. For proliferation assays, 1,000 cells were seeded into 96-well plates, and then Cell Counting Kit-8 (CCK8) Assay Kit (Dojindo Corp, Cat# CK04, Japan) was used to detect the proliferative ability of seeded cells from the second day after seeding. According to the protocol of the kit, a mixture of 10 µL kit reagent and 90 µL DMEM was added to each well, and the absorbance was subsequently measured at 450 nm 2 hours later.
In vitro cell migration and invasion assays
We evaluated the cell migration and invasion potential using a 24-well plate with 8-µm-pore size chamber (Corning, Cat# 354480) inserts, and 5×104 cells suspended in 200 µL serum-free culture medium were seeded into the upper chamber with the non-coated membrane in the migration assay. Meanwhile, 1×105 cells suspended in 200 µL serum-free culture medium were seeded into the upper chamber with a Matrigel-coated membrane in the invasion assay. After this, 800 µL DMEM containing 10% FBS was added to the lower chamber. After culturing for an appropriate time, the cells migrated across the membrane to the outside surface, were fixed for 30 minutes with 100% methanol, and stained for 30 minutes with 0.1% crystal violet. Then, pictures were imaged and counted by an optical microscope (Olympus, Japan).
Statistical analysis
GraphPad Prism 5 was used for all statistical analyses. Two-tailed Student’s t-test was used to determine differences between 2 means. The log-rank Cox test was conducted to assess the difference in survival with the data being presented as the mean ± standard error of the mean (SEM). Only values with P<0.05 were considered as significant.
Results
LncRNA screening using Gene Expression Omnibus (GEO) profiling datasets
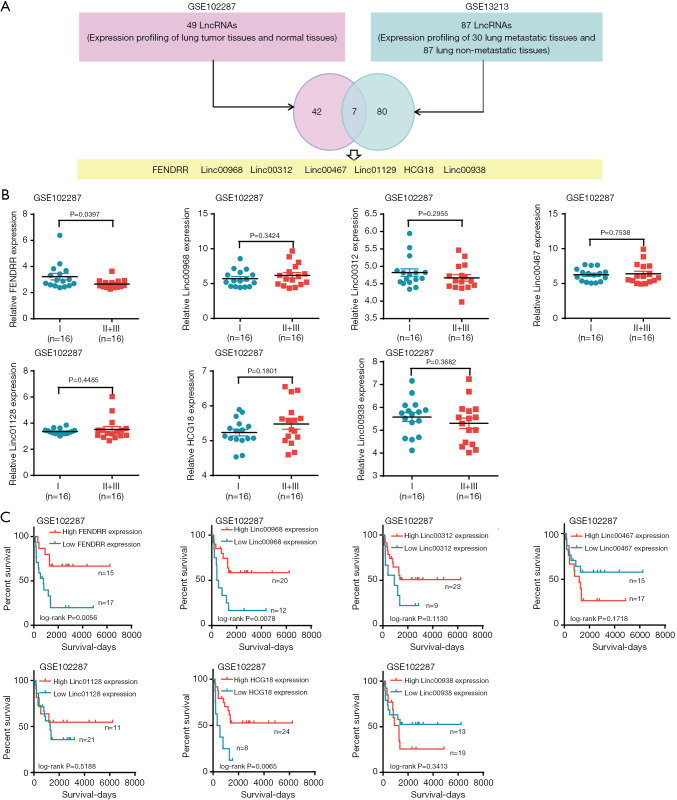
Two genomic profiling datasets (GSE102287, GSE13213) were analyzed with the online tool, “Analyze with 2R”. In total, 49 lncRNAs were screened from the expression profiling of lung tumor tissues and normal tissues in GSE102287, 87 lncRNAs were screened from the profiling of 30 metastatic lung tissues and 87 lung non-tumor tissues, and 7 lncRNAs present in both datasets were extracted for subsequent study (Figure 1A). To narrow the study scope, we further investigated the correlation of these lncRNAs with the advanced pathology and prognosis. The results showed that only FENDRR was negatively correlated with both advanced lung cancer pathology (Figure 1B) and high FENDRR expression related to good prognosis (Figure 1C).
Figure 1.
Long non-coding RNAs (lncRNAs) screening by genomics profiling datasets and clinical correlation analysis. (A) Seven lncRNAs extracted from GSE102287 and GSE13213 analyzed by online tool “Analyze with 2R”; (B) clinical advanced pathology correlation analysis of 7 lncRNAs in GSE102287; (C) Kaplan-Meier analysis of 7 lncRNAs in GSE102287.
FENDRR was negatively correlated with human NSCLC clinicopathologic features
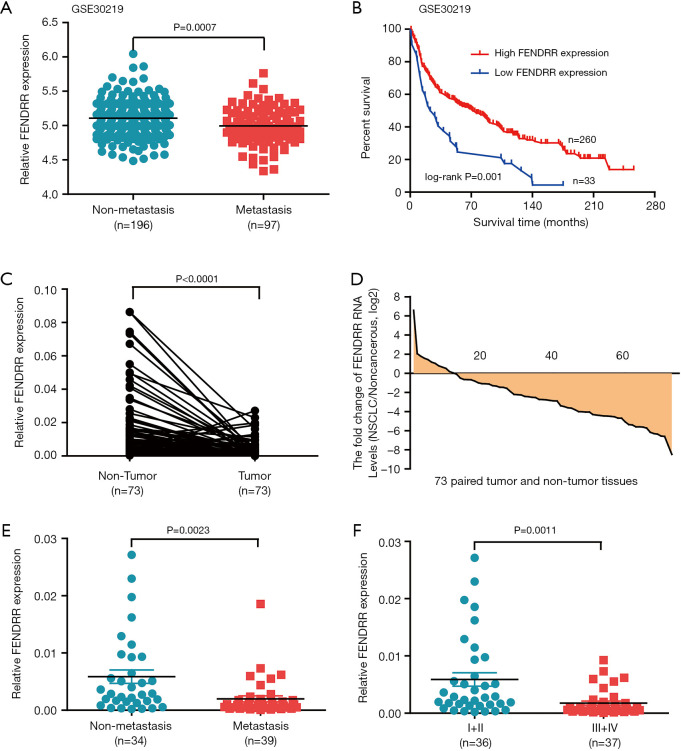
To further investigate the clinical correlation of FENDRR, we assessed the FENDRR expression in another GEO cohort (GSE30219). High FENDRR expression was negatively related to metastasis (Figure 2A), and, of particular note, elevated expression of FENDRR was significantly associated with good prognosis in GEO patient cohort (GSE30219) (Figure 2B). The correlation between lncRNA FENDRR RNA levels and human NSCLC clinicopathologic features were further confirmed in 73 pairs of clinical NSCLC tissues and matched non-tumorous tissues stored in our lab. FENDRR was shown to be remarkably reduced when compared to adjacent normal tissues (Figure 2C). Moreover, FENDRR was reduced in 83.6% (61/73) of NSCLC patient cases (Figure 2D), and the results indicated a negative association in FENDRR level and clinical metastasis and advanced pathological stages (Figure 2E,F). We further sought to determine whether FENDRR levels correlated with other clinicopathologic features of the NSCLC patients (Table 2), the results of which indicated no significant relationship between FENDRR expression and tumor size, gender, and age. As supplementary evidence, these results further support FENDRR as a suppressor of NSCLC.
Figure 2.
LncRNA FENDRR correlated with human non-small cell lung cancer (NSCLC) clinicopathologic features. (A,B) Low FENDRR expression is positively correlated with metastasis [lymph node metastasis and/or distal metastasis (A) and poor outcomes (B) in the 293-GEO-patient cohort of NSCLC tissues]; (C) FENDRR RNA levels were quantified in 73 pairs of NSCLC tissues and adjacent normal tissues using quantitative RT-PCR; (D) fold changes in 73 paired tissues (NSCLC/noncancerous: upregulated, >1; no change, −1 to 1; downregulated, <−1); (E,F) low FENDRR expression is positively correlated with metastasis [lymph node metastasis and/or distal metastasis (E) and TNM (tumor node metastasis) stage (F)]. Values represent mean ± SEM. Statistical analysis was performed using paired t-test (A) and Student’s t-test (A,E,F).
Table 2. The relationship between lncRNA FENDRR expression and their clinicopathologic parameters in 73 pairs of non-small cell lung cancer (NSCLC) patients.
| Clinicopathologic parameters | Number of cases | Median expression of FENDRR | |
|---|---|---|---|
| Mean ± SD | P value | ||
| Age | |||
| <60 years | 27 | 0.002609±0.0005843 | 0.1671 |
| ≥60 years | 46 | 0.004480±0.0009654 | |
| Gender | |||
| Male | 45 | 0.003879±0.0007820 | 0.8614 |
| Female | 28 | 0.003643±0.001160 | |
| Tumor size | |||
| ≤3 cm | 38 | 0.003528±0.0008352 | 0.6804 |
| >3 cm | 35 | 0.004070±0.001022 | |
| Degree of differentiation | |||
| Well and moderately | 48 | 0.002724±0.0006413 | 0.0225* |
| Poorly | 25 | 0.005831±0.001380 | |
| Local invasion | |||
| T1+T2 | 55 | 0.004486±0.0008257 | 0.0605 |
| T3+T4 | 18 | 0.001656±0.0005571 | |
| TNM stage | |||
| Stage I + II | 36 | 0.005882±0.001173 | 0.0011** |
| Stage III+IV | 37 | 0.001752±0.0003726 | |
| Metastasis | |||
| No | 36 | 0.005558±0.001128 | 0.0065** |
| Yes | 37 | 0.002066±0.0005507 | |
P value represents the probability from a student’s t-test for FENDRR expression between variable subgroups. *, P<0.05; **, P<0.01, which was considered to have a significant difference.
FENDRR was verified to be a lncRNA located in the nucleus
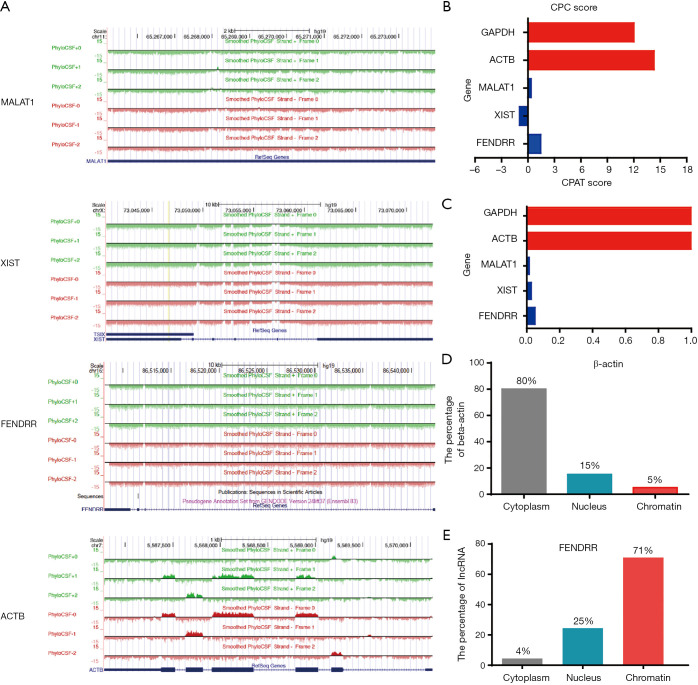
FENDRR is located in chromosomal locus 16q24 (1). Online coding ability analysis by PhyloCSF demonstrated low FENDRR coding potential (Figure 3A); meanwhile, CPC and CPAT result also corroborated this lower coding potential (Figure 3B,C). The subcellular fractionation experiment indicated that FENDRR was present mainly in the nucleus (Figure 3D,E). These data suggest that FENDRR is a nuclear lncRNA.
Figure 3.
The characteristics of lncRNA FENDRR in non-small cell lung cancer (NSCLC). (A) The coding potential of FENDRR was analyzed by the PhyloCSF CSF analysis. MALAT1 (metastasis associated lung adenocarcinoma transcript 1) and XIST (X inactive specific transcript) served as noncoding RNA controls, and ACTB served as coding RNA control. The regions with a score greater than 0 (above the baseline) were predicted to be coding, while regions with a score less than 0 (below the baseline) were predicted to be noncoding; (B,C) the coding potential of FENDRR was analyzed by Coding Potential Calculator (CPC) and coding potential Assessment Tool (CPAT). MALAT1 and XIST served as noncoding RNA controls, and ACTB and GAPDH served as coding RNA controls; (D,E) subcellular fractionation experiment to show the localization of FENDRR in cells.
FENDRR overexpression inhibited cell proliferation and growth
To clarify the function of FENDRR in NSCLC, quantitative RT-PCR was utilized to investigate FENDRR levels among 4 NSCLC cell lines (A549, H1299, H460, H358) (Figure 4A). Results showed a lower FENDRR RNA expression level in high metastatic lung cancer cell lines A549 and H1299, which preliminarily implicates FENDRR as a lung cancer metastasis suppressor. To determine how FENDRR functions in NSCLC progression, stable over-expressed cell lines (H1299-pWPXL-FENDRR and H1299-pWPXL-Control cell line) were established via lentiviral infection, and FENDRR was efficiently overexpressed in the H1299 cell line (Figure 4B). To verify whether cell proliferation was affected by FENDRR, we used clonogenicity assay and CCK8 assay to examine the proliferation ability. The H1299-pWPXL-FENDRR cells showed significantly reduced clonogenicity compared with the pWPXL-FENDRR cells (Figure 4C). From the second day, significant growth inhibition of H1299-pWPXL-FENDRR cells were observed when compared with H1299-pWPXL control cells (Figure 4D). Given the close correlation between FENDRR level and clinical metastasis, we sought to exploit the impact of FENDRR on NSCLC cell metastatic ability in vitro. A substantial inhibition of migration and invasion ability was present in the H1299-pWPXL-FENDRR cells compared with the pWPXL-FENDRR cells (Figure 4E,F). These observations suggest that FENDRR functions as an important tumor suppressor in NSCLC.
Figure 4.
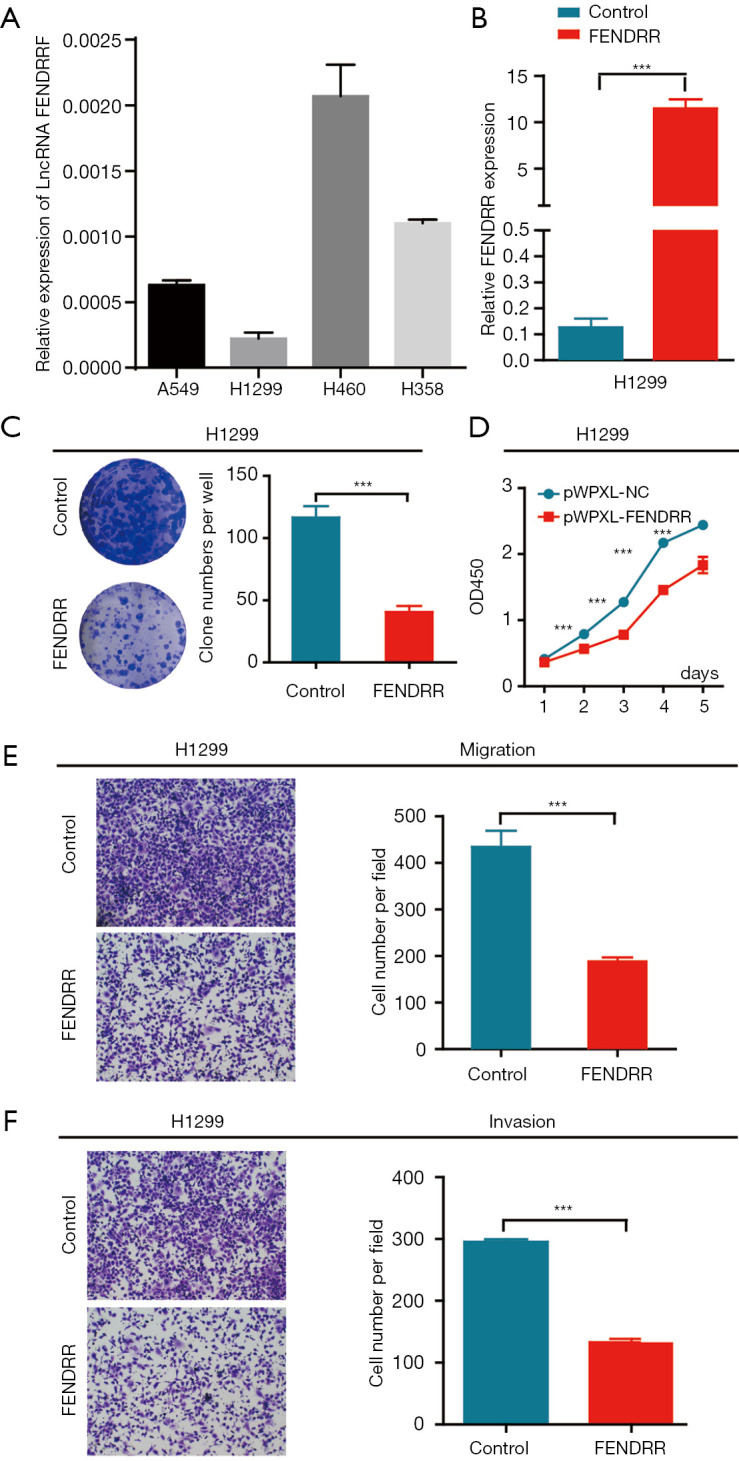
LncRNA FENDRR overexpression inhibited cell growth and aggressiveness in non-small cell lung cancer (NSCLC) cells in vitro. (A) FENDRR expression levels in 4 human lung cancer cell lines (A549, H1299, H460, H358) were measured by quantitative RT-PCR; (B) the quantitative RT-PCR assays quantified FENDRR level in H1299 cell with stable FENDRR over-expression; (C) FENDRR overexpression inhibited cell clonogenicity compared with the control transfectants in H1299 cell lines; (D) FENDRR overexpression led to cell growth arrest according to CCK8 assay compared with the control transfectants in H1299 cell lines; (E) Transwell migration assays in stable pWPXL-FENDRR H1299 cells and control; (F) Transwell invasion assays in stable pWPXL-FENDRR H1299 cells and control. The data are representative of 3 independent experiments. Values represent the mean ± SEM. ***, P<0.001 by Student’s t-test. The images in C were scanned and not enlarged. The images in E and F was magnified 100 times.
FENDRR expression caused aberrant immune tumor microenvironment
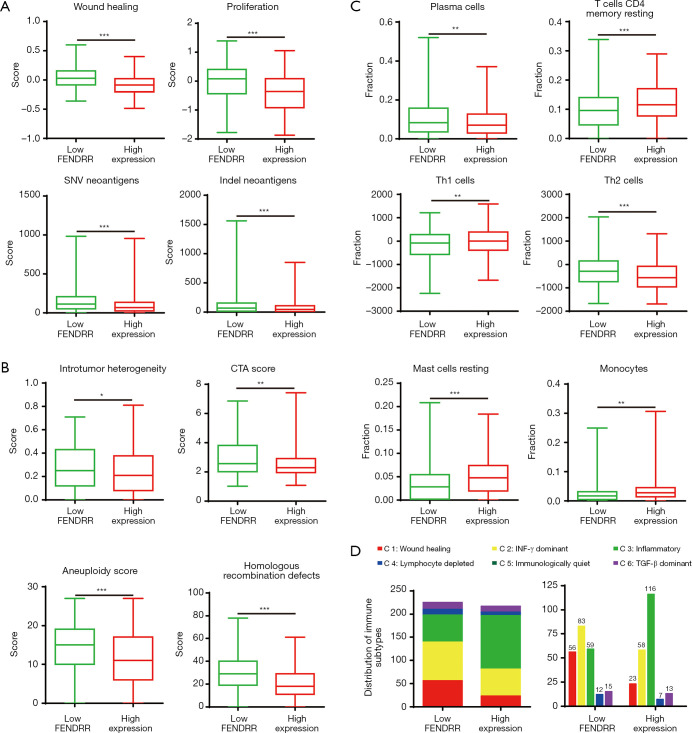
Several studies have demonstrated the relevance of lncRNA in many factors of cancer, so we further focused on the immune response in patients of the different FENDRR expression groups. Based on the data provided by Thorsson et al. (18), we performed an extensive immunogenomic analysis. High FENDRR expression showed decreased single-nucleotide variant (SNV) neoantigens, wound-healing ability, and proliferation ability. Furthermore, indel neoantigens of the tumor (Figure 5A) indicated that FENDRR was a lung cancer suppressor, which was in line with our results of cell proliferation assay. Moreover, patients in the high FENDRR expression group showed lower levels of intratumor heterogeneity, cancer-testis antigen score (CTA score) score, aneuploidy, and homologous recombination defects (HRD) (Figure 5B), suggesting the FENDRR might suppress DNA repair of deficiency, chromosomal instability, and spatially heterogeneous tumor structures to inhibit tumor progression. To characterize the tumor immune microenvironment between the patients in the low FENDRR expression and high FENDRR expression groups, we evaluated the elevation of plasma cells and Th2 cells in the low FENDRR expression group, along with the elevation of multiple immune cells, including resting CD4 memory T cells, Th1 cells, resting mast cells, and monocytes in the high FENDRR expression group (Figure 5C). Moreover, according to the immune states in intratumor mentioned by Thorsson et al. (18), we found a substantially varied proportion of immune subtypes (C1–C6) in these 2 groups (Figure 5D). As illustrated, patients of the low FENDRR expression group were richer in C1 than patients in the high FENDRR expression group.
Figure 5.
LncRNA FENDRR expression caused aberrant regulation of the tumor immune infiltrate. (A) The parameters mark tumor characteristics with low or high FENDRR expression; (B) four key immune expression signature scores; (C) proportion of major classes of immune cells; (D) numbers of patients and distribution of immune subtypes in the low or high FENDRR expression groups. The text above the bar show the specific number of patients in each immune subtype. Bar width reflects the number of tumor samples. Six immune subtypes: C1, wound healing; C2, IFN-γ dominant; C3, inflammatory; C4, lymphocyte-depleted; C5, immunologically quiet; C6, TGF-β dominant; values from min to max are plotted. *, P<0.05; **, P<0.01; ***, P<0.001.
Discussion
Malignant tumors have many characteristics, with migration, invasion, and metastasis being among the hallmarks (19). Cancer-related deaths are mostly due to metastases (20), and thus methods targeting metastasis mechanisms have become vital therapeutic strategies. Numerous investigations have discovered that lncRNAs are involved in important cellular functions in tumorigenesis and development (21). Lung cancer is emerging as the leading cause of cancer death among men and women worldwide (1). Studies involving lncRNAs have provided partial insights into the prominent role lncRNAs play in lung cancer progression; however, investigations into the function and underlying molecular mechanisms in this process are only in their infancy. Our results suggest that lncRNA FENDRR acts in the tumor-immune microenvironment as a tumor-suppressing lncRNA for NSCLC.
Substantial evidence has shown that lncRNAs participate in multiple cellular functions through complicated mechanisms (22). The function of lncRNAs in cancer cells has been closely linked to the localization in subcellular components. For example, cytoplasm-localized lncRNA Ptenp1 (phosphatase and tensin homolog pseudogene 1) was found to impact cancer progression by regulating Pten (phosphatase and tensin homolog) (23); in a similar fashion, nucleus-localized LncRNA F630028O10Rik displayed a tumor-suppressing effect through influencing angiogenesis (24), further confirming the significant correlation of lncRNA function with subcellular localization (25). Based on the above studies, we attempted to clarify the localization of FENDRR in subcellular components in NSCLC cells. It seemed that the nucleus was the main component where FENDRR localized, suggested a role of FENDRR in regulating the expression of other genes.
Zhang et al. discovered that FENDRR was significantly differentially expressed in lung adenocarcinoma (LUAD) samples compared to adjacent normal tissues (26). A genome-wide analysis by Li et al. in Xuanwei Lung Cancer Village revealed that FENDRR was among the most aberrantly regulated lncRNA between lung cancer patient tissue and adjacent normal samples (27). These findings are consistent with our present study, which found FENDRR to be significantly downregulated in human NSCLC tissues. More importantly, our results indicated that loss of function of FENDRR was correlated with poor prognosis and advanced pathologies, further indicating the tumor-suppressing role of lncRNA FENDRR in NSCLC progression. Similar results were obtained from research on FENDRR in other cancer types. For instance, data gathered by Zhang et al., suggested a reduction of FENDRR was associated with poor clinical outcomes in prostate cancer and its upregulation mediated in vitro growth and metastasis inhibition (28). Meanwhile, Xu et al.’s work on gastric cancer demonstrated that FENDRR is downregulated in cancerous tissues and negatively related to metastasis and prognosis, with vitro inhibition of metastatic potential also being connected to FENDER overexpression (29). Yang et al. found that survival time was elongated by FENDRR upregulation in Chinese patients (30), whereas a study performed by Kun-Peng et al. found that FENDRR exerted a promoting role in the progression of osteosarcoma (31). When considered in concert, the above studies suggest a phenomenon in which lncRNA FENDRR expression is differentially regulated in different cancer types, thus further complicating the understanding of the function and mechanism of lncRNAs.
A few articles exist which explore the mechanisms of FENDRR involvement in NSCLC. For example, Xu et al. found that lncRNA FOXF1 suppressed cell growth and chemotherapy resistance in A549 cells with no further mechanical study (32), Zhang et al. discovered that FENDRR inhibited NSCLC progression in A549 and H1975 cell lines by interacting with miR-761/TIMP2 (TIMP metallopeptidase inhibitor 2) (33), and Zhang et al. reported that lncRNA FENDRR sponged to miR-761 to exert a tumor-suppressing effect in NSCLC cells (34), which mostly involved microRNA as the downstream target of FENDRR. However, given the complexity and malignancy of lung cancer, we need to focus on a wide range of potential targets. Mowel et al. found lncRNAs could control immune cell homeostasis and function (35), Jiang et al. showed that the innate immune response was restricted by an inducible host lncRNA (36), and Li et al.’s study indicated hepatocellular carcinoma (HCC)-derived exosomal lncRNA TUC339 could regulate macrophage activation and polarization (37). Together, the above studies give evidence supporting the major role of lncRNAs in the regulation of immune response. In addition to this, Xu et al. found that colorectal cancer lncRNA SATB2-AS1 (SATB2 antisense RNA 1) could interact with SATB2 (SATB homeobox 2) to suppress tumor metastasis and influence the tumor immune microenvironment (38), Pei et al. observed regulatory T cell (Treg) differentiation and immune escape of breast cancer via the regulation of miR-448/IDO by LncRNA SNHG1 (39), and Huang et al. also found that lncRNA NKILA could sensitize T cells to activation-induced cell death to promote tumor immune evasion (40). These studies all confirm the significant influence of lncRNAs in cancer immune regulation and inspired us to more deeply investigate the mechanism underlying FENDRR involvement in NSCLC via the tumor immune microenvironment. We thus hypothesized that FENDRR might regulate immune homeostasis to inhibit NSCLC progression. Subsequently, we showed the increased infiltration of resting CD4 memory T cells, Th1 cells, resting mast cells, and monocytes in the high FENDRR expression groups accompanied by the down-regulated levels of plasma cells and th2 cells. Our findings are in line with several previous studies in regards to the tumor-regulating activity of immune cells. For instance, Dai et al. demonstrated that sustained Th1 response was important for tumor regression and cure (41), while Li et al. discovered that NSCLC prognosis was involved in a signature of tumor-immune microenvironment genes (42). Another study on ovarian cancer performed by Kovács et al. found the phagocytic function of monocytes and neutrophil granulocytes to be reduced (43). Furthermore, among the 6 immune subtypes, C1 had a high proliferation rate, with a higher percentage of C1 in cancer patients suggesting a tumor-promoting state, which is in line with our results. Our study also found that the low FENDRR expression group exhibited significantly higher levels of intratumor heterogeneity, homologous recombination deficiency, and aneuploidy compared with the high FENDRR expression group, suggesting different pathways for low FENDRR expression patients to develop carcinogenesis such as chromosomal instability probability (44) and impairment of DNA repair approaches (45), which are often related with drug resistance and poor prognosis in many cancer types.
In summary, we identified lncRNA FENDRR as a tumor suppressor of NSCLC and found it to be correlated with immune response regulation. FENDRR was negatively correlated with clinical metastasis and advanced pathologies in NSCLC. Moreover, elevated FENDRR expression inhibited NSCLC cell proliferation and metastatic potential in vitro. This work also provides new insights for understanding the function of FENDRR in the progression of NSCLC in terms of tumor-immune interactions, which is useful for identifying approaches towards conducting further research from an immunotherapy perspective.
Acknowledgments
We thank The Department of Lung Cancer at Shanghai Chest Hospital, Shanghai Jiao-tong University for kindly providing of the human tissues.
Funding: This work was supported by the National Natural Science Foundation of China (grant No. 81702846; grant No. 81972173), Shanghai Science and Technology Committee Foundation (grant No. 19140900800; grant No. 18140903700), and the Shanghai Health and Family Planning Commission Research Fund (grant No. 20174Y0183).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of The Department of Lung Cancer at Shanghai Chest Hospital, Shanghai Jiao Tong University (No. XS2017-007), and obtained the informed consent from all patients.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2147
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2147
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2147). The authors have no conflicts of interest to declare.
References
- 1.Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 2016;893:1-19. 10.1007/978-3-319-24223-1_1 [DOI] [PubMed] [Google Scholar]
- 2.Reck M, Rabe KF. Precision Diagnosis and Treatment for Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:849-61. 10.1056/NEJMra1703413 [DOI] [PubMed] [Google Scholar]
- 3.Ernani V, Steuer CE, Jahanzeb M. The End of Nihilism: Systemic Therapy of Advanced Non-Small Cell Lung Cancer. Annu Rev Med 2017;68:153-68. 10.1146/annurev-med-042915-102442 [DOI] [PubMed] [Google Scholar]
- 4.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell 2017;168:670-91. 10.1016/j.cell.2016.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y, Adjei AA. New strategies to develop new medications for lung cancer and metastasis. Cancer Metastasis Rev 2015;34:265-75. 10.1007/s10555-015-9553-5 [DOI] [PubMed] [Google Scholar]
- 6.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489:101-8. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145-66. 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013;193:651-69. 10.1534/genetics.112.146704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015;21:1253-61. 10.1038/nm.3981 [DOI] [PubMed] [Google Scholar]
- 11.He W, Zhong G, Jiang N, et al. Long noncoding RNA BLACAT2 promotes bladder cancer-associated lymphangiogenesis and lymphatic metastasis. J Clin Invest 2018;128:861-75. 10.1172/JCI96218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin A, Hu Q, Li C, et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol 2017;19:238-51. 10.1038/ncb3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Q, Shan S, Li Y, et al. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. FASEB J 2018;32:3957-67. 10.1096/fj.201701237RR [DOI] [PubMed] [Google Scholar]
- 14.Li K, Sun D, Gou Q, et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett 2018;420:80-90. 10.1016/j.canlet.2018.01.060 [DOI] [PubMed] [Google Scholar]
- 15.Yang B, Zhang L, Cao Y, et al. Overexpression of lncRNA IGFBP4-1 reprograms energy metabolism to promote lung cancer progression. Mol Cancer 2017;16:154. 10.1186/s12943-017-0722-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Sun M, Zang C, et al. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis 2016;7:e2225. 10.1038/cddis.2016.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miao L, Huang Z, Zengli Z, et al. Loss of long noncoding RNA FOXF1-AS1 regulates epithelial-mesenchymal transition, stemness and metastasis of non-small cell lung cancer cells. Oncotarget 2016;7:68339-49. 10.18632/oncotarget.11630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity 2019;51:411-2. 10.1016/j.immuni.2019.08.004 [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 20.Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer 2006;6:449-58. 10.1038/nrc1886 [DOI] [PubMed] [Google Scholar]
- 21.Jiang C, Li X, Zhao H, et al. Long non-coding RNAs: potential new biomarkers for predicting tumor invasion and metastasis. Mol Cancer 2016;15:62. 10.1186/s12943-016-0545-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang K, Shi ZM, Chang YN, et al. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene 2014;547:1-9. 10.1016/j.gene.2014.06.043 [DOI] [PubMed] [Google Scholar]
- 23.Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010;465:1033-8. 10.1038/nature09144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin L, Zhong M, Adah D, et al. A novel tumour suppressor lncRNA F630028O10Rik inhibits lung cancer angiogenesis by regulating miR-223-3p. J Cell Mol Med 2020;24:3549-59. 10.1111/jcmm.15044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayes EL, Lewis-Wambi JS. Mechanisms of endocrine resistance in breast cancer: an overview of the proposed roles of noncoding RNA. Breast Cancer Res 2015;17:40. 10.1186/s13058-015-0542-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Li S, Choi YL, et al. Systematic identification of cancer-related long noncoding RNAs and aberrant alternative splicing of quintuple-negative lung adenocarcinoma through RNA-Seq. Lung Cancer 2017;109:21-7. 10.1016/j.lungcan.2017.04.009 [DOI] [PubMed] [Google Scholar]
- 27.Li Q, Wu C, Song G, et al. Genome-Wide Analysis of Long Noncoding RNA Expression Profiles in Human Xuanwei Lung Cancer. Clin Lab 2015;61:1515-23. 10.7754/Clin.Lab.2015.150323 [DOI] [PubMed] [Google Scholar]
- 28.Zhang G, Han G, Zhang X, et al. Long non-coding RNA FENDRR reduces prostate cancer malignancy by competitively binding miR-18a-5p with RUNX1. Biomarkers 2018;23:435-45. 10.1080/1354750X.2018.1443509 [DOI] [PubMed] [Google Scholar]
- 29.Xu TP, Huang MD, Xia R, et al. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol 2014;7:63. 10.1186/s13045-014-0063-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L, Wu D, Chen J, et al. A functional CNVR_3425.1 damping lincRNA FENDRR increases lifetime risk of lung cancer and COPD in Chinese. Carcinogenesis 2018;39:347-59. 10.1093/carcin/bgx149 [DOI] [PubMed] [Google Scholar]
- 31.Kun-Peng Z, Chun-Lin Z, Xiao-Long M. Antisense lncRNA FOXF1-AS1 Promotes Migration and Invasion of Osteosarcoma Cells Through the FOXF1/MMP-2/-9 Pathway. Int J Biol Sci 2017;13:1180-91. 10.7150/ijbs.21722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu R, Han Y. Long non-coding RNA FOXF1 adjacent non-coding developmental regulatory RNA inhibits growth and chemotherapy resistance in non-small cell lung cancer. Arch Med Sci 2019;15:1539-46. 10.5114/aoms.2019.86707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang G, Wang Q, Zhang X, et al. LncRNA FENDRR suppresses the progression of NSCLC via regulating miR-761/TIMP2 axis. Biomed Pharmacother 2019;118:109309. 10.1016/j.biopha.2019.109309 [DOI] [PubMed] [Google Scholar]
- 34.Zhang MY, Zhang ZL, Cui HX, et al. Long non-coding RNA FENDRR inhibits NSCLC cell growth and aggressiveness by sponging miR-761. Eur Rev Med Pharmacol Sci 2018;22:8324-32. [DOI] [PubMed] [Google Scholar]
- 35.Mowel WK, Kotzin JJ, McCright SJ, et al. Control of Immune Cell Homeostasis and Function by lncRNAs. Trends Immunol 2018;39:55-69. 10.1016/j.it.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang M, Zhang S, Yang Z, et al. Self-Recognition of an Inducible Host lncRNA by RIG-I Feedback Restricts Innate Immune Response. Cell 2018;173:906-919.e13. 10.1016/j.cell.2018.03.064 [DOI] [PubMed] [Google Scholar]
- 37.Li X, Lei Y, Wu M, et al. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA TUC339. Int J Mol Sci 2018;19:2958. 10.3390/ijms19102958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu M, Xu X, Pan B, et al. LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol Cancer 2019;18:135. 10.1186/s12943-019-1063-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pei X, Wang X, Li H. LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. Int J Biol Macromol 2018;118:24-30. 10.1016/j.ijbiomac.2018.06.033 [DOI] [PubMed] [Google Scholar]
- 40.Huang D, Chen J, Yang L, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol 2018;19:1112-25. 10.1038/s41590-018-0207-y [DOI] [PubMed] [Google Scholar]
- 41.Dai M, Hellstrom I, Yip YY, et al. Tumor Regression and Cure Depends on Sustained Th1 Responses. J Immunother 2018;41:369-78. 10.1097/CJI.0000000000000231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Li X, Zhang C, et al. A signature of tumor immune microenvironment genes associated with the prognosis of nonsmall cell lung cancer. Oncol Rep 2020;43:795-806. [DOI] [PubMed] [Google Scholar]
- 43.Kovács AR, Pal L, Szucs S, et al. Phagocytic function of monocytes and neutrophil granulocytes in ovarian cancer. Orv Hetil 2018;159:1353-9. [DOI] [PubMed] [Google Scholar]
- 44.He Q, Au B, Kulkarni M. Chromosomal instability-induced senescence potentiates cell non-autonomous tumourigenic effects. Oncogenesis 2018;7:62. 10.1038/s41389-018-0072-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoppe MM, Sundar R, Tan DSP, et al. Biomarkers for Homologous Recombination Deficiency in Cancer. J Natl Cancer Inst 2018;110:704-13. 10.1093/jnci/djy085 [DOI] [PubMed] [Google Scholar]