Abstract
Background
Several studies have indicated that CXC chemokines influence the prognosis and therapy in patients with hepatocellular carcinoma (HCC). However, there are limited studies on the roles of CXC chemokines in HCC based on data acquired from various databases. This study aimed to conduct an in-depth and comprehensive bioinformatic analysis of the expression and functions of CXC chemokines in HCC.
Methods
Data was obtained from various databases including ONCOMINE, UALCAN, STRING, GeneMinia, DAVID, Kaplan-Meier plotter, TIMER, GSCALite and NetworkAnalyst for the analysis of the expression and functions of the CXC chemokines in HCC.
Results
Analysis of the differential expression levels of CXC chemokines between HCC and adjacent normal tissues revealed that the mRNA expression levels of CXCL1/2/5/6/7/12/14 were significantly lower in HCC tissues than those in adjacent normal tissues, whereas the mRNA expression levels of CXCL9/16/17 were significantly higher in HCC tissues. Analysis of the relationship between CXC chemokines and overall survival revealed that high mRNA expression levels of CXCL1/3/5/6/8 were associated with poor overall survival, whereas high mRNA expression levels of CXCL2/4/7/9/10/12 were associated with better overall survival. The functions of CXC chemokines and related genes were associated with cytokine-cytokine receptor interactions and chemokine signaling pathway. Analysis of the association between CXC chemokines and activity of cancer pathways indicated that the DNA damage response and hormone androgen receptor (AR) signaling pathways were inhibited, whereas apoptosis, epithelial-mesenchymal transition (EMT) and Ras/mitogen-activated protein kinase (MAPK) signaling pathways were activated. The expression of CXC chemokines was positively correlated with the infiltration of six types of immune cells (B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and dendritic cells).
Conclusions
This study has demonstrated that CXC chemokines can influence survival of patients with HCC by recruiting different types of immune cells into the tumor microenvironment.
Keywords: Hepatocellular carcinoma (HCC), chemokine, bioinformatics analysis, biomarker, prognosis
Introduction
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, and is the fifth most common cancer globally and the third leading cause of cancer-related mortality (1). Numerous studies have indicated that hepatitis virus infection is a principal risk factor of the occurrence and progression of HCC (2). Several patients are diagnosed at the advanced stages of HCC due to the undefined symptoms of early-stage HCC, thereby missing the opportunity to receive hepatectomy and liver transplantation (3,4). In addition, the high rates of recurrence and metastasis in patients after resection contribute to poor prognosis (5,6). Therefore, it is necessary to investigate the underlying molecular mechanisms of HCC to enhance precise diagnosis and targeted treatment.
Studies conducted in the recent past have revealed that various chemokines within the tumor microenvironment affect the initiation and progression of HCC by binding to their receptors (7,8). Chemokines are small secreted proteins (8–12 kDa), which can significantly affect various aspects of the development of HCC including recruitment and infiltration of immune cells, modulating function of immune cells, promoting metastasis of tumor cells, influencing effects of immunotherapies, and regulating angiogenesis (9-12). Chemokines are subdivided into four classes based on the location of the first two N-terminal cysteine residues: CXC chemokines, CC chemokines, C chemokines and CX3C chemokines (13). CXC chemokine-chemokine receptor signaling pathway can affect the progression and therapy of HCC both directly and indirectly (14-18).
In this study, we conducted an in-depth bioinformatic analysis of 16 CXC chemokines (CXCL1-CXCL17, excluding CXCL15) based on data extracted from several public databases. An analysis of the differential expression levels of CXC chemokines in HCC tissues in comparison with adjacent normal tissues was performed, and the association between the expression levels of CXC chemokines and overall survival of patients with HCC explored. In addition, we investigated the roles of CXC chemokines in recruiting immune cells and activation of the cancer pathway. Moreover, external data were downloaded from the Gene Expression Omnibus (GEO) database to validate the results obtained from The Cancer Genome Atlas (TCGA) database. With the development of second-generation sequencing technology and the establishment of online databases, this study will help to improve the accuracy of evaluating prognosis and the efficiency of adopting therapies for patients with HCC.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tcr-21-127).
Methods
ONCOMINE (www.oncomine.org) is a cancer microarray database that enables clinicians to study gene expression profiles of multiple cancer types (19). All the significant thresholds in the “Gene Summary” and “datasets” modules were set as follows: P value was set at 0.05, fold change was set at 2 and the gene rank was set in the top 10%. Student’s t-test was used for differential expression analysis.
UALCAN (http://ualcan.path.uab.edu/) is an easy-to-use web-portal that allows cancer researchers to easily conduct analysis of TCGA gene expression data (20). In this study, we analyzed the relative expressions of CXC chemokines between primary tumor and normal tissue samples in liver hepatocellular carcinoma (LIHC) dataset. Differential expression analyses were performed using Student’s t-test, and a P value <0.05 was considered statistically significant.
STRING (http://string-db.org/) is an online database with a collection of all available information on protein-protein interactions (PPIs), which aims to visualize a comprehensive PPI network (21). CXC chemokines were uploaded and the PPI network constructed based on an interaction score of 0.4.
GeneMinia (http://www.genemania.org) is a user-friendly website that characterizes the genes of interest based on functionally informative associations and displays the interaction network among genes (22). We identified CXC chemokine-related genes according to the networks using the following criteria: physical interactions, co-expression, predicted, pathway, co-localization, genetic interactions and shared protein domains.
DAVID (http://david.ncifcrf.gov/home.jsp) is a bioinformatic online tool that provides gene-annotation enrichment analysis for genes of interest, including the Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (23,24). The GO analysis consists of three categories: biological process (BP), cellular component (CC) and molecular function (MF). The annotation results were visualized using R software (https://www.r-project.org/) after completion of the functional enrichment analysis for CXC chemokines and related genes.
Kaplan-Meier plotter (http://kmplot.com/analysis) is an online tool that enables researchers to assess the effect of 54k genes on survival in 21 cancer types (25). The effect of CXC chemokines on overall survival of patients with HCC was analyzed, and patients were split according to the most significant cut-off value.
TIMER (http://cistrome.shinyapps.io/timer/) is a user-friendly website that allows researchers to analyze infiltration of immune cells (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) in various cancer types based on a TIMER algorithm (26). The association between CXC chemokines and six immune cell subsets was assessed. Additionally, we built a multivariable Cox model to investigate the correlation between survival and six immune cell subsets, and CXC chemokines.
GSCALite (http://bioinfo.life.hust.edu.cn/web/GSCALite/) is a web-based platform that allows researchers to analyze cancer gene sets (27). In this study, the association between expression of CXC chemokines and activity of ten cancer related pathways was analyzed.
NetworkAnalyst (http://www.networkanalyst.ca) is a visual analytics platform that enables users to analyze gene expression profiles and obtain biological networks (28). We integrated and analyzed three GEO datasets, including GSE121248, GSE60502 and GSE57957, using the Multiple Gene Expression Tables module to further validate the differential expression of CXC chemokines in HCC and normal tissues. The significant thresholds were set as follows: false discovery rate (FDR) was 0.05 and fold change was 2. Differential expression analyses were performed using Fisher’s method, and a combined adjusted P value <0.05 was considered statistically significant.
Ethical statement
TCGA and GEO belong to public databases. The patients involved in the database have obtained ethical approval. Users can download relevant data for free for research and publish relevant articles. Our study is based on open source data, so there are no ethical issues. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
R software was used for all statistical analyses. The Student’s t-test was used to perform the differential expression analysis between HCC tissues and adjacent normal tissues. The survival time in the high- and low-expression groups was estimated using Kaplan-Meier curves. Besides, the log-rank test was used to analyze the differences. The Cox proportional hazard model was performed to investigate the correlation between clinical outcomes of HCC patients and six immune cell subsets, and CXC chemokines. P values less than 0.05 were considered statistically significant, and all P values were two-tailed.
Results
The differential expression of CXC chemokines in HCC and adjacent normal tissues
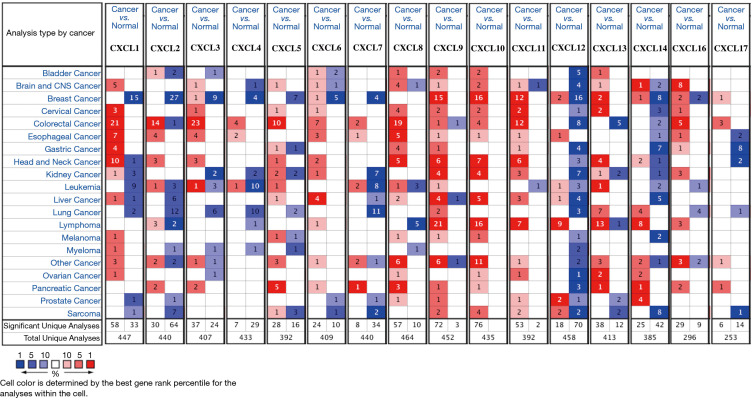
We identified 16 CXC chemokines (CXCL1-CXCL17, excluding CXCL15) using data from the ONCOMINE database. The mRNA expression levels of CXC chemokines in HCC and normal tissues were analyzed using the Gene Summary module of ONCOMINE (Figure 1). Results revealed that the mRNA expression levels of CXCL6, CXCL8, CXCL9, CXCL10 and CXCL11 in HCC tissues were significantly upregulated, whereas the mRNA expression levels of CXCL1, CXCL2, CXCL12 and CXCL14 were significantly downregulated. Table 1 displays four studies on the expression levels of CXC chemokines between HCC and normal tissues with significant differences. Chen et al. demonstrated that mRNA expression levels of CXCL1, CXCL2, CXCL12 and CXCL14 were significantly lower in HCC tissues than those in normal tissues (29). Similarly, Roessler et al. reported that mRNA expression levels of CXCL2, CXCL12 and CXCL14 were significantly lower in HCC tissues compared to normal tissues (30). Wurmbach et al. established that mRNA expression levels of CXCL2, CXCL12 and CXCL14 were significantly downregulated in HCC tissues, whereas the mRNA expression level of CXCL10 was significantly upregulated in HCC tissues (31). Mas et al. revealed that mRNA expression level of CXCL2 was significantly downregulated in HCC tissues, whereas the mRNA expression levels of CXCL6, CXCL8, CXCL9, CXCL10 and CXCL11 were significantly upregulated in HCC tissues (32).
Figure 1.
The transcription levels of CXC chemokines in different types of cancers (ONCOMINE). The figure shows the numbers of datasets with statistically significant mRNA high-expression (red) or low-expression (blue) of CXC chemokines.
Table 1. The significant changes of CXC chemokines expression in transcription level between hepatocellular carcinoma and normal tissues (ONCOMIINE).
| Gene | Type | Fold change | P value | t-test | Ref |
|---|---|---|---|---|---|
| CXCL1 | Hepatocellular carcinoma vs. Normal | −4.161 | 1.76E-20 | −10.489 | Chen |
| CXCL2 | Hepatocellular carcinoma vs. Normal | −6.540 | 8.33E-60 | −20.350 | Roessler |
| CXCL2 | Hepatocellular carcinoma vs. Normal | −3.601 | 4.65E-17 | −9.237 | Chen |
| CXCL2 | Hepatocellular carcinoma vs. Normal | −5.737 | 1.45E-7 | −6.632 | Wurmbach |
| CXCL2 | Hepatocellular carcinoma vs. Normal | −7.403 | 2.59E-7 | −6.414 | Roessler |
| CXCL2 | Hepatocellular carcinoma vs. Normal | −2.273 | 9.36E-7 | −5.344 | Mas |
| CXCL6 | Hepatocellular carcinoma vs. Normal | 3.749 | 3.15E-7 | 5.637 | Mas |
| CXCL8 | Hepatocellular carcinoma vs. Normal | 3.323 | 7.84E-6 | 4.972 | Mas |
| CXCL9 | Hepatocellular carcinoma vs. Normal | 5.334 | 6.20E-17 | 12.031 | Mas |
| CXCL10 | Hepatocellular carcinoma vs. Normal | 9.272 | 6.33E-17 | 11.988 | Mas |
| CXCL10 | Hepatocellular carcinoma vs. Normal | 5.928 | 1.78E-4 | 4.269 | Wurmbach |
| CXCL11 | Hepatocellular carcinoma vs. Normal | 2.188 | 2.26E-6 | 5.355 | Mas |
| CXCL12 | Hepatocellular carcinoma vs. Normal | −5.338 | 2.32E-93 | −26.987 | Roessler |
| CXCL12 | Hepatocellular carcinoma vs. Normal | −2.387 | 4.32E-31 | −14.115 | Chen |
| CXCL12 | Hepatocellular carcinoma vs. Normal | −4.036 | 4.43E-11 | −9.560 | Roessler |
| CXCL12 | Hepatocellular carcinoma vs. Normal | −4.233 | 3.82E-9 | −7.239 | Wurmbach |
| CXCL14 | Hepatocellular carcinoma vs. Normal | −10.940 | 6.98E-154 | −41.196 | Roessler |
| CXCL14 | Hepatocellular carcinoma vs. Normal | −9.667 | 1.24E-21 | −18.539 | Roessler |
| CXCL14 | Hepatocellular carcinoma vs. Normal | −12.903 | 1.96E-43 | −18.813 | Chen |
| CXCL14 | Hepatocellular carcinoma vs. Normal | −13.977 | 6.81E-10 | −8.892 | Wurmbach |
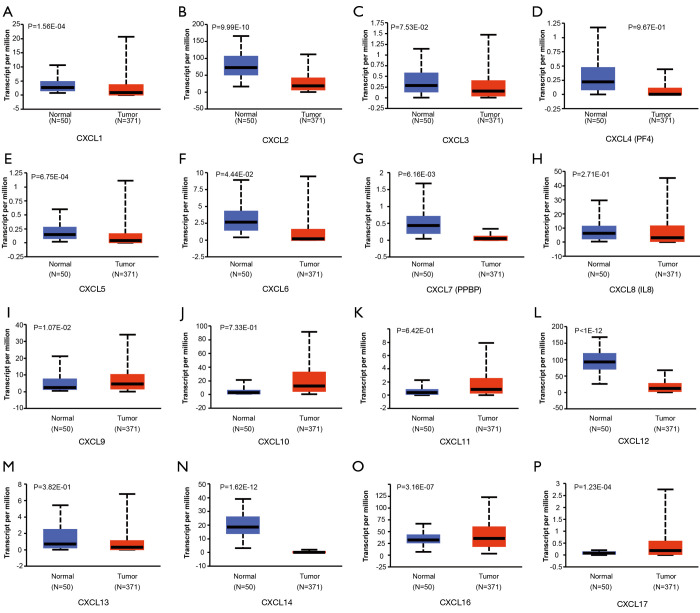
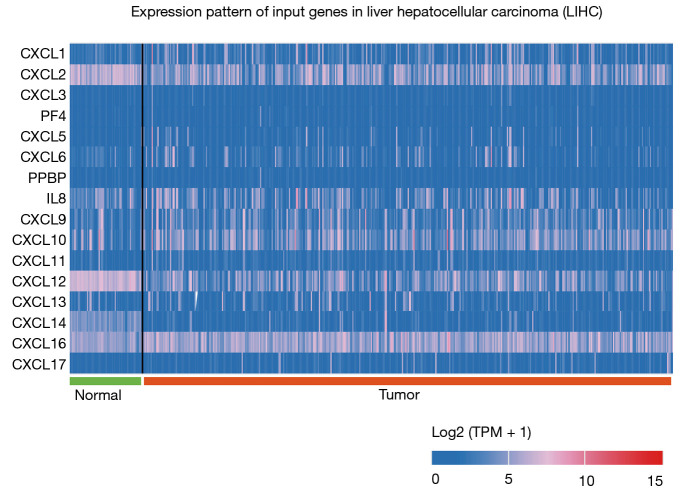
Additionally, the differential expression levels of CXC chemokines between HCC and normal tissues were analyzed using the UALCAN online tool, which was also used to analyze TCGA data. The analysis revealed that the mRNA expression levels of CXCL1, CXCL2, CXCL5, CXCL6, CXCL7 (PPBP), CXCL12 and CXCL14 were significantly lower in HCC tissues than those in adjacent normal tissues, whereas the mRNA expression levels of CXCL9, CXCL16 and CXCL17 were significantly higher in HCC tissues (Figure 2). A heatmap was constructed to illustrate the expression patterns of CXC chemokines in the LICH dataset (Figure 3).
Figure 2.
The expression levels of CXC chemokines between HCC and normal tissues (UALCAN). The mRNA expression levels of (A) CXCL1, (B) CXCL2, (E) CXCL5, (F) CXCL6, (G) CXCL7, (L) CXCL12, and (N) CXCL14 were significantly lower in HCC tissues than those in adjacent normal tissues, whereas the mRNA expression levels of (I) CXCL9, (O) CXCL16, and (P) CXCL17 were significantly higher in HCC tissues. Besides, six genes, including (C) CXCL3, (D) CXCL4, (H) CXCL8, (J) CXCL10, (K) CXCL11, and (M) CXCL13, do not show significantly differential expression between HCC tissues and adjacent normal tissues. HCC, hepatocellular carcinoma.
Figure 3.
Heatmap of CXC chemokines between HCC and normal tissues (UALCAN). HCC, hepatocellular carcinoma. TPM, transcripts per million.
Construction of protein-protein interaction networks and prediction of related genes of CXC chemokines
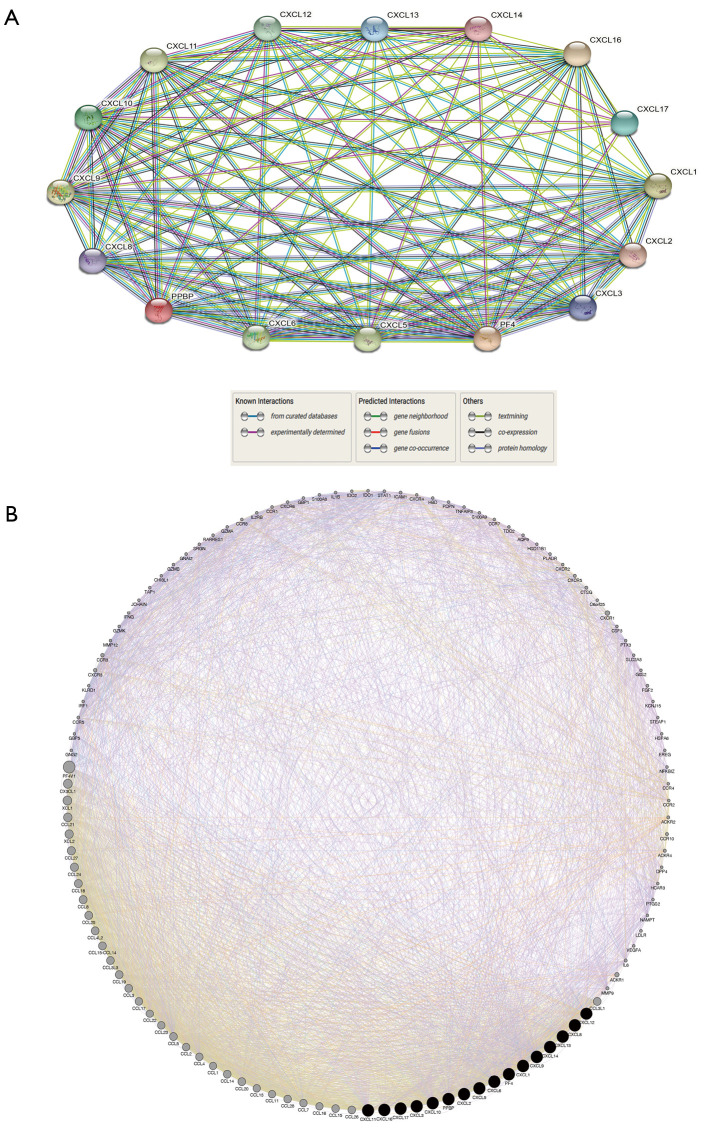
A protein-protein interaction network containing 16 nodes and 111 edges was constructed using data from the STRING database based on a minimum interaction score of 0.4 (Figure 4A). Furthermore, the top 100 related genes of CXC chemokines were predicted to further explore functional enrichment of genes using GeneMinia online tool (Figure 4B).
Figure 4.
Protein-protein interaction network of CXC chemokines and identification of CXC chemokines-related genes (STRING and GeneMinia). (A) protein-protein interaction network of CXC chemokines. Each network node represents the protein produced by a single, protein-coding gene. Edges represent protein-protein association. (B) Gene-gene interaction network of CXC chemokines and their related top 100 genes. The top 100 related genes meet these criteria: co-expression, co-localization, genetic interactions, pathway, physical interactions, predicted, and shared protein domains.
Functional enrichment analysis of CXC chemokines and related genes
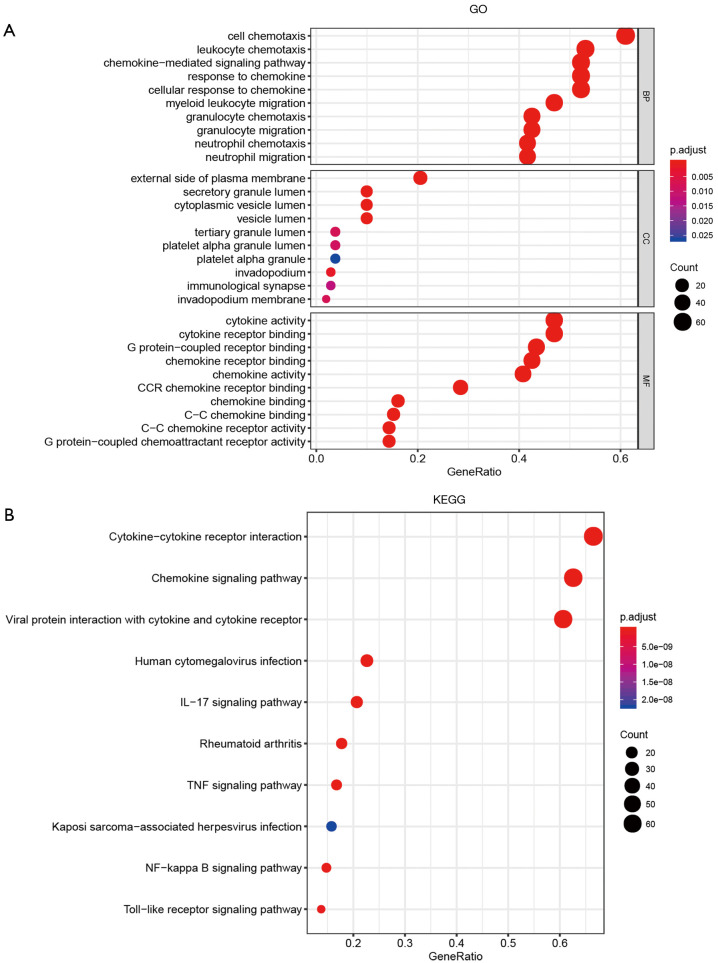
Functional enrichment analyses of the 116 genes including 16 CXC chemokines and 100 related genes were performed using DAVID database. Figure 5A presents the top 10 GO terms for three functional categories comprising BP, CC and MF, respectively. In the BP category, cell chemotaxis, leukocyte chemotaxis, chemokine-mediated signaling pathway, response to chemokines, cellular responses to chemokine, myeloid leukocyte migration, granulocyte chemotaxis, granulocyte migration, neutrophil chemotaxis and neutrophil migration were the top 10 terms. In the CC category, external side of plasma membrane, secretory granule lumen, cytoplasmic vesicle lumen, vesicle lumen, tertiary granule lumen, platelet alpha granule lumen, platelet alpha granule, invadopodium, immunological synapse and invadopodium membrane were the top 10 terms. Cytokine activity, cytokine receptor binding, G protein-coupled receptor binding, chemokine receptor binding, chemokine activity, CCR chemokine receptor binding, chemokine binding, C-C chemokine binding, C-C chemokine receptor activity and G protein-coupled chemoattractant receptor activity were the top 10 terms in the MF category. The top 10 KEGG pathways identified included the following: cytokine-cytokine receptor interaction, chemokine signaling pathway, viral protein interaction with cytokines and cytokine receptors, human cytomegalovirus infection, IL-17 signaling pathway, rheumatoid arthritis, TNF signaling pathway, Kaposi sarcoma-associated herpesvirus infection, NF-kappa B signaling pathway and toll-like receptor signaling pathway (Figure 5).
Figure 5.
Top 10 enrichment function and pathway terms of CXC chemokines and their related genes in hepatocellular carcinoma (DAVID). (A) Gene Ontology (GO) analysis including three categories: biological process (BP), cellular component (CC) and molecular function (MF). (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.
The prognostic value of CXC chemokines in patients with HCC
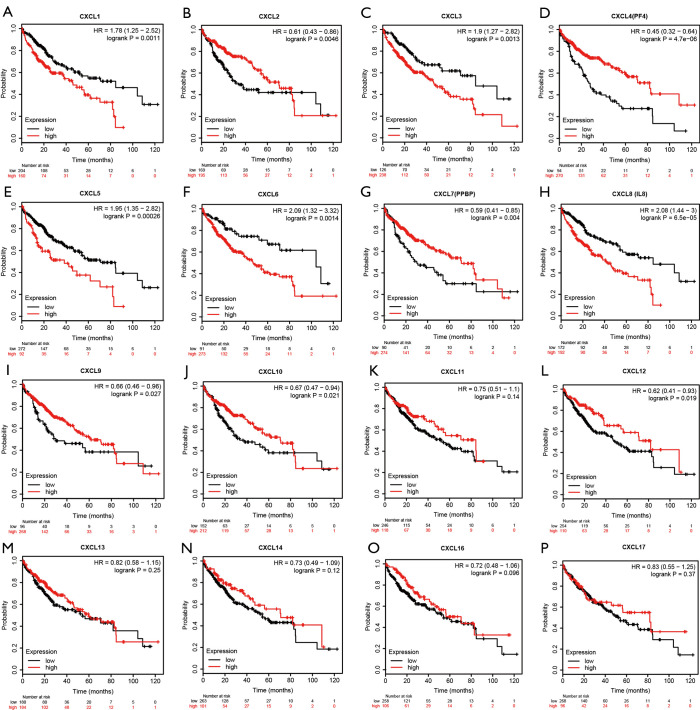
The association between the expression levels of CXC chemokines and overall survival was analyzed using Kaplan-Meier plotter database. Figure 6 illustrates 16 overall survival curves associated with CXC chemokines based on the most significant cut-off value. The analysis revealed that the high mRNA expression levels of CXCL1, CXCL3, CXCL5, CXCL6 and CXCL8 were correlated with poor overall survival, whereas high mRNA expression levels of CXCL2, CXCL4, CXCL7, CXCL9, CXCL10 and CXCL12 were correlated with better overall survival.
Figure 6.
Association between CXC chemokines and overall survival in patients with hepatocellular carcinoma (Kaplan-Meier plotter). The high mRNA expression levels of (A) CXCL1, (C) CXCL3, (E) CXCL5, (F) CXCL6, and (H) CXCL8 were associated with poor overall survival, whereas high mRNA expression levels of (B) CXCL2, (D) CXCL4, (G) CXCL7, (I) CXCL9, (J) CXCL10, and (L) CXCL12 were associated with better overall survival. Besides, the expression levels of five genes, including (K) CXCL11, (M) CXCL13, (N) CXCL14, (O) CXCL16, and (P) CXCL17, do not significantly correlate with the overall survival.
The interaction between CXC chemokines and ten cancer pathways
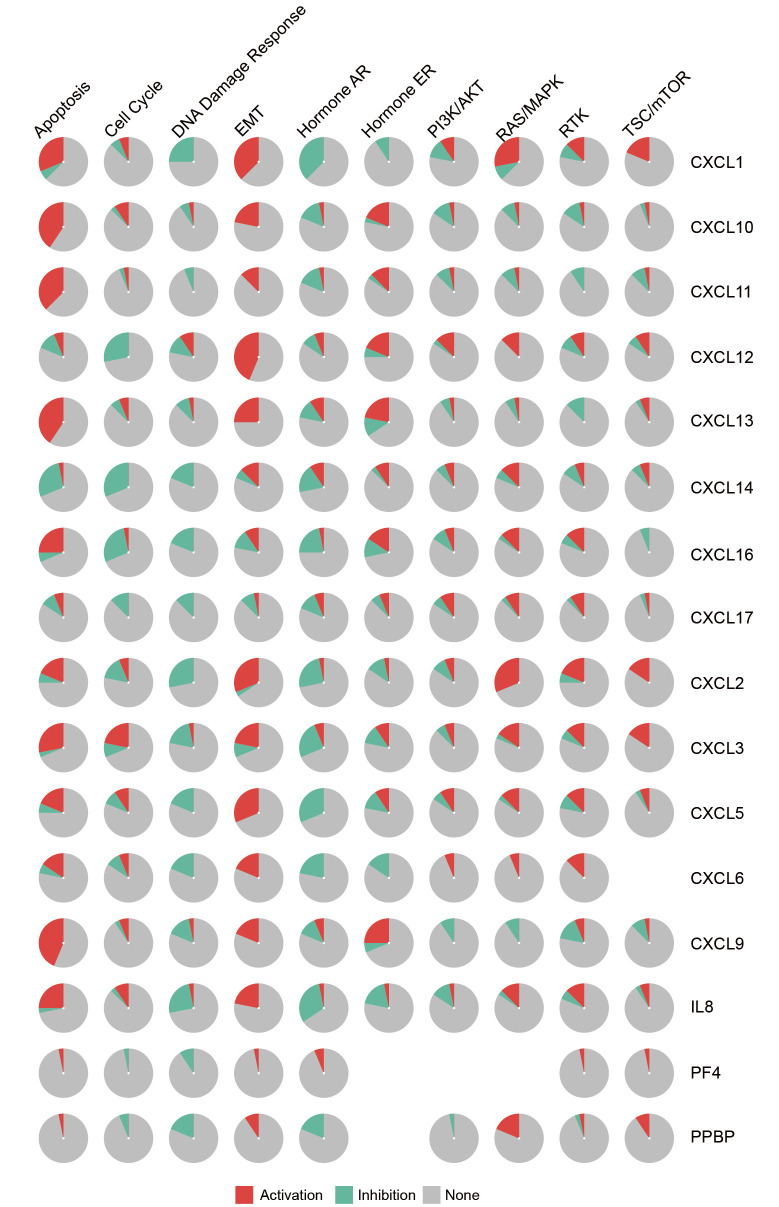
The association between CXC chemokines and activity of ten cancer pathways was analyzed using the GSCALite online tool (Figure 7). The analysis revealed that the DNA damage response and hormone AR signaling pathways were inhibited, whereas apoptosis, EMT and Ras/MAPK signaling pathways were activated.
Figure 7.
The association between CXC chemokines and activity of ten cancer pathways (GSCALite). Red indicates the activation percentage of CXC chemokines to a pathway. Green indicates the inhibition percentage of CXC chemokines to a pathway. Grey indicates the percentage of CXC chemokines that have no effect on a pathway.
The association between CXC chemokines and immune cell infiltration
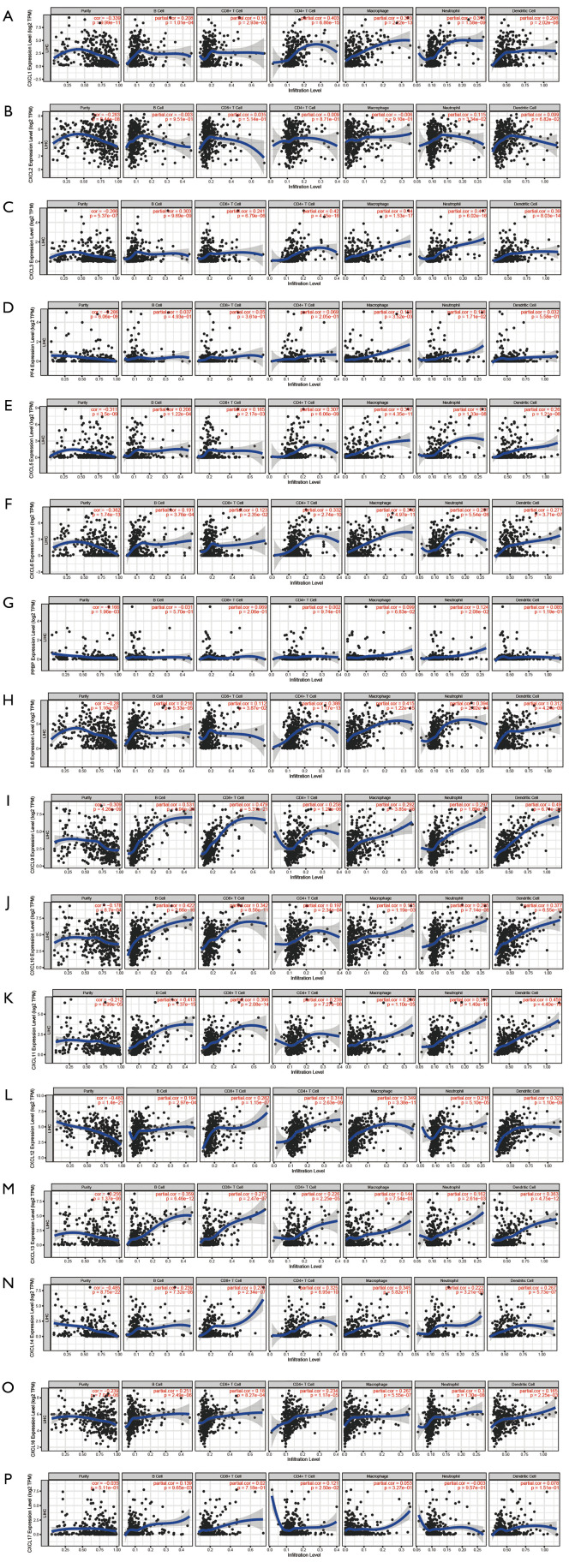
The association between CXC chemokines and infiltration of immune cells within HCC microenvironments was further evaluated using data from TIMER database (Figure 8). Results of the analysis demonstrated that the expression levels of CXCL1, CXCL3, CXCL5, CXCL6, CXCL8, CXCL9, CXCL10, CXCL11, CXCL12, CXCL13, CXCL14 and CXCL16 were positively correlated with six immune cell subsets. The expression levels of CXCL2 and CXCL7 were positively correlated with neutrophils. The expression level of CXCL4 was positively correlated with both macrophages and neutrophils. The expression level of CXCL17 was positively correlated with both B cells and CD4+ T cells.
Figure 8.
The correlation between CXC chemokines and immune cell infiltration abundance (TIMER). The x-axis represents the abundance of immune cell infiltration. The y-axis represents the expression levels of CXC chemokines. The blue line represents the trend of expression level. The scatter represents the distribution of the expression value for each sample.
Furthermore, the associations between the six immune cell subsets and CXC chemokines, and clinical outcomes of patients with HCC were analyzed using the Cox proportional hazard model. The analysis revealed that B cells, macrophages, dendritic cells, CXCL2, CXCL6, CXCL8 and CXCL12 can significantly affect the clinical outcomes of patients with HCC (Table 2).
Table 2. The Cox proportional hazard model of CXC chemokines and six immune cells in hepatocellular carcinoma (TIMER).
| Factor | coef | HR | 95% CI_low | 95% CI_up | P value | sig |
|---|---|---|---|---|---|---|
| B cell | −9.198 | 0.000 | 0.000 | 0.146 | 0.013 | * |
| CD8_Tcell | −5.388 | 0.005 | 0.000 | 1.095 | 0.054 | |
| CD4_Tcell | −3.968 | 0.019 | 0.000 | 15.100 | 0.245 | |
| Macrophage | 7.093 | 1,203.394 | 5.072 | 285,492.940 | 0.011 | * |
| Neutrophil | −4.923 | 0.007 | 0.000 | 5,375.505 | 0.475 | |
| Dendritic | 6.610 | 742.556 | 16.357 | 33,710.562 | 0.001 | ** |
| CXCL1 | 0.130 | 1.139 | 0.928 | 1.397 | 0.213 | |
| CXCL2 | −0.226 | 0.798 | 0.699 | 0.911 | 0.001 | ** |
| CXCL3 | −0.393 | 0.675 | 0.434 | 1.050 | 0.081 | |
| PF4 | −0.058 | 0.944 | 0.631 | 1.412 | 0.778 | |
| CXCL5 | 0.068 | 1.071 | 0.880 | 1.302 | 0.493 | |
| CXCL6 | −0.219 | 0.803 | 0.676 | 0.954 | 0.013 | * |
| PPBP | −0.095 | 0.909 | 0.484 | 1.710 | 0.768 | |
| IL8 | 0.292 | 1.340 | 1.126 | 1.594 | 0.001 | ** |
| CXCL9 | −0.174 | 0.840 | 0.671 | 1.051 | 0.127 | |
| CXCL10 | −0.002 | 0.998 | 0.797 | 1.249 | 0.984 | |
| CXCL11 | 0.039 | 1.040 | 0.758 | 1.427 | 0.810 | |
| CXCL12 | −0.173 | 0.841 | 0.725 | 0.975 | 0.022 | * |
| CXCL13 | 0.080 | 1.083 | 0.960 | 1.223 | 0.195 | |
| CXCL14 | −0.003 | 0.997 | 0.822 | 1.210 | 0.978 | |
| CXCL16 | −0.058 | 0.943 | 0.790 | 1.126 | 0.517 | |
| CXCL17 | 0.142 | 1.153 | 0.994 | 1.336 | 0.059 |
*P<0.05, **P<0.01.
Meta-analysis of external multiple gene sets for validating TCGA data
Three GSE datasets were integrated (a total of 221 samples) and meta-analysis performed using NetworkAnalyst online tool to validate mRNA expression of CXC chemokines in HCC and normal tissues. The meta-analysis revealed that the mRNA expression levels of CXCL10 and CXCL11 were significantly upregulated, whereas the mRNA expression levels of CXCL2, CXCL4, CXCL6, CXCL7, CXCL12 and CXCL14 were significantly downregulated. These results were consistent with results obtained from TCGA database (Table 3).
Table 3. The different expression levels of CXC chemokines between hepatocellular carcinoma and normal tissues by meta-analysis (NetworkAnalyst).
| Entrez ID | Gene | Type | CombinedTstat | CombinedPval |
|---|---|---|---|---|
| 2920 | CXCL2 | Hepatocellular carcinoma vs. Normal | −115.71 | 0 |
| 5196 | CXCL4 | Hepatocellular carcinoma vs. Normal | −62.788 | 7.23E-11 |
| 6372 | CXCL6 | Hepatocellular carcinoma vs. Normal | −28.223 | 1.88E-04 |
| 5473 | CXCL7 | Hepatocellular carcinoma vs. Normal | −78.391 | 7.25E-14 |
| 3627 | CXCL10 | Hepatocellular carcinoma vs. Normal | 25.257 | 6.12E-04 |
| 6373 | CXCL11 | Hepatocellular carcinoma vs. Normal | 29.874 | 9.70E-05 |
| 6387 | CXCL12 | Hepatocellular carcinoma vs. Normal | −165.13 | 0 |
| 9547 | CXCL14 | Hepatocellular carcinoma vs. Normal | −237.31 | 0 |
Discussion
The analysis of CXC chemokines in the HCC microenvironment has become more convenient and accurate with the establishment of several online databases. The present study explored the mRNA expression levels of CXC chemokines in HCC and normal tissues, and performed functional enrichment analyses for a total of 116 genes. Subsequently, the association between CXC chemokines and activity of ten cancer pathways was analyzed, and the correction between CXC chemokines and infiltration of six immune cell subsets was evaluated. Additionally, three GSE datasets were integrated as external data to validate mRNA expression levels of CXC chemokines.
CXCL1 was initially cloned from fibroblasts (33). Cao et al. demonstrated that the expression of CXCL1 significantly increased in HCC tissues compared to normal tissues, and high expression of CXCL1 could promote proliferation and invasion of HCC cells through the NF-kB-dependent pathway, which was associated with poor overall survival in comparison with low expression (34). Previous studies have demonstrated that the mechanisms of CXCL1 that induced tumor cell invitation were associated with stimulating the expression of epidermal growth factor (EGF) and increasing the rate of angiogenesis (35,36). The present study revealed that the expression of CXCL1 decreased in HCC tissues, and high expression of CXCL1 positively correlated with poor prognosis in patients with HCC.
CXCL2 is secreted by macrophages, endothelial, epithelial and tumor cells (37). Peng et al. revealed that monocyte-derived CXCL2 was responsible for the recruitment and survival of neutrophils in HCC environment, subsequently enhancing progression of HCC (15). Moreover, miRNA-532-5p can act as a tumor suppressor by downregulating the expression of its target, CXCL2 mRNA (38). By contrast, Ding et al. identified CXCL2 as a tumor suppressor for the first time, and established that CXCL2 was downregulated in HCC tissues compared to adjacent normal tissues, and upregulated CXCL2 indicated better overall survival, which is consistent with the findings of this study (39).
CXCL3 is an encoding growth factor protein that binds to CXCR2 and acts as a chemoattractant for neutrophils. Han et al. established that the mRNA and protein levels of CXCL3 were upregulated in HCC tissues compared to adjacent normal tissues in nude mice models (40). Similarly, Zhang et al. reported that the high expression of CXCL3 indicated poor prognosis, which enhanced maintenance and tumorigenesis of hepatocellular stem cells via phosphorylation of the ERK1/2 pathway (41). Results of this study indicated that the high expression of CXCL3 was associated with poor prognosis.
CXCL5 is believed to recruit neutrophils, promote angiogenesis and remodel connective tissues by binding to CXCR2. Moreover, CXCL5 participates in a positive feedback loop between tumor-associated neutrophils (TANs) and HCC stem-like cells induced by TANs (16). Zhou et al. demonstrated that the expression of CXCL5 increased in HCC tissues compared to adjacent normal tissues, and high expression of CXCL5 promoted the progression of HCC by activating the PI3K-Akt and ERK1/2 pathways (42). Results of the present study indicated that the expression of CXCL5 decreased in HCC tissues, and high expression of CXCL5 positively correlated with poor prognosis in patients with HCC.
CXCL6, which functions as an angiogenic gene, is also a chemotactic for neutrophils in HCC. In our research, Table 1 indicated that the expression level of CXCL6 was increased in HCC tissues compared with adjacent normal tissues. This finding was from a small sample size. On the contrary, based on TCGA database (N=374) and integrated three GEO datasets (Table 3) (N=221), we found that CXCL6 was decreased in HCC tissues compared with adjacent normal tissues. This conclusion was from a large sample size. Additionally, according to information from the TCGA database, Table 2 indicated that CXCL6 was an independent protective factor (HR =0.803) of OS in patients with HCC. Therefore, from sample size perspective, we think that CXCL6 may be decreased in HCC tissues compared with adjacent normal tissues. This conclusion was in accordance with a previous study. Zhao et al. demonstrated that circ-HOMER1 facilitated the growth and aggressiveness of HCC by enhancing the inhibition of miR-1322 on CXCL6 (43). However, Kaplan-Meier analysis in Figure 6 showed that increased expression of CXCL6 positively correlated with poor prognosis of patients with HCC, which caused confusing among researchers and clinicians. Although Tian et al. reported that HIF-1α induced migration and invasion of HCC cells in a CXCL6-dependent manner, and that high expression of CXCL6 was positively correlated with poor prognosis (44). For this inconsistent finding in our study, we consider that this is primary due to the different choices of cut-off value. In this study, we chose the most significant cut-off value not median value to perform Kaplan-Meier analysis, which leaded to inconsistent results. Therefore, the relationship between the expression level of CXCL6 and prognosis of HCC warrants further research.
CXCL8 is a chemotactic for neutrophils and is secreted by macrophages, neutrophils, eosinophils, T lymphocytes, epithelial cells, and fibroblasts. A study by Yin et al. revealed that tumor associated macrophages (TAMs) triggered growth and metastasis of HCC cells by expressing high levels of CXCL8 (45). Sun et al. reported that the expression of CXCL8 was upregulated in highly metastatic HCC cell lines compared to low metastatic cell lines, and that CXCL8 promoted invasion of HCC cells through activation of the PI3K/Akt pathway (46). Huang et al. explored the association between long non-coding RNAs (lncRNAs) and proliferation, and invasion of HCC cells (47). The study demonstrated that the lncRNA, MALAT1 can upregulate the expression of CXCL6 and CXCL8 to stimulate HCC progression. In this study, we established that high expression of CXCL8 positively correlated with poor survival in patients with HCC.
CXCL9, which binds to CXCR3, is a chemoattractant for lymphocytes. Ding et al. demonstrated that the CXCL9-CXCR3 axis can promote metastasis of HCC cells by activating the ERK1/2-MMP2/MMP9 pathway (48). Moreover, a previous study revealed that high expression of CXCL9 was positively correlated with poor prognosis in patients with HCC (49). Additionally, Liu et al. reported that CXCR3+ B cells induced M2b macrophage polarization resulting in early recurrence of HCC, in which recruitment and maturation were associated with high expression of CXCL9 stimulated by proinflammatory IL-17+ cells (50). By contrast, the present study demonstrates that high expression of CXCL9 is positively correlated with better prognosis in patients with HCC.
CXCL10 can activate monocytes, natural killer cells and T-cell migration by binding to CXCR3. Huang et al. established that the expression of circular RNA, circMET increased in HCC tissues compared to adjacent normal tissues, which subsequently induced immunosuppression within the HCC microenvironment through miR-30-5p/Snail/DPP4/CXCL10 axis (14). Li et al. reported that IL-25-induced M2 macrophages activated EMT in HCC cells, which was associated with secretion of CXCL10 and phosphorylation of ERK (51). Ren et al. demonstrated that upregulated CXCL10 enhanced invasion and metastasis of HCC cells by activating MMP-2 expression (52). The present study established that the high expression of CXCL10 is associated with better prognosis in patients with HCC.
CXCL11 is a dominant ligand for CXCR3, and is a chemoattractant for T-cells. Zhang et al. reported that the high expression of CXCL10 promoted acquisition and maintenance of the properties of HCC stem cells by binding to CXCR3, which was associated with activation of the ERK1/2 pathway (18). Monnier et al. indicated that mRNA and protein expression levels of CXCL10 in HCC tissues were higher than those in adjacent normal tissues, resulting in the growth and metastasis of HCC cells (53). In addition, CXCL11 facilitated M2b macrophage polarization and caused early recurrence in patients with HCC (50).
CXCL12 is a ligand for CXCR4 and is responsible for inflammatory responses. Tsai et al. established that CXCL12 activated by SOX4 can modulate neovascularization and cause distant metastasis of HCC (17). Yang et al. also demonstrated that the CXCL12-CXCR4 axis participated in angiogenesis of HCC, and was positively correlated with poor survival of patients with HCC (54). Lu et al. reported that high expression of miR-342 could suppress the Wnt/β-catenin pathway by inhibiting CXCL12 expression, which subsequently inhibited proliferation and metastasis of HCC cells (55). In this study, we established that the expression of CXCL12 decreased in HCC tissues, and high expression of CXCL12 positively correlated with better prognosis in patients with HCC.
CXCL13, which is a chemoattractant for B lymphocytes, is strongly expressed in the spleen, lymph nodes, and Peyer’s patches. Li et al. reported that the serum levels of CXCL13 were significantly higher in patients with HCC compared to healthy controls, and that the serum level of CXCL13 was positively correlated with large tumor size, HCC metastasis and advanced stage of HCC (56). Similarly, a previous study demonstrated that high mRNA and serum levels of CXCL13 contributed to the progression of HCC by activating the Wnt/β-catenin pathway (57).
CXCL14 is a chemoattractant for monocytes and has been identified as an essential tumor suppressor in HCC (58). Two previous studies have indicated that the expression of CXCL14 decreased in HCC tissues compared to adjacent normal tissues, and that high expression of CXCL14 can inhibit proliferation and invasion of HCC cells, which is consistent with the findings of the present study (59,60).
CXCL16 is a ligand for CXCR6, which plays a role in tumorigenesis. Two previous studies have indicated that the expression of CXCL16-CXCR6 axis increased in HCC tissues compared to adjacent normal tissues, which was associated with early recurrence and poor survival of patients with HCC (61,62). In addition, Cai et al. reported that the expression of CXCL16 was upregulated in M2-polarized macrophages, which triggered migration and invasion of HCC cells (63). Results of this study also revealed that the expression of CXCL16 increased in HCC tissues.
CXCL17 is a chemoattractant for monocytes and dendritic cells, and it promotes tumorigenesis by inducing angiogenic activity. Previous studies have revealed that the expression of CXCL17 increased in HCC tissues compared to adjacent normal tissues (64,65). This may play a role in promoting progression of HCC in patients. Furthermore, Wang et al. established that CXCL17 could regulate proliferation, metastasis and autophagy of HCC cells via the LKB1-AMPK axis (66). Results obtained in this study on the expression of CXCL16 in HCC tissues are consistent with those of previous studies.
The roles of CXC chemokines in the occurrence and progression of HCC was analyzed using several online databases. These databases can be used to analyze several aspects conveniently including mRNA expression levels, survival of patients, activity of cancer pathways, and infiltration of immune cells. Nevertheless, we identified disparities with reference to mRNA levels and prognostic values for certain CXC chemokines after reviewing and comparing previous studies. The contrasting findings observed in CXC chemokines may be associated with several reasons. First, different clinical samples and experimental conditions may result in the difference. Second, these online analysis tools used in this study may adopt different data cleaning methods. Third, different RNA sequencing technologies could lead to inconsistent findings. Therefore, further clinical trials are required to validate the potential roles and mechanisms of the CXC chemokines evaluated by this study. This study provides a better understanding of the roles of CXC chemokines in HCC and the information will help clinicians to predict prognosis and select suitable therapies for patients with HCC.
Conclusions
This study has demonstrated that CXC chemokines can modulate the interactions between immune and HCC cells in a tumor microenvironment. This consequently influences the prognosis and therapy of patients with HCC. Analysis of differential expression levels of CXC chemokines between HCC and adjacent normal tissues revealed that high mRNA expression levels of CXCL1/3/5/6/8 were associated with poor overall survival, whereas high mRNA expression levels of CXCL2/4/7/9/10/12 were associated with better overall survival. Analysis of the association between CXC chemokines and activity of cancer pathways indicated that the DNA damage response and hormone AR signaling pathways were inhibited, whereas apoptosis, EMT and Ras/MAPK signaling pathways were activated. In addition, the expressions of CXC chemokines were positively correlated with infiltration of six types of immune cells (B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and dendritic cells).
Acknowledgments
Funding: Beijing Hospitals Authority Youth Programme (QMS20200803 to CZ). National Natural Science Foundation of China (No. 81800483).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-21-127
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-21-127). The authors have no conflicts of interest to declare.
References
- 1.Marrero JA, Kulik LM, Sirlin CB, et al. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018;68:723-50. 10.1002/hep.29913 [DOI] [PubMed] [Google Scholar]
- 2.Tu T, Bühler S, Bartenschlager R. Chronic viral hepatitis and its association with liver cancer. Biol Chem 2017;398:817-37. 10.1515/hsz-2017-0118 [DOI] [PubMed] [Google Scholar]
- 3.Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma . Cell 2017;169:1327-41.e23. 10.1016/j.cell.2017.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singal AG, Nehra M, Adams-Huet B, et al. Detection of hepatocellular carcinoma at advanced stages among patients in the HALT-C trial: where did surveillance fail? Am J Gastroenterol 2013;108:425-32. 10.1038/ajg.2012.449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roayaie S, Obeidat K, Sposito C, et al. Resection of hepatocellular cancer ≤2 cm: results from two Western centers. Hepatology 2013;57:1426-35. 10.1002/hep.25832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-14. 10.1016/S0140-6736(18)30010-2 [DOI] [PubMed] [Google Scholar]
- 7.Hernandez-Gea V, Toffanin S, Friedman S, et al. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology 2013;144:512-27. 10.1053/j.gastro.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greten FR, Grivennikov SI. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019;51:27-41. 10.1016/j.immuni.2019.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer 2004;4:540-50. 10.1038/nrc1388 [DOI] [PubMed] [Google Scholar]
- 10.Crespo J, Sun H, Welling T, et al. T cell anergy, exhaustion, senescence, and stemness in the tumor microenvironment. Curr Opin Immunol 2013;25:214-21. 10.1016/j.coi.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol 2014;32:659-702. 10.1146/annurev-immunol-032713-120145 [DOI] [PubMed] [Google Scholar]
- 12.Zou W, Wolchok J, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med 2016;8:328rv4. 10.1126/scitranslmed.aad7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zlotnik A, Yoshie O. The chemokine superfamily revisited. Immunity 2012;36:705-16. 10.1016/j.immuni.2012.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang XY, Zhang PF, Wei CY, et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer 2020;19:92. 10.1186/s12943-020-01213-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng ZP, Jiang ZZ, Guo HF, et al. Glycolytic activation of monocytes regulates the accumulation and function of neutrophils in human hepatocellular carcinoma. J Hepatol 2020;73:906-17. 10.1016/j.jhep.2020.05.004 [DOI] [PubMed] [Google Scholar]
- 16.Zhou SL, Yin D, Hu ZQ, et al. A Positive Feedback Loop Between Cancer Stem-Like Cells and Tumor-Associated Neutrophils Controls Hepatocellular Carcinoma Progression. Hepatology 2019;70:1214-30. 10.1002/hep.30630 [DOI] [PubMed] [Google Scholar]
- 17.Tsai CN, Yu SC, Lee CW, et al. SOX4 activates CXCL12 in hepatocellular carcinoma cells to modulate endothelial cell migration and angiogenesis in vivo. Oncogene 2020;39:4695-710. 10.1038/s41388-020-1319-z [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Zhao W, Li S, et al. CXCL11 promotes self-renewal and tumorigenicity of α2δ1 liver tumor-initiating cells through CXCR3/ERK1/2 signaling. Cancer Lett 2019;449:163-71. 10.1016/j.canlet.2019.02.016 [DOI] [PubMed] [Google Scholar]
- 19.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-D613. 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warde-Farley D, Donaldson S, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214-20. 10.1093/nar/gkq537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang da W , Sherman B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 24.Huang da W , Sherman B, Lempicki R. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci 2018;5:181006. 10.1098/rsos.181006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-e10. 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu CJ, Hu FF, Xia MX, et al. GSCALite: a web server for gene set cancer analysis. Bioinformatics 2018;34:3771-2. 10.1093/bioinformatics/bty411 [DOI] [PubMed] [Google Scholar]
- 28.Zhou G, Soufan O, Ewald J, et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 2019;47:W234-W41. 10.1093/nar/gkz240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, Cheung S, So S, et al. Gene expression patterns in human liver cancers. Mol Biol Cell 2002;13:1929-39. 10.1091/mbc.02-02-0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roessler S, Jia HL, Budhu A, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res 2010;70:10202-12. 10.1158/0008-5472.CAN-10-2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wurmbach E, Chen YB, Khitrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 2007;45:938-47. 10.1002/hep.21622 [DOI] [PubMed] [Google Scholar]
- 32.Mas VR, Maluf DG, Archer KJ, et al. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med 2009;15:85-94. 10.2119/molmed.2008.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haskill S, Peace A, Morris J, et al. Identification of three related human GRO genes encoding cytokine functions. Proc Natl Acad Sci U S A 1990;87:7732-6. 10.1073/pnas.87.19.7732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao Z, Fu B, Deng B, et al. Overexpression of Chemokine (C-X-C) ligand 1 (CXCL1) associated with tumor progression and poor prognosis in hepatocellular carcinoma. Cancer Cell Int 2014;14:86. 10.1186/s12935-014-0086-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyake M, Goodison S, Urquidi V, et al. Expression of CXCL1 in human endothelial cells induces angiogenesis through the CXCR2 receptor and the ERK1/2 and EGF pathways. Lab Invest 2013;93:768-78. 10.1038/labinvest.2013.71 [DOI] [PubMed] [Google Scholar]
- 36.Warner KA, Miyazawa M, Cordeiro MMR, et al. Endothelial cells enhance tumor cell invasion through a crosstalk mediated by CXC chemokine signaling. Neoplasia 2008;10:131-9. 10.1593/neo.07815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vansaun MN, Mendonsa AM, Lee Gorden D. Hepatocellular proliferation correlates with inflammatory cell and cytokine changes in a murine model of nonalchoholic fatty liver disease. PLoS One 2013;8:e73054. 10.1371/journal.pone.0073054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang ZT, Han Y, Liu XY, et al. Tripterine Restrains the Aggressiveness of Hepatocellular Carcinoma Cell via Regulating miRNA-532-5p/CXCL2 Axis. Onco Targets Ther 2020;13:2973-85. 10.2147/OTT.S238074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding J, Xu K, Zhang J, et al. Overexpression of CXCL2 inhibits cell proliferation and promotes apoptosis in hepatocellular carcinoma. BMB Rep 2018;51:630-5. 10.5483/BMBRep.2018.51.12.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han KQ, Han H, He XQ, et al. Chemokine CXCL1 may serve as a potential molecular target for hepatocellular carcinoma. Cancer Med 2016;5:2861-71. 10.1002/cam4.843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Zhang L, Li H, et al. CXCL3 contributes to CD133(+) CSCs maintenance and forms a positive feedback regulation loop with CD133 in HCC via Erk1/2 phosphorylation. Sci Rep 2016;6:27426. 10.1038/srep27426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou SL, Dai Z, Zhou ZJ, et al. Overexpression of CXCL5 mediates neutrophil infiltration and indicates poor prognosis for hepatocellular carcinoma. Hepatology 2012;56:2242-54. 10.1002/hep.25907 [DOI] [PubMed] [Google Scholar]
- 43.Zhao M, Dong G, Meng Q, et al. Circ-HOMER1 enhances the inhibition of miR-1322 on CXCL6 to regulate the growth and aggressiveness of hepatocellular carcinoma cells. J Cell Biochem 2020;121:4440-9. 10.1002/jcb.29672 [DOI] [PubMed] [Google Scholar]
- 44.Tian H, Huang P, Zhao Z, et al. HIF-1α plays a role in the chemotactic migration of hepatocarcinoma cells through the modulation of CXCL6 expression. Cell Physiol Biochem 2014;34:1536-46. 10.1159/000366357 [DOI] [PubMed] [Google Scholar]
- 45.Yin Z, Huang J, Ma T, et al. Macrophages activating chemokine (C-X-C motif) ligand 8/miR-17 cluster modulate hepatocellular carcinoma cell growth and metastasis. Am J Transl Res 2017;9:2403-11. [PMC free article] [PubMed] [Google Scholar]
- 46.Sun F, Wang J, Sun Q, et al. Interleukin-8 promotes integrin β3 upregulation and cell invasion through PI3K/Akt pathway in hepatocellular carcinoma. J Exp Clin Cancer Res 2019;38:449. 10.1186/s13046-019-1455-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang M, Wang H, Hu X, et al. lncRNA MALAT1 binds chromatin remodeling subunit BRG1 to epigenetically promote inflammation-related hepatocellular carcinoma progression. Oncoimmunology 2019;8:e1518628. 10.1080/2162402X.2018.1518628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding Q, Xia Y, Ding S, et al. An alternatively spliced variant of CXCR3 mediates the metastasis of CD133+ liver cancer cells induced by CXCL9. Oncotarget 2016;7:14405-14. 10.18632/oncotarget.7360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lan X, Xiao F, Ding Q, et al. The effect of CXCL9 on the invasion ability of hepatocellular carcinoma through up-regulation of PREX2. J Mol Histol 2014;45:689-96. 10.1007/s10735-014-9593-0 [DOI] [PubMed] [Google Scholar]
- 50.Liu RX, Wei Y, Zeng QH, et al. Chemokine (C-X-C motif) receptor 3-positive B cells link interleukin-17 inflammation to protumorigenic macrophage polarization in human hepatocellular carcinoma. Hepatology 2015;62:1779-90. 10.1002/hep.28020 [DOI] [PubMed] [Google Scholar]
- 51.Li Q, Ma L, Shen S, et al. Intestinal dysbacteriosis-induced IL-25 promotes development of HCC via alternative activation of macrophages in tumor microenvironment. J Exp Clin Cancer Res 2019;38:303. 10.1186/s13046-019-1271-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ren T, Zhu L, Cheng M. CXCL10 accelerates EMT and metastasis by MMP-2 in hepatocellular carcinoma. Am J Transl Res 2017;9:2824-37. [PMC free article] [PubMed] [Google Scholar]
- 53.Monnier J, Boissan M, L'Helgoualc'h A, et al. CXCR7 is up-regulated in human and murine hepatocellular carcinoma and is specifically expressed by endothelial cells. Eur J Cancer 2012;48:138-48. 10.1016/j.ejca.2011.06.044 [DOI] [PubMed] [Google Scholar]
- 54.Yang J, Zhang L, Jiang Z, et al. TCF12 promotes the tumorigenesis and metastasis of hepatocellular carcinoma via upregulation of CXCR4 expression. Theranostics 2019;9:5810-27. 10.7150/thno.34973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu C, Jia S, Zhao S, et al. MiR-342 regulates cell proliferation and apoptosis in hepatocellular carcinoma through Wnt/β-catenin signaling pathway. Cancer Biomark 2019;25:115-26. 10.3233/CBM-192399 [DOI] [PubMed] [Google Scholar]
- 56.Li B, Su H, Cao J, et al. CXCL13 rather than IL-31 is a potential indicator in patients with hepatocellular carcinoma. Cytokine 2017;89:91-7. 10.1016/j.cyto.2016.08.016 [DOI] [PubMed] [Google Scholar]
- 57.Li C, Kang D, Sun X, et al. The Effect of C-X-C Motif Chemokine 13 on Hepatocellular Carcinoma Associates with Wnt Signaling. Biomed Res Int 2015;2015:345413. 10.1155/2015/345413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu X, Wang H, Wang A, et al. An intronic polymorphism rs2237062 in the CXCL14 gene influences HBV-related HCC progression in Chinese population. Mol Biol Rep 2012;39:797-803. 10.1007/s11033-011-0801-7 [DOI] [PubMed] [Google Scholar]
- 59.Lin Y, Chen B, Yu X, et al. Suppressed Expression of CXCL14 in Hepatocellular Carcinoma Tissues and Its Reduction in the Advanced Stage of Chronic HBV Infection. Cancer Manag Res 2019;11:10435-43. 10.2147/CMAR.S220528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang W, Huang P, Zhang L, et al. Antitumor efficacy of C-X-C motif chemokine ligand 14 in hepatocellular carcinoma in vitro and in vivo. Cancer Sci 2013;104:1523-31. 10.1111/cas.12279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao Q, Zhao Y, Wang X, et al. CXCR6 upregulation contributes to a proinflammatory tumor microenvironment that drives metastasis and poor patient outcomes in hepatocellular carcinoma. Cancer Res 2012;72:3546-56. 10.1158/0008-5472.CAN-11-4032 [DOI] [PubMed] [Google Scholar]
- 62.Peddibhotla S, Hershberger P, Jason Kirby R, et al. Discovery of small molecule antagonists of chemokine receptor CXCR6 that arrest tumor growth in SK-HEP-1 mouse xenografts as a model of hepatocellular carcinoma. Bioorg Med Chem Lett 2020;30:126899. 10.1016/j.bmcl.2019.126899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cai H, Zhu X, Ao J, et al. Colony-stimulating factor-1-induced AIF1 expression in tumor-associated macrophages enhances the progression of hepatocellular carcinoma. Oncoimmunology 2017;6:e1333213. 10.1080/2162402X.2017.1333213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li L, Yan J, Xu J, et al. CXCL17 expression predicts poor prognosis and correlates with adverse immune infiltration in hepatocellular carcinoma. PLoS One 2014;9:e110064. 10.1371/journal.pone.0110064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou Z, Lu X, Zhu P, et al. VCC-1 over-expression inhibits cisplatin-induced apoptosis in HepG2 cells. Biochem Biophys Res Commun 2012;420:336-42. 10.1016/j.bbrc.2012.02.160 [DOI] [PubMed] [Google Scholar]
- 66.Wang L, Li H, Zhen Z, et al. CXCL17 promotes cell metastasis and inhibits autophagy via the LKB1-AMPK pathway in hepatocellular carcinoma. Gene 2019;690:129-36. 10.1016/j.gene.2018.12.043 [DOI] [PubMed] [Google Scholar]