Abstract
Based on the variability of 43 spacers within the direct repeat (DR) locus of Mycobacterium tuberculosis complex organisms, spoligotyping is a rapid method that aids in the study of the epidemiology of tuberculosis. It was recently hypothesized that despite its presence in the DR locus, spacer 31 could not be amplified in M. tuberculosis clinical isolates belonging to spoligotype 50 due to the insertion of an extra copy of IS6110 between spacers 31 and 32 that could lead to an asymmetrical split of the primer targets (I. Filliol, C. Sola, and N. Rastogi, J. Clin. Microbiol. 38:1231–1234, 2000). In the present investigation, previous observations were extended to 25 clinical isolates of type 50 showing that the primer set IS6-DRb that selectively amplified the left and central DR regions was indeed able to demonstrate the presence of spacer 31. IS6110-restriction fragment length polymorphism (RFLP) and DR-RFLP showed that type 50 isolates were characterized by the presence of two copies of IS6110 associated with the DR locus and an additional double IS6110 band of 1.4 kb. The primer set IS3-IS6 was then used to selectively amplify a 750-bp inter-IS6110 fragment within the DR locus. The sequencing of the central DR region corroborated our previous findings and showed that the absence of spacer 31 among the type 50 isolates was due to the asymmetric insertion of an extra copy of IS6110 between spacers 31 and 32, leading to an unequal split of the DRa-DRb target into two portions, of 6 and 30 bp, respectively. These results show that the DR locus constitutes an ideal IS6110 preferential locus (ipl), permitting the insertion of two or more copies of IS6110, and provide new clues for epidemiological and phylogenetic interpretation of changes in IS6110-RFLP and spoligotyping profiles.
Among DNA fingerprinting techniques that permit the investigation of patient-to-patient transmission of tubercle bacilli, both IS6110-restriction fragment length polymorphism (RFLP) and spoligotyping constitute important molecular tools to study tuberculosis epidemiology (15). However, in contrast to IS6110-RFLP, which remains a cumbersome technique requiring large amounts of bacterial DNA, PCR-based spoligotyping is an easy and rapid method that can be easily applied in a routine microbiological laboratory and is useful for both detection and strain differentiation within the Mycobacterium tuberculosis complex (11). Based on the variability of the inter-direct repeat (DR) spacers, this methodology was originally described for a set of 43 spacers (37 from M. tuberculosis H37Rv and 6 from Mycobacterium bovis BCG [11]). Among 51 new spacers evaluated recently, 26 were found to be specific to Mycobacterium canettii (17), which is a newly recognized species in the M. tuberculosis complex (13, 19).
In a previous work we showed that two of the most prevalent spoligotypes in the world differed only by the absence or presence of spacer 31 (spoligotype patterns 50 and 53, respectively); we hypothesized that despite its presence among the isolates of type 50, spacer 31 may not have been amplified with the primer set DRa-DRb because of the insertion of an extra copy of IS6110 within the DR locus leading to its asymmetrical split and the subsequent lack of the DRa target (6). Using appropriate primer combinations and subsequent sequencing of the central DR region within the DR locus, the present investigation confirms the postulated mechanism by which asymmetric insertion of IS6110 in a DR can eliminate a primer site for the adjacent spacer and thus alter the spoligotype.
Spoligotyping.
All 25 strains (isolates 94072, 94098, 94126, 95013, 95048, 95069, 96020, 96027, 96042, 96060, 96106, 96110, 96121, 96128, 96135, 96136, 96137, 96139, 98042, 98045, 98049, IPC14, IPC22, IPC26, and IPC107) used in this study were isolated at the Institut Pasteur from clinical specimens from patients residing in Guadeloupe or were received for identification and drug-susceptibility testing from neighboring Martinique and French Guiana. The isolates were grown on Löwenstein-Jensen slants at 37°C, and DNA was prepared using the cetyl-trimethyl-ammonium bromide method (18). Standard spoligotyping was performed by using biotinylated (biot) DRa and DRb. The left-right (LR) spoligotyping was performed by using primer sets (biot)DRa-IS3, (biot)IS6-DRb, and (biot)IS6-IS3, which resulted in the amplification of the right and central DR spacers, the left and central DR spacers, and the central DR spacers, respectively. The primers (sequences) used were DRa (GGTTTTGGGTCTGACGAC), DRb (CCGAGAGGGGACGGAAAC), IS3 (GCTGCCTACTACGCTCAAC), and IS6 (CAAGTAGACGGGCGACCTC).
IS6110- and DR-RFLP.
The separation of PvuII-digested DNA restriction fragments was performed on a 0.8% agarose gel (Gibco-BRL Life Technologies, Cergy-Pontoise, France) in 1× Tris-acetate-EDTA buffer (40 mM Tris-acetate, 1 mM EDTA, pH 8.0) followed by Southern blot hybridization using the IS6110 and the DR-right (DR-r) probes as described previously (10, 18). The chemiluminescence-based detection of bands was performed using the ECL RPN 3000 kit for IS6110-RFLP and the ECL RPN 2130 kit for DR-RFLP (Amersham, Buckinghamshire, United Kingdom), according to the manufacturer's instructions.
Inter-IS6110 PCR and sequencing.
Inter-IS6110 PCR using primers IS3 and IS6 was performed as previously described (3, 4). The PCR product was loaded on a 2% (wt/vol) low-melting-temperature agarose gel (Gibco-BRL Life Technologies), run in 1× Tris-acetate-EDTA buffer, and detected by ethidium bromide staining. A 100-bp ladder (Pharmacia-Biotech, Uppsala, Sweden) served as an external molecular size marker. The images were video captured and analyzed using Gel-Analyst software (Bioprobe Systems, Montreuil, France). A specific IS3-IS6 PCR-amplified fragment of about 750 bp was purified on Qiagen columns (QIAquick Gel Extraction Kit; Qiagen S.A., Courtaboeuf, France) and used for both standard and LR spoligotyping. Direct sequencing of the purified 100-ng PCR product was performed using the IS3 and IS6 primers and Thermo-Sequenase dye terminator kits Cy5.0 and Cy5.5 (AP-Biotech, Piscataway, N.J.) on an Opengene Long Read Tower sequencing system (Visible Genetics Inc., Toronto, Canada) or, alternatively, by using the BigDye Terminators premix on an Applied Biosystems 373XL sequencer according to the manufacturer's instructions (Applied Biosystems, Foster City, Calif.). In the latter case, the sequencing was subcontracted to Genome-Express (Genome-Express, Grenoble, France).
Results and discussion.
First described for M. bovis and later for the members of the M. tuberculosis complex (10), the DR locus is composed of multiple 36-bp DR copies interspersed by nonrepetitive short sequences (inter-DR or spacers) of about equal length (11). The DR locus may contain one or two IS6110 sequences that usually occur in tandem (2, 8, 9). A careful examination of the DR locus sequencing data by Groenen et al. (8) revealed that an IS6110 copy may be inserted either symmetrically, with a resulting split of the DR flanking IS6110 into two equal portions, or asymmetrically, with portions of 6 and 30 bp. As we will see below, this little detail may have important consequences on the spoligotyping patterns of one of the most widely represented groups of M. tuberculosis clinical isolates (type 50) which is ubiquitous throughout the world (16).
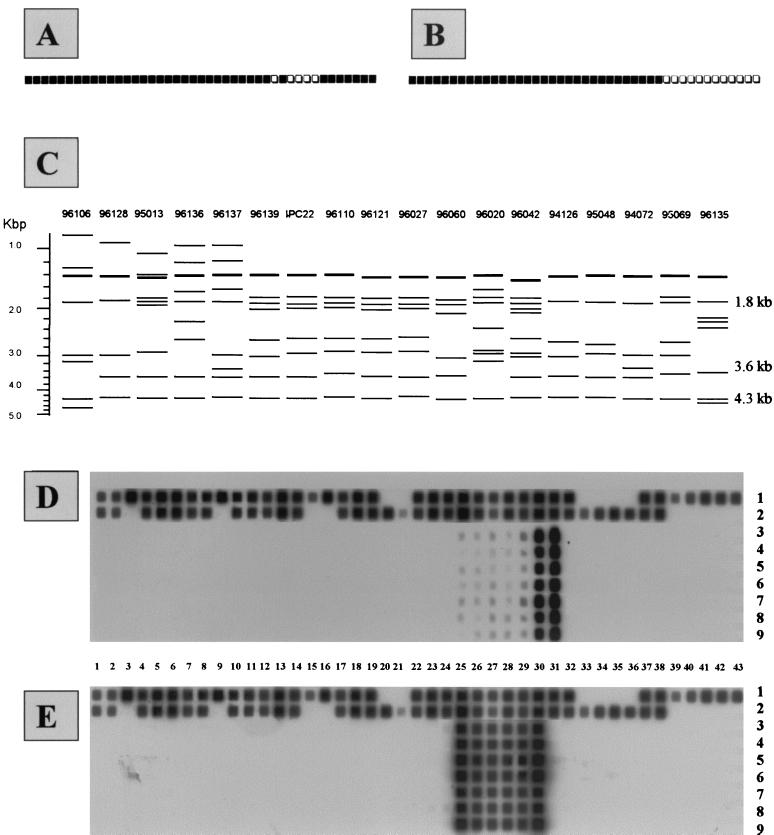
All of the type 50 isolates were characterized by the absence of spacer 31 after routine spoligotyping using primers (biot)DRa and DRb (Fig. 1A); however, spacer 31 was effectively revealed by LR spoligotyping using primers (biot)IS6 and DRb (Fig. 1B). IS6110-RFLP showed that all of the type 50 isolates were characterized by the presence of a double band of 1.4 kb, a single copy at 1.8 kb, and nearly always two other copies, at 3.6 and 4.3 kb (Fig. 1C). To investigate the number of IS6110 elements within or near the DR locus, the PvuII-IS6110 blots were rehybridized with oligonucleotide probe DR-r as described previously (10). In all the strains tested, the DR-r probe uniformly hybridized with the 1.8-, 3.6-, and 4.3-kb PvuII fragments. We conclude that the type 50 isolates do not differ with regard to the chromosomal position of the insertion element and the copy number of the DRs. Since the restriction of a sequence at two digestion sites provides three fragments, these results showed that the presence of three PvuII fragments giving a uniform DR hybridization was indeed compatible with two PvuII sites being located within the DR locus of type 50 isolates. Thus, the two IS6110 copies should allow us to selectively amplify the central part of the DR locus that contains the direct variable repeats (DVRs) numbered 25 to 31, a DVR being defined as the association of one DR and its adjacent spacer (8, 17). Lastly, despite the fact that all of the strains of type 50 represented a unique clade of organisms on the basis of spoligotyping and also the fact that they did not differ in regard to the chromosomal position of the IS6110 element (as revealed by the DR-r probing of the PvuII-digested DNAs), there was significant variation in the IS6110-RFLP patterns obtained (Fig. 1C). Indeed, except for 5 isolates that were clonal (isolates 96139, IPC22, 96110, 96121, and 96027) and 2 isolates that were closely related (94126 and 95048), the remaining 11 isolates were unrelated by IS6110-RFLP (Fig. 1C). This observation suggests that the type 50 isolates belong to a spoligotyping-defined ubiquitous family whose members are not necessarily epidemiologically related.
FIG. 1.
Spoligotyping and IS6110-RFLP results. (A) Schematic representation of spoligotyping using (biot)DRa and DRb primers for type 50 isolates of M. tuberculosis. (B) Schematic representation of spoligotyping pattern using (biot)IS6 and DRb primers. (C) Schematic representation of the IS6110-RFLP results. The 1.4-kb band drawn in bold indicates the double band observed, whereas the three bands at 1.8, 3.6, and 4.3 kb show the PvuII fragments that hybridized with the DR-r probe in strains 94072, 94126, 95013, 95048, 95069, 96027, and IPC22 (shown in the figure) and also in strains 94098, 98049, 98042, 98045, IPC14, IPC26, and IPC107 (not represented here). (D) Spoligotyping of the 750-bp inter-IS6110 fragment and whole DNA controls using primers (biot)IS6 and -IS3. Numbers 1 to 43 show the 43 spacers used for spoligotyping; numbers 1 and 2 on the right-hand side show the results for total DNA from control isolates M. tuberculosis H37Rv and M. bovis BCG, whereas numbers 3 to 9 show the results for the 750-bp inter-IS6110 fragment from the following strains: 3, isolate 94072; 4, isolate 94126; 5, isolate 95013; 6, isolate 95048; 7, isolate 95069; 8, isolate 96027; 9, isolate IPC22. (E) Spoligotyping of the 750-bp inter-IS6110 fragment and whole DNA controls using primers (biot)DRa and -DRb.
Based on the above observations and previous DR sequencing data (8), we decided to selectively amplify the central DR region within the DR locus using primers IS3 and IS6 in order to use the resulting PCR product for both routine and LR spoligotyping as well as for sequencing. The parallel controls included primer sets IS3-IS3 and IS6-IS6 and a negative control. The results obtained with seven randomly selected isolates showed no amplification with the negative control and primers IS3 and IS3, whereas many bands were obtained with primers IS6 and IS6, indicating that inversed IS6110 copies may exist elsewhere in the bacterial choromosome (results not shown). The PCR product obtained using primers IS3 and IS6 showed bands similar to those obtained using IS6 and IS6; however, a single specific band of about 750 bp was found to be exclusively present with primers IS3 and IS6 and corresponded to the theoretical length expected for the central DR region (6, 8). When this fragment was purified and used directly for LR spoligotyping using (biot)IS6-IS3, it revealed the presence of the previously missing spacer 31 as well as spacers 25 to 30 (Fig. 1D). When the same fragment was used for the routine spoligotyping method using (biot)DRa-DRb, only spacers 25 to 30 (and not spacer 31) were revealed (Fig. 1E). One intriguing observation was the fact that this 750-bp fragment always resulted in a high hybridization signal for spacers 30 and 31 and a low hybridization signal for spacers 25 to 29 (Fig. 1D). We have no valid explanation for this observation for the time being.
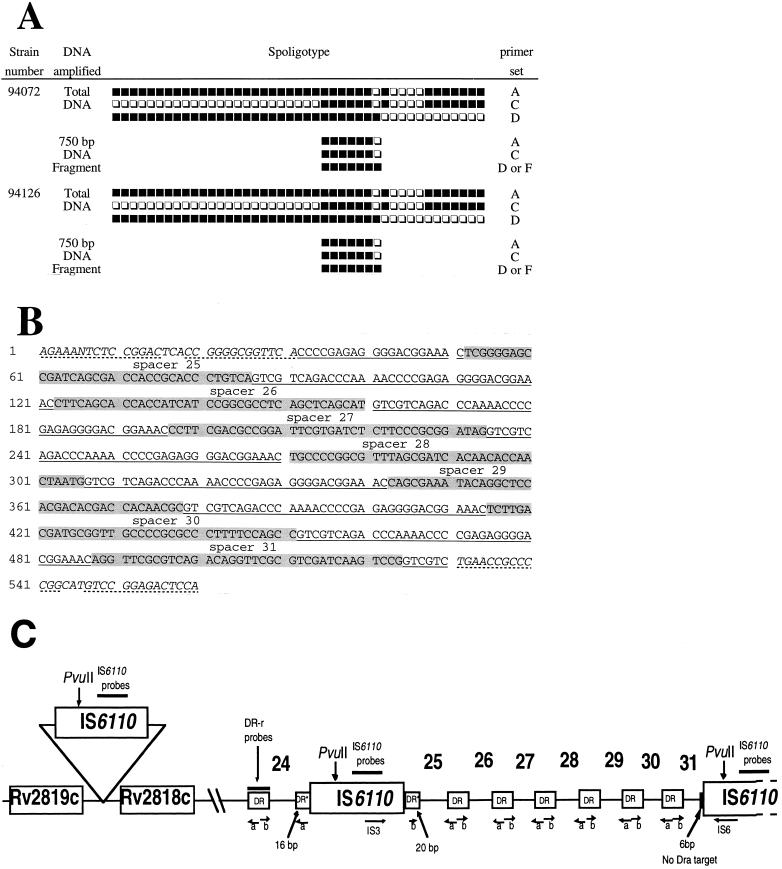
Following these results, two clinical isolates (94072 and 94126) were selected for sequencing experiments. Before sequencing, both the total DNA and the 750-bp fragment from these two isolates were subjected to spoligotyping using all the primer combinations tested; the results obtained (Fig. 2A) unambiguously confirmed that the 750-bp fragment did correspond to the central DR region within the DR locus and strengthened our previous assumption that spacer 31, although present, is not detected among type 50 M. tuberculosis isolates due to an asymmetric insertion of a second copy of IS6110 between spacers 31 and 32 (6).
FIG. 2.
Spoligotyping and inter-IS6110 sequencing results and schematic representation of the central region of the DR locus. (A) Spoligotyping and LR spoligotyping of the total bacterial DNA versus the 750-bp inter-IS6110 fragment for isolates 94072 and 94126. Primer sets: A, (biot)DRa-DRb; C, (biot)DRa-IS3; D, (biot)IS6-DRb; F, (biot)IS6-IS3. (B) Sequencing results of the 750-bp inter-IS6110 fragment obtained using primers IS3 and IS6; the sequence shown was identical for isolates 94072 and 94126, and the illustration shown is limited to the 560-bp portion within the IS6110-flanking region that comprises spacers 25 to 31 as shown below. The shaded areas show the corresponding spacer sequences, and the underlined area corresponds to the DR repeats. The 3′ side of the first copy of IS6110 and the 5′ side of the second copy of IS6110 are shown in italics, with the inverted repeats shown with dotted lines. (C) Schematic representation of the central region of the DR locus for spoligotype 50 isolates of M. tuberculosis. In this figure, spacers 25 to 31 are flanked by two tandem copies of IS6110. Note that on the 3′ flanking side of the first IS6110 copy, 20 bases of one DR are present in DVR 25, whereas on the 5′ flanking side of the second IS6110 copy, DVR 31 is split into two asymmetrical portions, resulting in only a 6-bp portion of the DR bordering the second IS6110 copy, which is not sufficient to allow hybridization with the 18-mer DRa primer. Arrows on IS3, IS6, a (DRa), and b (DRb) show the sense of the PCRs, whereas Rv2818c and Rv2819c show the hypothetical presence of an additional ipl region adjacent to the DR locus. The targets for hybridization with IS6110 and DR-r probes are shown with bold lines; note that the DR-r probe hybridizes with all the DRs. DR* represents target DR flanking the first copy of IS6110.
As expected, the sequencing results showed the presence of spacers 25, 26, 27, 28, 29, 30, and 31 flanked by two tandem copies of IS6110 (Fig. 2B); on the 3′ flanking side of the first IS6110 copy, 20 bases of one DR are present before spacer 25, whereas on the 5′ flanking side of the second IS6110 copy, DVR 31 is split into two asymmetrical portions, resulting in only a 6-bp portion of the DR bordering the second IS6110 copy (Fig. 2C). However, the 6-bp portion of the DR bordering the second IS6110 copy is not sufficient to allow hybridization with the 18-mer DRa primer, hence creating the inability to detect spacer 31 by routine spoligotyping using primers DRa and DRb (Fig. 2C). In our opinion, this is the first definitive evidence showing that IS6110 transposition may alter the spoligotyping profile, and it provides new clues on one of the potential evolutionary mechanisms of the DR locus. In an independent investigation, similar results were recently reported by van Embden et al. (17) for explaining single spacer differences between two M. bovis strains that were isolated during an epidemic of multidrug-resistant tuberculosis in Spain.
In a phylogenetic context, LR spoligotyping will permit discrimination between isolates that really lack spacer 31 and those in which it is present but not revealed due to the asymmetric disruption of the DRa target due to IS6110 insertion. As shown in this paper, LR spoligotyping may also constitute an interesting tool to study IS6110 transpositional events. These results show that the DR locus constitutes an ideal IS6110 preferential locus (ipl), permitting the insertion of two or more copies of IS6110, and provide new clues for epidemiological and phylogenetic interpretation of changes in IS6110-RFLP and spoligotyping profiles. Indeed, other mycobacterial ipl loci have been reported, e.g., the ipl locus, which has been reported by Fang et al. (3, 4, 5) to form part of the recently identified IS1547 insertion sequence, mapping to two genomic locations represented by Rv0797 and Rv3327 within the M. tuberculosis H37Rv genome (1). Another insertional hot-spot region described in clinical isolates from South Africa spans a 9.4-kb region containing seven IS6110 insertion points (14) and maps to different positions, in region Rv1754c to Rv1765c (7). The presence of IS6110 copies in tandem in the case of these South African isolates may hypothetically be detected by PCR using primers IS3 and IS6 (as shown in the present investigation to selectively amplify the inter-insertion sequence region), e.g., sequencing of the region Rv1754c to Rv1758c in M. tuberculosis H37Rv shows the presence of two IS6110 copies in tandem separated by a 621-bp portion, with the second copy being inserted in plcD (Rv1755c), a phospholipase C homologue (7). Using insertion site mapping, another study recently identified a 537-bp dnaA-dnaN intergenic region as an IS6110 hot spot (12).
Recently, in a study on the genome plasticity of M. tuberculosis that focused on IS6110-flanking regions, an insertion site between the Rv2818c and Rv2819c genes was found to lie close to the DR locus (20). A calculation based on the genome sequence of M. tuberculosis (1) suggested that it may correspond to the additional band of 4.3 kb observed in the present investigation after DR-r probing of the PvuII-IS6110 blots (Fig. 1C). On the other hand, we do not yet have an explanation for the presence of a double band at 1.4 kb in all the type 50 isolates that we studied. Further studies on the relative importance of IS6110 insertion, homologous recombination, and replication slippage in guiding evolutionary changes of the M. tuberculosis genome may help us to understand the link between spoligotyping patterns and IS6110-RFLP.
Acknowledgments
This work was supported through grants from the “Délégation Générale au Réseau International des Instituts Pasteur et Instituts Associés,” Institut Pasteur, and Fondation Française Raoul Follereau, Paris, France.
REFERENCES
- 1.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Bary III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 2.Fang Z, Doig C, Kenna D T, Smittipat N, Palittapongarnpim P, Watt B, Forbes K J. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J Bacteriol. 1999;181:1014–1020. doi: 10.1128/jb.181.3.1014-1020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang Z, Forbes K J. A Mycobacterium tuberculosis IS6110 preferential locus (ipl) for insertion into the genome. J Clin Microbiol. 1997;35:479–481. doi: 10.1128/jcm.35.2.479-481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang Z, Morrison N, Watt B, Doig C, Forbes K J. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J Bacteriol. 1998;180:2102–2109. doi: 10.1128/jb.180.8.2102-2109.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang Z, Doig C, Morrison N, Watt B, Forbes K J. Characterization of IS1547, a new member of the IS900 family in the Mycobacterium tuberculosis complex, and its association with IS6110. J Bacteriol. 1999;181:1021–1024. doi: 10.1128/jb.181.3.1021-1024.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filliol I, Sola C, Rastogi N. Detection of a previously unamplified spacer within the DR locus of Mycobacterium tuberculosis: epidemiological implication. J Clin Microbiol. 2000;38:1231–1234. doi: 10.1128/jcm.38.3.1231-1234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon S V, Heym B, Parkhill J, Barrell B, Cole S T. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosis H37Rv. Microbiology. 1999;145:881–892. doi: 10.1099/13500872-145-4-881. [DOI] [PubMed] [Google Scholar]
- 8.Groenen P M A, Bunschoten A E, van Soolingen D, van Embden J D A. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol Microbiol. 1993;10:1057–1065. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 9.Gutierrez M C, Galan J C, Blazquez J, Bouvet E, Vincent V. Molecular markers demonstrate that the first described multidrug-resistant Mycobacterium bovis outbreak was due to Mycobacterium tuberculosis. J Clin Microbiol. 1999;37:971–975. doi: 10.1128/jcm.37.4.971-975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermans P W M, van Soolingen D, Bik E M, De Haas P E W, Dale J W, van Embden J D A. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamerbeek J, Schouls L, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J D A. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurepina N E, Sreevatsan S, Plikaytis B B, Bifani P J, Connell N D, Donnelly R J, van Soolingen D, Musser J M, Kreiswirth B N. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuber Lung Dis. 1998;79:31–42. doi: 10.1054/tuld.1998.0003. [DOI] [PubMed] [Google Scholar]
- 13.Pfyffer G E, Auckenthaler R, van Embden J D A, van Soolingen D. Mycobacterium canettii, the smooth variant of M. tuberculosis, isolated from a Swiss patient exposed in Africa. Emerg Infect Dis. 1998;4:631–634. doi: 10.3201/eid0404.980414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sampson S L, Warren R M, Richardson M, van der Spuy G D, van Helden P D. Disruption of coding regions by IS6110 insertion in Mycobacterium tuberculosis. Tuber Lung Dis. 1999;79:349–359. doi: 10.1054/tuld.1999.0218. [DOI] [PubMed] [Google Scholar]
- 15.Small P M, van Embden J D A. Molecular epidemiology of tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection and control. Washington, D.C.: American Society for Microbiology; 1994. pp. 569–582. [Google Scholar]
- 16.Sola C, Devallois A, Horgen L, Maïsetti J, Filliol I, Legrand E, Rastogi N. Tuberculosis in the Caribbean: using spacer oligonucleotide typing to understand strain origin and transmission. Emerg Infect Dis. 1999;5:404–414. doi: 10.3201/eid0503.990311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Embden J D A, van Gorkom T, Kremer K, Jansen R, van der Zeijst B A M, Schouls L M. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J Bacteriol. 2000;182:2393–2401. doi: 10.1128/jb.182.9.2393-2401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Soolingen D, Hoogenboenzem T, de Haas P E W, Hermans P W M, Koedam M A, Teppema K S, Brennan P J, Besra G S, Portaels F, Top J, Schouls L M, van Embden J D A. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int J Syst Bacteriol. 1997;47:1236–1245. doi: 10.1099/00207713-47-4-1236. [DOI] [PubMed] [Google Scholar]
- 20.Warren R M, Sampson S L, Richardson M, van der Spuy G D, Lombard C J, Victor T C, van Helden P D. Mapping of IS6110 flanking regions in clinical isolates of Mycobacterium tuberculosis demonstrates genome plasticity. Mol Microbiol. 2000;37:1405–1416. doi: 10.1046/j.1365-2958.2000.02090.x. [DOI] [PubMed] [Google Scholar]