Abstract
Background
In cancer radiotherapy, microbeam is an advanced and effective tool in investigating radiobiology. Currently, evidence to support the radiobiology of proton beam radiotherapy for prostate cancer is limited. This study aimed to investigate the DNA damage response of proton microbeam irradiation in prostate cancer.
Methods
Single-particle irradiation system to cells (SPICE) was used to perform the proton microbeam radiation-induced DNA damage response. The SPICE can deliver defined number of protons (3.4 MeV) to the cell nucleus. Different quantities of protons were irradiated to observe differential dose responses in prostate cancer cells. A total of 500 protons or defined proton doses were applied to PC-3 cell nucleus to investigate the kinetics of DNA double-strand breaks (DSB) repair after different time intervals; between 1 and 24 h post-irradiation. Subsequently, immunofluorescent staining of γ-H2AX was performed to detect DSB, and images were captured by immunofluorescence microscopy. Finally, γ-H2AX fluorescence intensity in each nucleus was quantified with Image J software.
Results
Proton microbeam radiation-induced DSB were dependent on proton dose applied. After irradiated with 500 protons, relative expression levels of γ-H2AX were time dependent during DSB repair process. The γ-H2AX fluorescence intensity was maximum at 1 h post-irradiation. However, a gradual decrease was observed from 4 to 24 h.
Conclusions
Microbeam is a valuable tool for the exploration of DSB response. The findings of the present study show that microbeam irradiation targeted the nucleus with precision. This study is the first to reveal that immune-stained γ-H2AX assay with proton microbeam irradiation could predict DSB repair kinetics in PC-3 cells.
Keywords: DNA damage response, irradiation, microbeam irradiation, proton radiotherapy, prostate cancer
Introduction
Radiotherapy is one of the main treatment options for localized prostate cancer (1,2). Radiotherapy, specifically proton radiotherapy, has the advantage of high-dose deposition and precise tumor target thus less adverse effects (3,4). Therefore, many prostate cancer patients in American, Europe, and Asia opt to receive proton radiotherapy (5-7).
Proton radiotherapy is considered the most advanced technology in radiation field (8). Currently, to improve the precision of proton beam radiotherapy, pencil-beam scanning irradiation with a small beam can be applied in the clinic. However, information about radiobiology of proton radiotherapy is scanty. With an increasing demand for proton radiotherapy, it is necessary to find the right tool to study the radiobiology of proton radiotherapy. We found microbeam is valuable tool in the study of radiobiology. Microbeam is a beam with micrometer or sub-micrometer diameter and has been extensively applied in the exploration of radiation-induced biological responses at sub-cellular targets (9-11). For example, studies have reported the average diameter of the cell nucleus to be 5–10 µm, and the proton microbeam is about 2 µm (12,13). To investigate cellular radiobiological effects, microbeam can be applied to selectively irradiate individual cells within a cell population (14). Therefore, microbeam irradiates with high spatial accuracy to the cell nucleus. Currently, few studies have focused on radiobiology of proton microbeam (15,16), especially in prostate cancer (17). Therefore, this study was conducted to investigate DNA damage response to proton microbeam in prostate cancer.
Radiation-induced cell killing usually involves targeting the cell nucleus and causing DNA double-strand breaks (DSB). The phosphorylated member of the H2A protein family, γ-H2AX, is a biomarker of DSB. It is useful in the detection of DNA damage and repair (18). Previous studies have indicated that γ-H2AX can predict the efficacy of cancer therapy treatments, especially some forms of chemotherapy and radiotherapy that induce DNA damage (19). Therefore, γ-H2AX induced to detect proton microbeam irradiation-induced DSB in this research. In this study, proton microbeam was performed to irradiate prostate cancer cells with a defined number of protons at various time intervals. Proton microbeam delivered doses precisely to the cell nucleus, which induced DSB in selected cell populations.
Methods
Cell culture
Prostate cancer PC-3 cells were provided by Cell Bank, Chinese Academy of Sciences. The F12K medium mixed with 10% fetal bovine serum (Gibco, Grand Island, NY, USA) was used for cell culture. The cells were cultured at 37 °C in a humidified atmosphere with 5% CO2 to maintain physiological pH.
Sample preparation and microbeam irradiation
This was performed according to the method described in literature with few modifications (13,20). Briefly, cells were plated on a dish that was specifically designed for proton microbeam irradiation. A special 6 µm polypropylene film (Chemplex Industries, FL, USA) was attached to the dish (diameter, 24 mm). To make sure that the cells were attached to the dish, 1 mL of 100 ng/mL fibronectin solutions (Wako, Chemical Industries, Osaka, Japan) was added to the film and incubated for 2 h, then rinsed with phosphate-buffered saline (PBS) twice before seeding cells. A total of 5×104 PC-3 cells in 1 mL medium were seeded two days before irradiation.
The irradiation was performed by proton microbeam system, Single-Particle Irradiation System to Cells (SPICE) at the National Institute of Radiological Sciences (NIRS) in Japan. The SPICE is a focused vertical microbeam system composed of a tandem accelerator and operation system (13). The PC-3 cells were irradiated by proton microbeam (3.4 MeV, LET 11.7 keV/µm) with a beam diameter of 2 µm. Instructions on irradiation procedures were obtained from literature (13). The SPICE dish was covered with mylar film before irradiation to protect against desiccation. With the advantage of microbeam, only the cell nucleus was irradiated using the online SPICE system. To verify the dose-response of proton microbeam in prostate cancer cells, 100, 250, 500 protons were delivered to the target area of X-Y coordinates, and the results were observed 1 h post-irradiation. To verify whether DNA damage response was dependent on duration of exposure, 500 protons were delivered to the cells and to observe the results after 1, 4, 8, 24 h irradiation.
Immunofluorescence staining for γ-H2AX
To detect DSB, γ-H2AX was used as a representative biomarker. Cells for the dose-response experiment were prepared for γ-H2AX staining 1 h post-proton microbeam irradiation, whereas those for time-dependent experiments were prepared for the same staining after defined time of irradiation. To begin the staining process, the medium was moved, and the cells were washed thrice with cold PBS. Cells were then fixed with 4% paraformaldehyde at room temperature for 10 min. After fixation, 1 mL blocking solution containing 1% bovine serum albumin (BSA), 5% goat serum, and 0.1% Tween 20 (Wako, Chemical Industries, Osaka, Japan) in PBS was added to the cells then incubated for 1 h at 37 °C. Subsequently, the blocking solution was removed, and the dish was rinsed twice with PBS, after which 300 µL of primary antibody, anti-γ-H2AX [anti-Phospho-Histone H2A.X (ser 139) Antibody, clone JBW301, Merk Millipore, Germany] was added to the dish and incubated at 37 °C for 1 h. The antibody was diluted to 1:500 with antibody buffer (0.1% BSA, 0.3% Triton X-100 in PBS) [Wako, Chemical Industries, Osaka, Japan] before use. The dish was washed twice with PBS buffer to remove the antibody. The secondary antibody was diluted to 1:1,000 in antibody buffer (goat anti-Mouse IgG, Alexa Fluor 555, Thermo Fisher Scientific, Waltham, MA, USA) and 300 µL of the antibody solution was added to the SPICE dish then incubated at 37 °C for 1 h. The solution was removed by washing the dish twice with PBS, then 1 mL of Hoechst 33342 (1:2,000 dilution, DOJINDO, Japan) was added to the SPICE dish followed by incubation at 37 °C for 30 minutes. Finally, to remove the Hoechst solution, the dish was washed twice with PBS then 2 mL of PBS was added to the dish before irradiation. The images were captured with offline SPICE system.
Quantification of fluorescence intensity
Detection and quantification of γ-H2AX were performed on immune-stained cells using a fluorescent microscope, offline SPICE system. The microscope was equipped with 40× water immersion objective lens (LUMPLFLN-W 40× W, NA: 0.8, Olympus, Japan) and a C-MOS camera; the light source was provided by a mercury lamp (130W, U-HGLGPS, Olympus, Japan). The detail of this microscope is described elsewhere (13). To capture the images, the filter was changed for various fluorescence; for example, the filter for Hoechst 33342 was U-MNUA2. The same procedure was used to capture images of the control group. After adjusting the parameters to focus on the target area, all images were automatically captured using the microscope.
To analyze these images, Image J software (21) was used to quantify fluorescence. The cell nucleus was stained with Hoechst 33342, whereas γ-H2AX was stained with Alexa Fluor 555. All the images were imported as a tile into Image J software from an image sequence menu. Cell nuclei were distinguished from the background according to threshold button, and the corresponding information including area, size and mean fluorescence intensity per pixel were obtained from the region of interest (ROI). These ROIs applied to Alexa Fluor 555 images, which indicated the fluorescence intensity of γ-H2AX. And the intensity measurements per cell nucleus were presented as average of each category of cells. The mean fluorescence intensity of γ-H2AX for each duration of exposure was normalized against the non-irradiated control group.
Statistical analysis
All data from this study are represented as the mean ± standard deviation. The values are averages of three independent experiments. To compare means of two different groups, Student’s t-test was performed at significance level of P<0.05. We used SPSS version 23.0 software for statistical analysis.
Results
Precision irradiation with proton microbeam
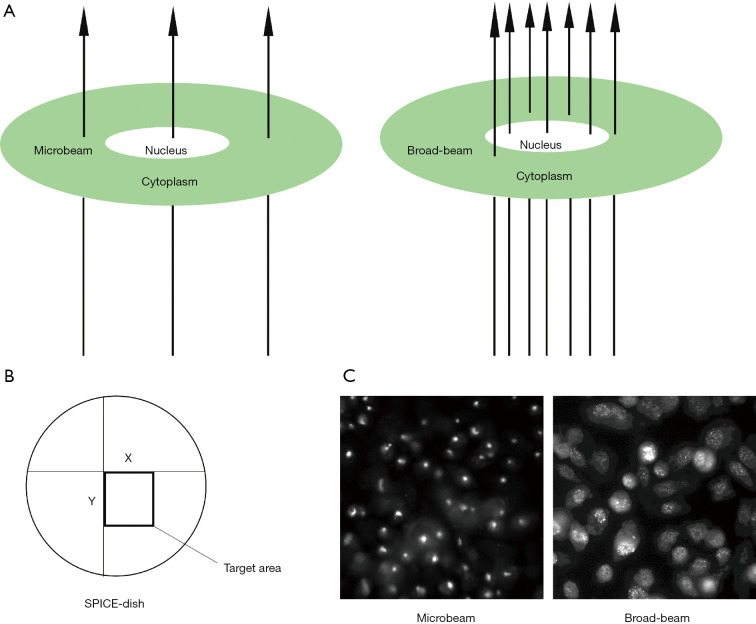
In this study, we investigated the DNA damage response of prostate cancer cells with microbeam irradiation. Because the beam diameter was about 2 µm, it was possible to target the cell nucleus or the cell cytoplasm with precision (Figure 1A). However, the accuracy of broad-beam irradiation was not the same as microbeam; thus, some doses scattered near the nucleus (Figure 1A). Figure 1B shows the target area of the cell nucleus by microbeam with X-Y coordinates. And Figure 1C demonstrates the representation of γ-H2AX expression at the DSB site by microbeam and broad-beam in PC-3 cells. The bright field or foci shows stained γ-H2AX antibody, which highlights the effect of DSB. Microbeam radiation-induced DNA damage was focused on the cell nucleus, and all the dose deposition targeted the nucleus. On the contrary, broad-beam radiation-induced dose distribution was scattered.
Figure 1.
The model of microbeam and broad-beam irradiation. (A) Proton microbeam targeted the cell nucleus or cytoplasm precisely, whereas broad-beam radiation lacked precision, and got scattered in cells; (B) schematic representation of SPICE-dish target areas by microbeam irradiation with X-Y coordinates; (C) representative image of γ-H2AX fluorescence with microbeam and broad-beam irradiation (40×); γ-H2AX foci collected only at the irradiation site with microbeam irradiation; but broad-beam induced γ-H2AX expression scattered near the nucleus. The contrast of the photos was enhanced by software, but the other parameters remained the same. SPICE, single-particle irradiation system to cells.
Induction of γ-H2AX in PC-3 cells with dose response
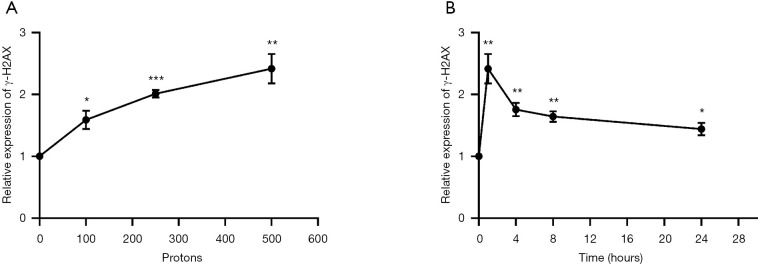
To investigate the function of proton microbeam in prostate cancer, dose variation response was evaluated. The experiments were conducted with 0 (control group), 100, 250, and 500 protons to irradiate PC-3 cells. The results demonstrated that γ-H2AX expression level rose with increased irradiation (Figure 2A). This was highly significant (*, P<0.05; **, P<0.01; ***, P<0.001) compared to the control group.
Figure 2.
Dose and time responses of γ-H2AX expression levels after irradiation. (A) Response of γ-H2AX expression levels after proton microbeam irradiation of different doses; (B) response of γ-H2AX expression levels after proton microbeam irradiation at different exposure durations. Significance levels of *, P<0.05; **, P<0.01; ***, P<0.001 compared to control group. All data were normalized to control.
Induction of γ-H2AX after different times of exposure
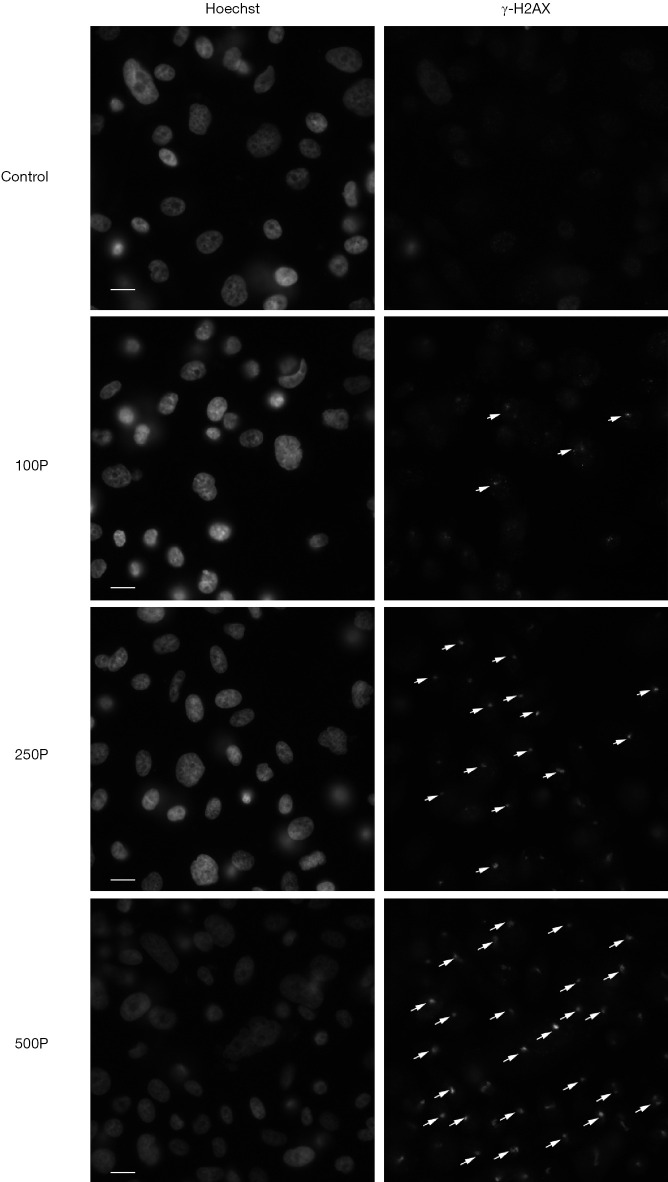
The expression level of γ-H2AX increased after 1 h of proton microbeam irradiation. However, after 4, 8, and 24 h of irradiation, the γ-H2AX level decreased because of the repair process (Figure 2B). The γ-H2AX level was higher than in the control group at 24 h post-irradiation. These results indicated that the cells could have recovered gradually from DSB, and the γ-H2AX expression level was time-dependent. The representative image of γ-H2AX expression level with microbeam irradiation was provided (Figure 3). Also, the γ-H2AX expression level was showed after irradiated by different number of proton (Figure 4).
Figure 3.
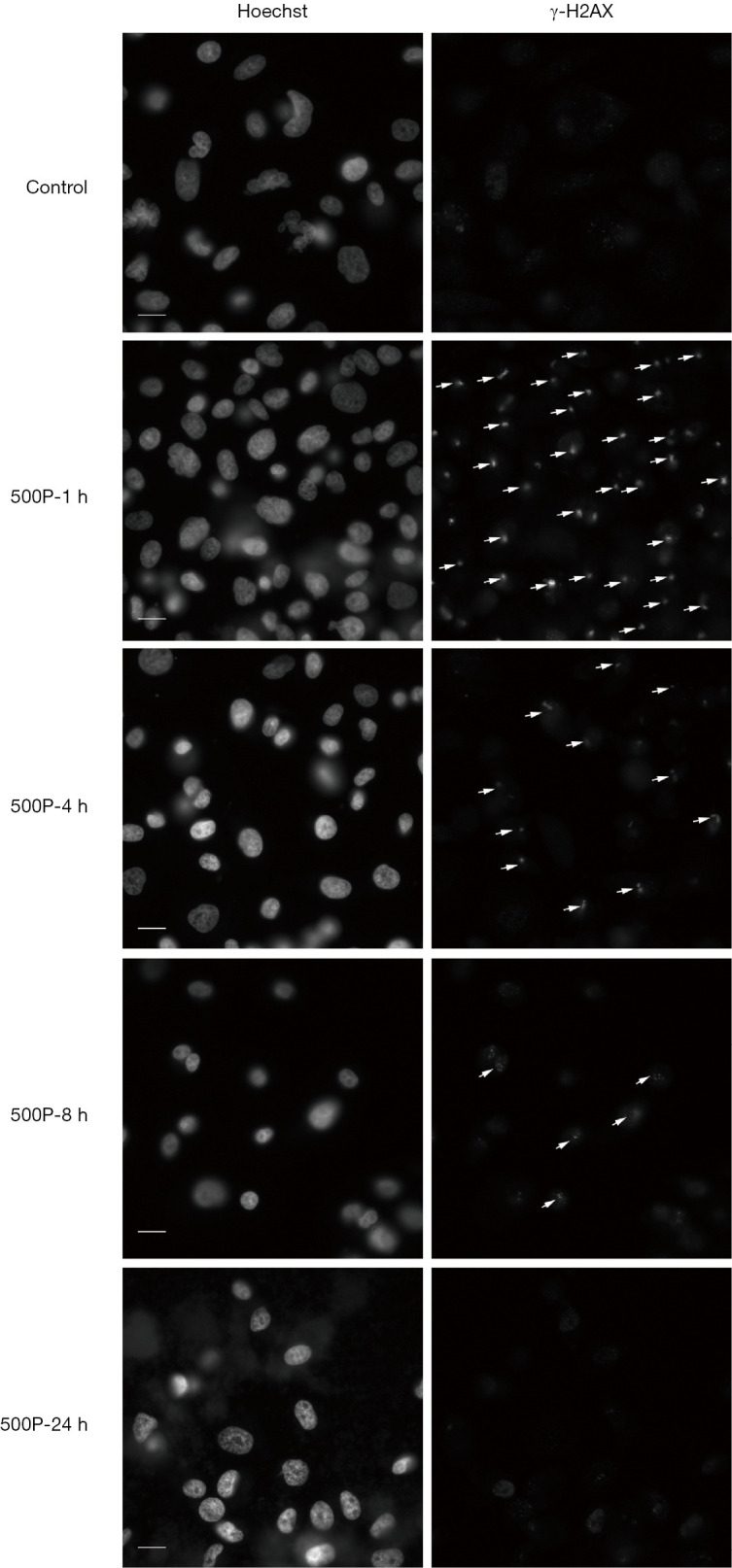
Representative image of γ-H2AX expression level after irradiated by proton microbeam at different time intervals. The bright-field illumination indicates DSB. The white arrow sign indicates γ-H2AX foci. The left photos were stained by Hoechst 33342, and the right-side photos were stained by γ-H2AX antibodies. Scale bar: 20 µm. DSB, DNA double-strand breaks.
Figure 4.
Representative image of γ-H2AX expression level after irradiated by different number of protons. Cells were irradiated with 100, 250 and 500 protons. The bright-field illumination indicates DSB. The white arrow sign indicates γ-H2AX foci. The left photos were stained by Hoechst 33342, and the right-side photos were stained by γ-H2AX antibodies. Scale bar: 20 µm. DSB, DNA double-strand breaks.
Discussion
Prostate cancer has been the main cause of morbidity among men in the United States in the past decade. A total of 174,650 estimated new cases were reported in 2019, and this was the highest among other cancer cases (22). The prostate cancer incidence rate is sixth in China and is increasing steadily (23). To arrest the progression of prostate cancer, radiotherapy is recommended as the standard treatment for localized prostate cancer. Accumulating evidence from clinical trials demonstrates that proton beam radiotherapy not only targets tumors precisely but also spare the surrounding normal tissues from unintended damage. Proton beam radiotherapy has salient features that aid in killing tumors, thus improving the quality of life. Many outstanding results have been achieved against many cancers by application of proton radiotherapy, including prostate cancer. However, the mechanism of radiobiology in proton beam therapy is still unclear. What’s more, the appropriate tool for studying proton therapy radiobiology is unidentified.
To investigate the radiobiological effect of the proton beam irradiation, we found microbeam was a valuable and effective tool. Microbeam was used to target cellular compartment (the cell nucleus) in selected cell populations. The main aim was to investigate the association between proton microbeam and DNA damage response and repair. Increasing evidence indicates that γ-H2AX is highly expressed during DNA damage response and DSB repair. Rogakou et al. and his colleagues reported that H2AX is phosphorylated at serine 139, and γ-H2AX is rapidly recruited to DNA lesion within seconds following DSB induced by ionizing radiation. Based on their findings, they concluded that γ-H2AX formation is sensitive to DNA damage and rapidly responds to DNA double-strand breaks (18). The DNA damage response transfer signals to phosphorylation of H2AX to γ-H2AX.
To investigate proton microbeam radiation-induced response, a dose-escalation experiment was performed with a defined number of protons. The immune-stained γ-H2AX expression levels increased 1 h post-irradiation (Figure 2A). The current results indicated that proton microbeam induced DNA damage in PC-3 cells was dependent on the amount of proton dose applied. The γ-H2AX fluorescence assay is a sensitive and precise indicator of DSB, γ-H2AX are recruited immediately to the irradiation site after proton microbeam induced DNA damage. Paull et al. reported that DNA repair factors BRCA1, Rad50, Rad51 colocalize with γ-H2AX after DNA damage by irradiation; their DNA repair-deficient cells model suggested that γ-H2AX formation play a vital role in DNA repair (24). Similar findings have indicated that repair protein XRCC1 and 53BP1 colocalize with γ-H2AX at the targeted site, which was useful in monitoring DNA damage at early stages (25). These results reveal that γ-H2AX could have a role in the repair process, which is consistent with this study. In the present study, the change in expression levels of γ-H2AX as a result of irradiation at different time intervals was further investigated to help in understanding the repair process. Results indicated that γ-H2AX expression level reached a maximum value at 1 h post-irradiation (Figure 2B). And then decreased at 4 h post-irradiation. This could mean that the DNA repair kinetic was faster than damage response. A further decrease in γ-H2AX expression level was observed from 4 to 24 h post-irradiation. The gradual appearance and disappearance of γ-H2AX foci suggested that the DNA repair pathway was already activated. These findings demonstrate that microbeam irradiation is capable of inducing DSB and activated repair pathway.
In the present study, microbeam irradiation induced cell death precisely. More time is needed to repair DSB in cases where most of the irradiation doses met their target. Microbeam irradiation deposited all the proton doses into the cell nucleus. During the DNA repair process, γ-H2AX fluorescence intensity gradually disappeared. The kinetics of DNA repair process showed different radiosensitivity. In general,proton microbeam irradiation was precise and efficient in targeting tumor cells and could be useful in investigating radiobiology. The biomarker (γ-H2AX) was sensitive in predicting DSB and DNA repair process in prostate cancer after proton microbeam irradiation.
Many previous studies have revealed two main DNA repair pathways, that is, homologous recombination (HR) and non-homologous end-joining (NHEJ) (26,27). However, DNA repair mechanisms are still unclear. Many factors could trigger the DNA damage response and repair. Signaling pathways including bystander effect, adaptive response, and genomic instability affect DNA response to irradiation (26,28). Therefore, in this study, we focus on verifying whether microbeam irradiation could monitor DSB and DNA repair via γ-H2AX expression. The details of DNA damage response and repair mechanisms are beyond the scope of this study.
Despite the many advantages of using a proton microbeam model to estimate the DNA damage and repair process in prostate cancer cells, this study has some limitations. Two main DNA repair pathways have not been discussed in detail. Thus, further studies are needed to explain these processes further. This study focused more on the association of γ-H2AX with proton microbeam induced DSB response and DNA repair. This was the first step in the investigation of proton microbeam radiobiological response with γ-H2AX. However, multiple cell lines should be considered in future studies on the same.
Conclusions
In summary, microbeam is a valuable and effective tool to investigate radiobiology. The results showed that proton microbeam irradiation is more capable of precisely delivering doses to the cell nucleus. The phenomenon of appearance and disappearance of γ-H2AX fluorescence was sensitive to proton microbeam irradiation-induced DNA damage response. Microbeam radiation-induced DSB response and repair was dose and time-dependent and was based on γ-H2AX fluorescence intensity.
Acknowledgments
We highly thank Professor Teruaki Konishi and his team members (Dr. Yoshiya Furusawa, Dr. Alisa Kobayashi, Dr. Daisuke Ohsawa) at the National Institute of Radiological Science (NIRS) for their technical help and excellent support.
Funding: This work was supported by the National Key Research and Development Program of China (No. 2017YFC0107600), the National Natural Science Foundation of China (No. 81773225).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-19-2915
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2915). The authors have no conflicts of interest to declare.
References
- 1.Martin NE, D'Amico AV. Progress and controversies: Radiation therapy for prostate cancer. CA Cancer J Clin 2014;64:389-407. 10.3322/caac.21250 [DOI] [PubMed] [Google Scholar]
- 2.Zietman AL, Bae K, Slater JD, et al. Randomized trial comparing conventional-dose with high-dose conformal radiation therapy in early-stage adenocarcinoma of the prostate: long-term results from proton radiation oncology group/american college of radiology 95-09. J Clin Oncol 2010;28:1106-11. 10.1200/JCO.2009.25.8475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sidaway P. Proton therapy delays progression. Nat Rev Urol 2016;13:181. 10.1038/nrurol.2016.50 [DOI] [PubMed] [Google Scholar]
- 4.Mohan R, Grosshans D. Proton therapy - Present and future. Adv Drug Deliv Rev 2017;109:26-44. 10.1016/j.addr.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The LO . Proton therapy for prostate cancer: time for evidence. Lancet Oncol 2014;15:775. 10.1016/S1470-2045(14)70304-3 [DOI] [PubMed] [Google Scholar]
- 6.Iwata H, Ishikawa H, Takagi M, et al. Long-term outcomes of proton therapy for prostate cancer in Japan: a multi-institutional survey of the Japanese Radiation Oncology Study Group. Cancer Med 2018;7:677-89. 10.1002/cam4.1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Royce TJ, Efstathiou JA. Proton therapy for prostate cancer: A review of the rationale, evidence, and current state. Urol Oncol 2019;37:628-36. 10.1016/j.urolonc.2018.11.012 [DOI] [PubMed] [Google Scholar]
- 8.Terasawa T, Dvorak T, Ip S, et al. Systematic review: charged-particle radiation therapy for cancer. Ann Intern Med 2009;151:556-65. 10.7326/0003-4819-151-8-200910200-00145 [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Hei TK. Focus small to find big - the microbeam story. Int J Radiat Biol 2018;94:782-8. 10.1080/09553002.2017.1364801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heiss M, Fischer BE, Jakob B, et al. Targeted irradiation of Mammalian cells using a heavy-ion microprobe. Radiat Res 2006;165:231-9. 10.1667/RR3495.1 [DOI] [PubMed] [Google Scholar]
- 11.Hable V, Greubel C, Bergmaier A, et al. The live cell irradiation and observation setup at SNAKE. Nucl Instrum Methods Phys Res B 2009;267:2090-7. 10.1016/j.nimb.2009.03.071 [DOI] [Google Scholar]
- 12.Prise KM, Schettino G, Folkard M, et al. New insights on cell death from radiation exposure. Lancet Oncol 2005;6:520-8. 10.1016/S1470-2045(05)70246-1 [DOI] [PubMed] [Google Scholar]
- 13.Konishi T, Oikawa M, Suya N, et al. SPICE-NIRS Microbeam: a focused vertical system for proton irradiation of a single cell for radiobiological research. J Radiat Res 2013;54:736-47. 10.1093/jrr/rrs132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghita M, Fernandez-Palomo C, Fukunaga H, et al. Microbeam evolution: from single cell irradiation to pre-clinical studies. Int J Radiat Biol 2018;94:708-18. 10.1080/09553002.2018.1425807 [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi A, Tengku Ahmad TAF, Autsavapromporn N, et al. Enhanced DNA double-strand break repair of microbeam targeted A549 lung carcinoma cells by adjacent WI38 normal lung fibroblast cells via bi-directional signaling. Mutat Res 2017;803-805:1-8. 10.1016/j.mrfmmm.2017.06.006 [DOI] [PubMed] [Google Scholar]
- 16.Autsavapromporn N, Liu C, Kobayashi A, et al. Emerging Role of Secondary Bystander Effects Induced by Fractionated Proton Microbeam Radiation. Radiat Res 2019;191:211. 10.1667/RR15155.1 [DOI] [PubMed] [Google Scholar]
- 17.Lipiec EW, Wiecheć A, Dulińska-Litewka J, et al. Changes in cellular response to the damage induced in PC-3 prostate cancer cells by proton microbeam irradiation. Gen Physiol Biophys 2012;31:11. 10.4149/gpb_2012_001 [DOI] [PubMed] [Google Scholar]
- 18.Rogakou EP, Pilch DR, Orr AH, et al. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998;273:5858-68. 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- 19.Ivashkevich A, Redon CE, Nakamura AJ, et al. Use of the γ-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett 2012;327:123-33. 10.1016/j.canlet.2011.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi A, Autsavapromporn N, Ahmad TAFT, et al. Bystander WI-38 cells modulate DNA double-strand break repair in microbeam-targeted A549 cells through gap junction intercellular communication. Radiat Prot Dosimetry 2019;183:142-6. 10.1093/rpd/ncy249 [DOI] [PubMed] [Google Scholar]
- 21.Rasband WS, ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA. Available online: https://imagej.nih.gov/ij/, 1997-2018.
- 22.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: A Cancer Journal for Clinicians 2019;69:7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 23.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 24.Paull TT, Rogakou EP, Yamazaki V, et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 2000;10:886-95. 10.1016/S0960-9822(00)00610-2 [DOI] [PubMed] [Google Scholar]
- 25.Jakob B, Splinter J, Conrad S, et al. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res 2011;39:6489-99. 10.1093/nar/gkr230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clouaire T, Rocher V, Lashgari A, et al. Comprehensive Mapping of Histone Modifications at DNA Double-Strand Breaks Deciphers Repair Pathway Chromatin Signatures. Mol Cell 2018;72:250-262.e6. 10.1016/j.molcel.2018.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mateo J, Boysen G, Barbieri CE, et al. DNA Repair in Prostate Cancer: Biology and Clinical Implications. Eur Urol 2017;71:417-25. 10.1016/j.eururo.2016.08.037 [DOI] [PubMed] [Google Scholar]
- 28.van Oorschot B, Uitterhoeve L, Oomen I, et al. Prostate Cancer Patients with Late Radiation Toxicity Exhibit Reduced Expression of Genes Involved in DNA Double-Strand Break Repair and Homologous Recombination. Cancer Res 2017;77:1485-91. 10.1158/0008-5472.CAN-16-1966 [DOI] [PubMed] [Google Scholar]