Abstract
Background
Regulatory T (Treg) cells are a major component of the microenvironment of hepatocellular carcinoma (HCC) contributing to immunosuppression. The present study aimed to evaluate the effects of Treg cells on the invasion potential of HCC.
Methods
Infiltrating Treg cells were isolated from fresh HCC tissues by immunomagnetic bead separation and detected by flow cytometry. Circulating tumor cells (CTCs) were detected using the CellSearch platform. The cell migration and invasion potentials were evaluated by Transwell assays. The cell viability was tested by the cell counting kit-8 (CCK8) approach, and the apoptosis rates were determined by flow cytometry. The concentrations of active transforming growth factor-β1 (TGFβ1) were measured by enzyme-linked immunosorbent assay.
Results
Infiltrating Treg cells significantly correlated with the number of CTCs and vascular invasion (both P<0.05). Moreover, these cells could greatly promote HCC migration, invasion, and proliferation, and inhibit HCC apoptosis. Polymerase chain reaction and Western blot assays revealed that Treg cells significantly decreased the expression levels of epithelium-related molecules and increased the expression levels of mesenchyme-related molecules. Treg cells could activate Smad2/3 via secreting TGFβ1, and these effects could be impaired by knocking down the expression of TGFβ1 in Treg cells.
Conclusions
The involvement of infiltrating Treg cells in triggering the TGFβ1 signaling pathway and promoting the epithelial-mesenchymal transition (EMT) of cancer cells during tumor hematogenous dissemination is presumably responsible for increasing the invasiveness potential of HCC cells. Targeting Treg cells in microenvironments can be a promising therapeutic strategy to improve the prognosis for patients with HCC undergoing resection.
Keywords: Circulating tumor cells, epithelial-mesenchymal transition (EMT), hepatocellular carcinoma (HCC), regulatory T cells, transforming growth factor-β1 (TGFβ1)
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide, with recently escalating incidence and generally dismal outcomes (1,2). By far, surgical resection is still considered as the first option for HCC cure (3). However, postoperative survival remains unsatisfactory because of high recurrence and metastasis rates due to the high invasiveness potential of HCC cells (4-6). Therefore, a better understanding of HCC cell spread may provide novel insights into HCC management in the future. The transforming growth factor-β1 (TGFβ1)-induced TGFβ signaling pathway has been reported to play a vital role in HCC progression and spread (7). Active TGFβ1 could trigger epithelial–mesenchymal transition (EMT) in a Smad2/3-dependent manner, greatly promoting the invasiveness of tumor cells (8-10). Previous studies reported that regulatory T (Treg) cells were a critical resource of TGFβ1, and Treg-derived TGFβ1 could also promote tumor invasion (11,12). However, whether infiltrating Treg was an important origin of TGFβ1 in HCC, and its role in promoting HCC progression, remained elusive.
Recent data have provided solid evidence that the generation of circulating tumor cells (CTCs) was an early event in tumor metastasis, and these CTCs were considered as the “seeds” of metastasis (13,14). Accumulating reports have proven the role of CTCs in HCC metastasis cascade (15). CTCs expressing an epithelial cell adhesion molecule (EpCAM) could be detected in about half of patients with HCC, and patients with high EpCAM-CTCs have been proved to encounter poor outcomes, including a high incidence of recurrence and tumor-related death (15,16). However, the mechanism underlying the release of these cells from tumor bulks is extremely complicated and needs to be further investigated. Recent data revealed that the acquisition of invasiveness by interaction with mesenchymal cells, including Tregs, was essential for tumor cells, suggesting a novel strategy for investigating HCC progression (17).
In light of the aforementioned considerations, the correlation between infiltrating Treg cells and CTCs and the effects of Treg cells on HCC cells were further investigated via in vitro experiments. The data demonstrated that in situ Treg cells could significantly promote the invasiveness potentials of HCC cells via TGFβ1-induced Smad2/3 phosphorylation. Therefore, the findings indicated that infiltrating Treg cells served as a critical regulator of HCC invasion, and future therapeutic approaches targeting Treg cells might pave the novel way for curing HCC.
Methods
Cell lines and cell culture
The Huh7 cell line was purchased from the Cell Bank, Institute of Biochemistry and Cell Biology, China Academy of Science (Shanghai, China). The MHCC-97H cell line was a gift from the Liver Cancer Institute of Zhongshan Hospital, Fudan University (Shanghai, China). Both cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), supplemented with 100 IU/mL penicillin and 100 µg/mL streptomycin and incubated at 37 °C under a humidified atmosphere with 5% CO2. All cell culture reagents were obtained from Life Technology (Thermo Fisher Scientific, MA, USA).
Fresh clinical tissue specimens
A total of 40 fresh HCC tissues were collected at Liver Cancer Institute of Zhongshan Hospital, Fudan University (Shanghai, China). No previous local or systemic treatment had been conducted for these patients before treatment. Surgical specimens were obtained from all patients at the time of resection. All samples were received in the laboratory within 2 h, immediately mechanically disaggregated and digested with type 4 collagenase (Thermo Fisher Scientific), and then resuspended in DMEM. Single-cell suspensions were obtained by filtration through a 40-µm filter. Red blood cells were lysed with ACK buffer (Thermo Fisher Scientific). The viable cells were counted and analyzed using trypan blue. The approval for the use of human participants was obtained from the research ethics committee of Zhongshan Hospital, and informed consent was obtained from each individual.
Infiltrating lymphocyte isolation and detection
Human lymphocytes were isolated from fresh tissues by centrifugation through a discontinuous Percoll (GE Healthcare, Piscataway, New Jersey, USA) gradient with densities of 1.06 and 1.08 g/mL according to a previous study (18). Isolated lymphocytes were harvested for further experiments. Treg cells were detected by flow cytometry using a human CD4+CD25+FoxP3+ Treg detection kit (eBioscience, CA, USA) according to the manufacturer’s protocols. Briefly, 20 µL of anti-CD4-fluorescein isothiocyanate (FITC)/anti-CD25-APC (allophycocyanin) and the relevant isotype control antibody (IgG1Ƙ-FITC and IgG1Ƙ-APC, respectively) were added into two separate flow cytometry tubes. Then, 100 µL of the suspension containing 106 freshly isolated lymphocytes was added, followed by incubation at 4 °C for 30 min in the dark. Afterward, each tube was supplemented with 1 mL of permeabilization reagent and incubated at 4 °C for 60 min. After washing with phosphate-buffered saline (PBS), 100 µL of mouse serum was added and the mixture was incubated at room temperature (RT) in the dark for 15 min. Further, 20 µL of intracellular antibody anti-Foxp3-PE and isotype control IgG2a-K-PE were each added, and the resulting solution was incubated at RT in the dark for another 30 min. After resuspension in PBS, the sample was loaded for flow cytometry analysis. The measurement included proportions of CD4+CD25+FoxP3+ T cells (Tregs) in total lymphocytes and CD4+ T cells.
Co-culture of Treg and tumor cells
Treg cells were separated using a human CD4+CD25+ regulatory T cell isolation kit (Miltenyi, Germany) according to the manufacturer’s protocols. The purity of Treg cells was verified by flow cytometry. Afterward, 1×104 Huh7 or 97H cells were seeded with 1×105 Treg cells in 96-well plates with DMEM containing 10% FBS, using Huh7 or 97H cells without any lymphocytes as controls. The co-cultures were incubated for 72 h, and the tumor cell viability on each day was evaluated by CCK-8 assays (Dojindo Inc., Japan) according to the manufacturer’s protocols. All experiments were conducted in triplicate.
Migration, invasion, cell cycle, and apoptosis assays
For detecting migration and invasion, Transwell assays were used. Briefly, 1×105 Huh7 or 97H cells in 100 µL of serum-free DMEM were added to the well of Boyden chamber with an 8-µm-pore membrane (Corning, Inc., NY, USA). The bottom chamber contained 10% FBS in DMEM as a chemoattractant. Further, 1×106 Treg cells were added to the upper chambers. For evaluating migration, the cells were cultured without MatriGel, while for evaluating invasion, the cells were cultured with MatriGel. The cells were allowed to invade for 24 h (migration evaluation) or 72 h (invasion evaluation); the cells that had not penetrated the filters were then removed by scrubbing with cotton swabs. The untransfected cells were similarly treated and included as a negative control. The chambers were fixed for 20 min at RT with 4% formaldehyde in PBS, stained with 0.1% crystal violet for 30 min, and rinsed in water. The cells that migrated to the bottom surface of the filter were considered to have invaded through the matrix and were counted under a light microscope. The assays were performed three times using triplicate wells.
The cell cycle assays were conducted with flow cytometry, briefly, 3×105 Huh7 or 97H cells were seeded in 6-well plates with 3×106 Treg cells and co-cultured for 48 h. Then, the tumor cells were harvested and washed with PBS three times and suspended in cold PBS. The cells were stained with propidium iodide (PI)/RNase staining solution at 37 °C in the dark for 30 min. The cell cycle was performed in three replicates using flow cytometry (BD Biosciences, CA, USA).
Cell apoptosis was analyzed by flow cytometry using Annexin V–FITC. Apoptosis Detection Kits (BD Biosciences) were employed according to the manufacturer’s protocol. Briefly, 1×105 Huh7 or 97H cells were seeded in 24-well plates with 1×106 Treg cells; these cells were co-cultured for 48 h and then harvested and suspended in a binding buffer (1×). The cells were similarly treated without Treg cells as a negative control. An aliquot of 100 µL was incubated with 5 µL of Annexin V–FITC and 5 µL of PI for 15 min in the dark, and 400 µL of binding buffer (1×) was added to each sample. The stained cells were analyzed by flow cytometry within 1 h. All in vitro experiments were conducted in triplicate. Treg cells separated from HCC tissues were used for a single function assay in the present study.
Detection of CTCs
CTCs were detected using the CellSearch system, as previously described (16). In brief, the semiautomated CellSearch platform (Janssen Diagnostics, NJ, USA) enriched the sample for cells expressing the EpCAM with ferromagnetic beads. Afterward, fluorescently labeled monoclonal antibodies specific for leukocytes (CD45) and cytokeratins were used to distinguish epithelial cells from leukocytes. The identification and enumeration of CTCs were performed using the CellSpotter Analyzer, and the results were expressed as the number of cells per 7.5 mL of blood.
RNA isolation and quantitative reverse transcription–polymerase chain reaction
Total RNA was extracted from cell lines using the RNeasy mini kit (Qiagen, Germany) according to the manufacturer’s protocols. mRNA was reverse transcribed into cDNA using a QuantiTect Reverse Transcription Kit (Qiagen). The mRNA expression level in HCC cell lines was measured by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) using a LightCycle480 instrument (Roche Diagnostics, Germany). qRT-PCR was conducted using a LightCycle480 SYBR I Master Mix (Roche Diagnostics) according to the manufacturer’s protocols. qRT-PCR was performed with an initial denaturation at 95 °C for 5 min followed by 40 cycles of denaturation at 95 °C for 15 s, annealing, and extension at 60 °C for 30 s. GAPDH was used as an internal control. The relative mRNA expression levels were calculated based on the Ct values and normalized using GAPDH expression, according to the equation: 2−ΔCt [ΔCt = Ct (Target) – Ct (GAPDH)]. All experiments were performed in triplicate.
Cell transfection
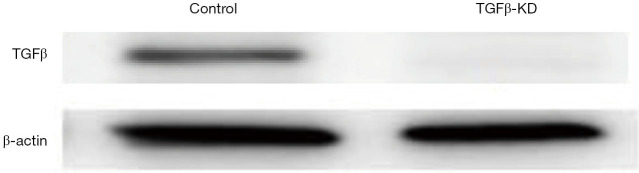
The expression of KNPN3 was knocked down using a retroviral vector pMSCV. The retrovirus was produced by 293T packaging cells. The pLKO.1-shRNAs targeting TGFβ oligonucleotide were TGCTGTT GC -GAAGACATAGAAATGAATAGTGAAGCCACAGATGTATTCATTTCTATGTCTTCGCATTTGCCTACTGCCTCGGA. The cells were infected with these viruses or control virus and then selected against puromycin for 3 days before being split for further assays. The reporter plasmid and luciferase promoter were purchased from Promega (WI, USA). The results of the validation of TGFβsilencing are displayed in Figure S1.
Figure S1.
The validation of TGF-β knockdown was confirmed by Western-blot analysis.
Enzyme-linked immunosorbent assay
The concentration of TGFβ1 in the conditioned supernatant from the co-culture experiments and in the peripheral blood of patients or healthy individuals was determined using a sandwich enzyme-linked immunosorbent assay (ELISA) kit according to the manufacturer’s protocols (R&D systems, Minneapolis, USA).
Western blot analysis
Total protein of Huh7 or 97H cells was extracted in lysis buffer for 30 min on ice. Equal amounts of proteins were separated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrophoretically transferred to polyvinylidene difluoride membranes (Millipore, MA, USA) using a mini trans-blot apparatus (Bio-Rad,CA, USA). The membranes were blocked with Tris-buffered saline with Tween (TBST) containing 5% nonfat dry milk for 1 h and incubated with primary antibodies, E-cadherin (Cell Signaling Technology CST, Boston, USA,, 1:1,000), Vimentin (CST, 1:1,000), Twist (CST, 1:1,000), α-SMA (CST, 1:1,000), Smad2/3 (CST, 1:500), pSmad2/3 (CST, 1:500), and β-Actin (CST, 1:1,000), overnight at 4 °C. The membranes were then washed three times with TBST and incubated with horseradish peroxidase–conjugated immunoglobulin G (Chemicon, MA, USA) at a 1:5,000 dilution for 1 h at RT. The blots were developed using an enhanced chemiluminescence kit (Pierce, WA, USA). Each experiment was repeated three times.
Statistical analysis
Statistical analyses were performed using SPSS 21.0 software (IBM, USA). The experimental values for continuous variables were expressed as the mean ± standard error of the mean. The chi-square test, Fisher’s exact probability tests, and the Student’s t-test were used as appropriate to evaluate the significance of differences in data between the groups. If variances within the groups were not homogeneous, the nonparametric Mann-Whitney test or the Wilcoxon signed-rank test was used. A P value less than 0.05 was considered statistically significant.
Results
Infiltrating Treg cells positively correlated with HCC invasiveness
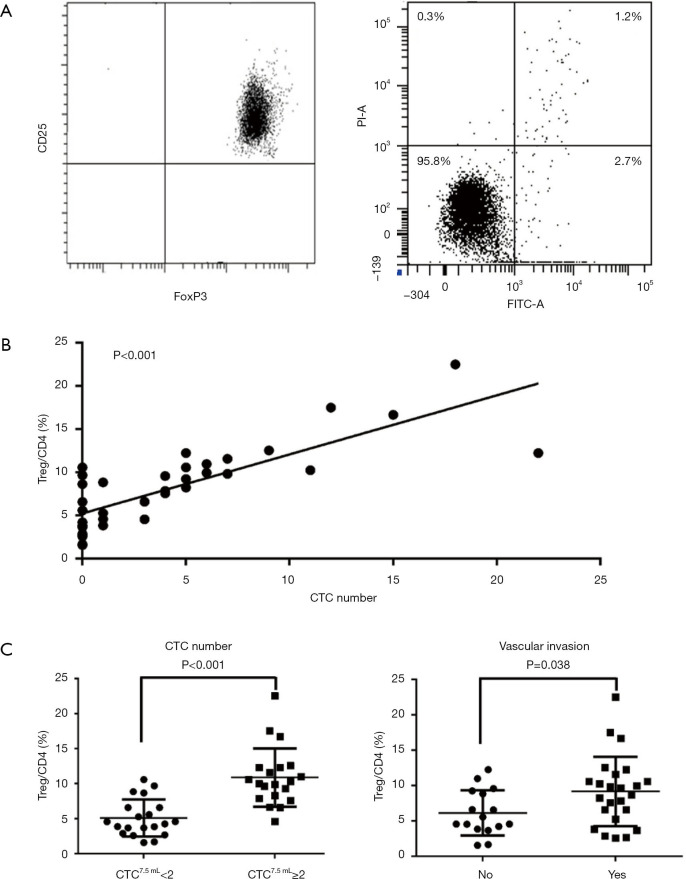
To investigate the effect of infiltrating Tregs on HCC invasiveness, the correlation between Treg cells and CTCs was evaluated in patients with HCC. First, the purity and viability of the separated Treg cells were assessed. The result showed that almost all separated cells were Treg cells, and the apoptosis rate of these cells was lower than 5.0% (Figure 1A). Next, Treg contents were found to be significantly correlated with the number of CTCs (P<0.001, Figure 1B). Subsequently, the Treg cell contents were compared between high (CTC ≥2 per 7.5 mL) and low (CTC <2 per 7.5 mL) CTC load subgroups. The result showed that Treg contents significantly increased in patients with high CTC loads (P<0.001, Figure 1C). Vascular invasion was a typical symbol of invasiveness. Therefore, the infiltrating Treg cell contents were also compared between patients with or without vascular invasion. Similarly, patients with vascular invasion exhibited higher Treg cell contents (P=0.038, Figure 1C). Collectively, these findings indicated that infiltrating Treg cells positively correlated with HCC invasiveness.
Figure 1.
Infiltrating Treg cells positively correlated with circulating tumor cells and invasiveness phenotype of HCC. (A) Assessment of the purity (left) and apoptosis rate (right) of Treg cells using FACS. (B) Treg cell contents significantly correlated with the numbers of CTCs (P<0.001). (C) Treg cell contents significantly increased in patients with high CTC loads (CTC ≥2 per 7.5 mL) (P<0.001), and patients with vascular invasion exhibited higher Treg cell contents (P=0.038).
Effects of infiltrating Treg cells on HCC cell motility, proliferation, cell cycle, and apoptosis in vitro
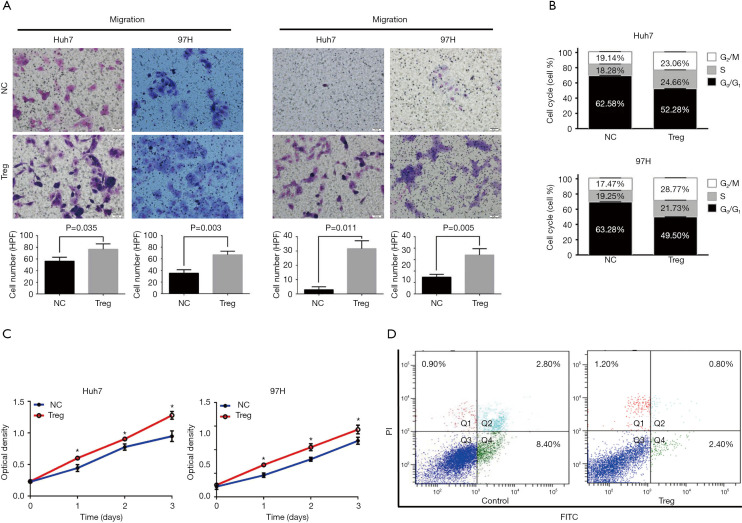
To further explore the effects of Treg cells on HCC cells, infiltrating Treg cells were isolated via magnetic-based approaches and then co-cultured with HCC cell lines. Transwell assays showed that the migration and invasion capacities were significantly promoted when co-cultured with Treg cells in both Huh7 and 97H cells (all P<0.050, Figure 2A). The effects of Treg cells on proliferation were also evaluated. Both Huh7 and 97H cells proliferated faster when co-cultured with Treg cells according to CCK-8 assays (Figure 2B). Whether Treg cells could promote cell cycle transition was further explored considering their proliferation-promoting ability. Based on the flow cytometry results, the cells co-cultured with Treg cells showed a higher proportion of S phase, indicating higher proliferation rates (Figure 2C). Finally, the effects of Treg cells on apoptosis were investigated, and the results showed that cell apoptosis was significantly inhibited when co-cultured with Treg cells (Figure 2D). Taken together, the results demonstrated that infiltrating Treg cells could promote cell motility and proliferation and inhibit cell apoptosis, inevitably resulting in HCC progression.
Figure 2.
In vitro effects of infiltrating Treg cells on HCC cells. (A) Migration and invasion potentials of HCC cells were significantly promoted when co-cultured with Treg cells in both Huh7 and 97H cells (Crystal Violet Staining, Magnification, ×100. P<0.050). (B) Effects of infiltrating Tregs on the proliferation potentials of HCC cells detected using CCK8 assays. (C) Effects of infiltrating Treg cells on the cell cycle of HCC cells detected using FACS. Tregs showed a higher proportion of S phase. (D) Cell apoptosis was significantly inhibited when co-cultured with Treg cells.
Infiltrating Treg cells induced EMT in HCC cells
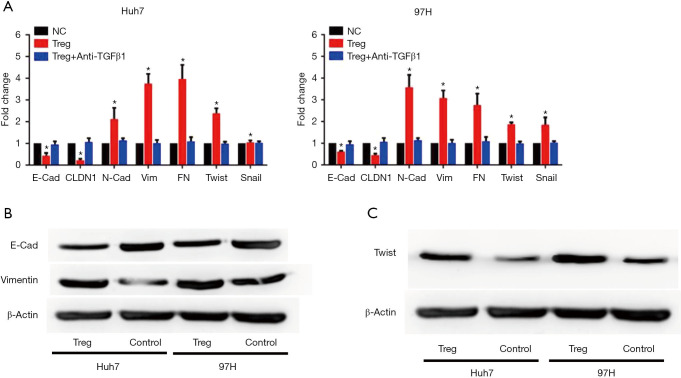
EMT was considered as a key contributor for cell invasiveness. Thus, whether infiltrating Treg cells could induce EMT was investigated. When co-cultured with infiltrating Tregs, HCC cells showed significantly lower expression levels of E-cadherin and claudin1 mRNA, while the mRNA expression levels of mesenchymal biomarkers, including N-cadherin, Vimentin, and fibronectin increased (Figure 3A). Moreover, two critical molecules involved in EMT, Twist and Snail, also showed increased mRNA expression levels after Treg stimulation (Figure 3A). To validate these findings, Western blot (WB) assays were conducted, and the results showed that the expression level of E-cadherin decreased while that of Vimentin increased at the protein level when co-cultured with Treg cells (Figure 3B). Importantly, the expression level of Twist also increased by Treg stimulation according to the WB assay (Figure 3C).
Figure 3.
Infiltrating Treg cells triggered EMT in HCC cells. (A) mRNA expression of EMT-related molecules evaluated using RT-PCR assays. The expression levels of mesenchymal biomarkers, including N-cadherin, Vimentin, and fibronectin increased (*P<0.05 vs. NC). Protein expression levels of E-cadherin and Vimentin (B) and Twist (C) in Huh7 and MHCC97H cells evaluated using WB assays. The expression levels of E-cadherin decreased while the expression levels of Vimentin and Twist increased at the protein level when co-cultured with Treg cells.
Infiltrating Treg cells induced EMT via secreting TGFβ1
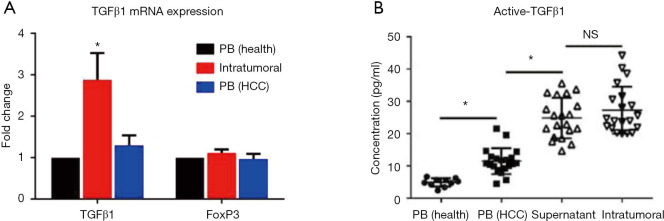
TGFβ1-induced Smad2/3 activation was reported to a major cause for EMT, while Treg cells were conventionally considered as an important resource of TGFβ1. Hence, it was speculated that Treg cells might trigger EMT via secreting TGFβ1. To confirm this, the TGFβ1 mRNA expression levels of Treg cells were detected in the peripheral blood (either healthy donors or patients with HCC) and intratumoral environment (infiltrating Treg cells). The infiltrating Treg cells exhibited the highest expression levels of TGFβ1 mRNA compared with Treg cells from the peripheral blood of both healthy donors (n=10) and patients with HCC (n=10) (Figure 4A). Meanwhile, the expression levels of Foxp3 mRNA showed no difference among the three groups (Figure 4A). Further, ELISA revealed that the supernatant of infiltrating Treg cells contained significantly higher concentrations of TGFβ1 compared with that of Treg cells from the peripheral blood of both healthy donors (n=10) and patients with HCC (n=20), while intratumoral TGFβ1 concentrations showed no significant difference compared with those in the supernatant of infiltrating Treg cells (Figure 4B). These findings indicated that the infiltrating Treg cells exhibited impressive TGFβ1 secretion ability and might serve as a major resource of TGFβ1 within the tumor microenvironment.
Figure 4.
Infiltrating Treg cells secreted high levels of TGFβ1 and had high mRNA expression. Active TGFβ1 concentrations were detected by ELISA. mRNA expression levels of TGFβ1 and FoxP3 were detected by RT-PCR. Treg cells were obtained from the intratumoral environment and the peripheral blood of patients with HCC, and Treg cells from the peripheral blood of healthy donors were selected as control. (A) Treg cells exhibited the highest TGFβ1 mRNA expression levels compared with Tregs from the peripheral blood of healthy donors. Foxp3 mRNA expression levels showed no difference among the three groups. (B) Supernatant of infiltrating Treg cells contained significantly higher concentrations of TGFβ1 compared with those of Treg cells from the peripheral blood of healthy donors while no significant difference compared with intratumoral TGFβ1 (*P<0.01 vs. peripheral blood).
To validate the role of Treg cells in activating Smad2/3, WB assays were performed. Treg cells could greatly increase the phosphorylation levels of Smad2/3 and promote the expression of α-SMA, confirming the effect of Treg cells on Smad2/3 activation (Figure 5). Next, to verify the role of TGFβ1 in Treg-mediated Smad2/3 activation and EMT, the expression of TGFβ1 was knocked down via siRNA on isolated infiltrating Treg cells, which were then co-cultured with HCC cells. RT-PCR showed that Treg cells lost their function to induce EMT (Figure 3A). Meanwhile, WB assays indicated that TGFβ1-KD impaired the effect of Treg cells on Smad2/3 activation (Figure 5). Collectively, these data demonstrated that infiltrating Treg cells promoted cell invasiveness by secreting TGFβ1.
Figure 5.
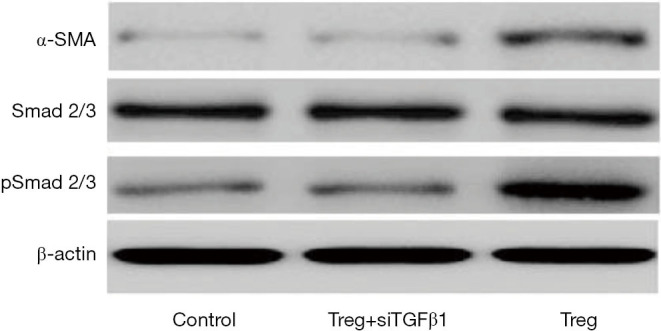
Western blot assays were conducted to detect the expression level of α-SMA and phosphorylation level of Smad2/3; β-actin was selected as a control. The TGFβ1 signaling pathway was activated by Treg-derived TGFβ1. Treg cells could greatly increase the phosphorylation levels of Smad2/3 and promote the expression of α-SMA.
Discussion
Previous studies revealed that Treg cells significantly correlated with the prognosis of HCC (19,20). In addition, they correlated with the invasiveness potential of HCC (21-23). The present study found that infiltrating Treg cells were positively correlated with the number of CTCs and vascular invasion. In vitro experiments revealed that Treg cells could promote the invasiveness potential of HCC cells through secreting TGFβ1, thus triggering EMT. Moreover, Treg cells also significantly promoted cell proliferation and hindered cell apoptosis. The present data were consistent with previous findings and could serve as a translational basis for future anti-Treg therapeutic strategy.
Invasiveness is a major characteristic of HCC that contributes to the high incidence of metastasis or recurrence after treatment (24,25). The acquisition of invasion potential is a complex process, and EMT is currently considered as the critical step for this biological transformation (26-28). The infiltrating Treg cells induced a mesenchyme-like, phenotype, while knocking down TGFβ1 in Treg cells reversed previous changes. Moreover, co-culture with Treg cells directly affected the invasiveness potential of HCC cells, as demonstrated by Transwell assays. Moreover, intratumoral Treg cells secreted more TGFβ1 compared with Treg cells from the peripheral blood of patients with HCC, in line with previous studies showing that tumor cells could activate Treg cells so that more TGFβ1 was secreted from Treg cells (20). These observations showed that Treg cells served as a key regulator that positively regulated EMT to promote tumor progression, and targeting Treg cells in HCC might be a promising therapeutic approach for preventing HCC progression.
The activation of Smad2/3 is a hallmark for invasiveness and HCC progression, while reversing this abnormal activation is considered a promising therapeutic approach for HCC. Previous studies demonstrated that TGFβ1 was a major stimulator for activating Smad2/3 signaling, including HCC (29). The present study found that infiltrating Treg cells could activate Smad2/3 via secreting TGFβ1, thus greatly triggering the TGFβ1 signaling pathway. Importantly, Treg cells were identified as the major source of intratumoral TGFβ1. The results clarified the mechanism by which infiltrating Treg cells positively controlled EMT and suggested a novel, promising target for inhibiting TGFβ1 signaling to improve the prognosis of patients with HCC.
Hematogenous spread is an important cause of recurrence and metastasis in HCC, and CTCs in the bloodstream plays an important role in HCC metastasis (30). Meanwhile, the release of tumor cells into circulation is a multiple-step process, and Treg acts as one of the major regulators involved in this process, which could affect the viability of HCC cells. A positive correlation was observed between the infiltrating Treg cells and the number of peripheral CTCs in patients with HCC, indicating that Treg cells might be a critical switch for CTC release. Considering the role of Treg cells in mediating TGFβ1 signaling activation, intratumoral Tregs could be a promising therapeutic target to hinder CTC release and directly prevent HCC metastasis and recurrence. However, the clinical data of the correlation between Treg cells and metastasis/recurrence was not provided in the present study due to the lack of follow-up data. A prospective cohort study is ongoing in the center to address this issue. Although a previous study indicated that TGFβ1 might act as a suppressor in HCC (31), several studies revealed the pro-tumor effects of TGFβ1 in HCC development (32,33). Meanwhile, considering its critical role in promoting cell invasiveness (34), TGFβ1 might serve as a key regulator of invasion in HCC, which was consistent with previous findings. The present study also showed that TGFβ1 secreted from Treg cells could significantly promote the invasiveness of HCC cells.
This study was novel in demonstrating the significance of infiltrating Treg cells in mediating HCC motility and progression. The data indicated that Treg cells could serve as a novel target for preventing CTC release and HCC metastasis. The involvement of infiltrating Treg cells in triggering the TGFβ1 signaling pathway and promoting EMT of cancer cells during tumor hematogenous dissemination is presumably responsible for increasing the invasiveness potentials of HCC cells. Furthermore, targeting infiltrating Treg cells in the microenvironments of patients with HCC might be a promising therapeutic strategy to reduce the recurrence and metastasis for patients with HCC undergoing resection.
Acknowledgments
The authors thank Jie-Yi Shi (Liver Cancer Institute, Zhongshan Hospital) for his expert technical assistance.
Funding: This study was sponsored by the Natural Science Foundation of Shanghai (grant no. 16ZR1405400).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Zhongshan Hospital (Reference number Y2014–028), and written informed consent was obtained from the patients for the publication of this manuscript and any accompanying images.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.54). The authors have no conflicts of interest to declare.
References
- 1.Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol 2018;15:137-51. 10.1038/nrgastro.2017.169 [DOI] [PubMed] [Google Scholar]
- 2.Ma XL, Zhou JY, Gao XH, et al. Application of the albumin-bilirubin grade for predicting prognosis after curative resection of patients with early-stage hepatocellular carcinoma. Clin Chim Acta 2016;462:15-22. 10.1016/j.cca.2016.08.005 [DOI] [PubMed] [Google Scholar]
- 3.Ringelhan M, Pfister D, O'Connor T, et al. The immunology of hepatocellular carcinoma. Nat Immunol 2018;19:222-32. 10.1038/s41590-018-0044-z [DOI] [PubMed] [Google Scholar]
- 4.Nault JC, Galle PR, Marquardt JU. The role of molecular enrichment on future therapies in hepatocellular carcinoma. J Hepatol 2018;69:237-47. 10.1016/j.jhep.2018.02.016 [DOI] [PubMed] [Google Scholar]
- 5.Iñarrairaegui M, Melero I, Sangro B. Immunotherapy of Hepatocellular Carcinoma: Facts and Hopes. Clin Cancer Res 2018;24:1518-24. 10.1158/1078-0432.CCR-17-0289 [DOI] [PubMed] [Google Scholar]
- 6.Finn RS, Zhu AX, Farah W, et al. Therapies for advanced stage hepatocellular carcinoma with macrovascular invasion or metastatic disease: A systematic review and meta-analysis. Hepatology 2018;67:422-35. 10.1002/hep.29486 [DOI] [PubMed] [Google Scholar]
- 7.Pu W, Li J, Zheng Y, et al. Targeting Pin1 by inhibitor API-1 regulates microRNA biogenesis and suppresses hepatocellular carcinoma development. Hepatology 2018;68:547-60. 10.1002/hep.29819 [DOI] [PubMed] [Google Scholar]
- 8.Hansen MT, Forst B, Cremers N, et al. A link between inflammation and metastasis: serum amyloid A1 and A3 induce metastasis, and are targets of metastasis-inducing S100A4. Oncogene 2015;34:424-35. 10.1038/onc.2013.568 [DOI] [PubMed] [Google Scholar]
- 9.Wittmann P, Grubinger M, Groger C, et al. Neuropilin-2 induced by transforming growth factor-beta augments migration of hepatocellular carcinoma cells. BMC Cancer 2015;15:909. 10.1186/s12885-015-1919-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ali A, Zhang P, Liangfang Y, et al. KLF17 empowers TGF-beta/Smad signaling by targeting Smad3-dependent pathway to suppress tumor growth and metastasis during cancer progression. Cell Death Dis 2015;6:e1681. 10.1038/cddis.2015.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joly AL, Seitz C, Liu S, et al. Alternative Splicing of FOXP3 Controls Regulatory T Cell Effector Functions and Is Associated with Human Atherosclerotic Plaque Stability. Circ Res 2018;122:1385-94. 10.1161/CIRCRESAHA.117.312340 [DOI] [PubMed] [Google Scholar]
- 12.Geng L, Tang X, Zhou K, et al. Micro RNA-663 induces immune dysregulation by inhibiting TGF-beta1 production in bone marrow-derived mesenchymal stem cells in patients with systemic lupus erythematosus. Cell Mol Immunol 2019;16:260-74. 10.1038/cmi.2018.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma XL, Gao XH, Gong ZJ, et al. Apolipoprotein A1: a novel serum biomarker for predicting the prognosis of hepatocellular carcinoma after curative resection. Oncotarget 2016;7:70654-68. 10.18632/oncotarget.12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo W, Yang XR, Sun YF, et al. Clinical significance of EpCAM mRNA-positive circulating tumor cells in hepatocellular carcinoma by an optimized negative enrichment and qRT- PCR-based platform. Clin Cancer Res 2014;20:4794-805. 10.1158/1078-0432.CCR-14-0251 [DOI] [PubMed] [Google Scholar]
- 15.Guo W, Sun YF, Shen MN, et al. Circulating Tumor Cells with Stem-Like Phenotypes for Diagnosis, Prognosis, and Therapeutic Response Evaluation in Hepatocellular Carcinoma. Clin Cancer Res 2018;24:2203-13. 10.1158/1078-0432.CCR-17-1753 [DOI] [PubMed] [Google Scholar]
- 16.Hu B, Yang XR, Xu Y, et al. Systemic immune-inflammation index predicts prognosis of patients after curative resection for hepatocellular carcinoma. Clin Cancer Res 2014;20:6212-22. 10.1158/1078-0432.CCR-14-0442 [DOI] [PubMed] [Google Scholar]
- 17.Zhou SL, Zhou ZJ, Hu ZQ, et al. Tumor-Associated Neutrophils Recruit Macrophages and T-Regulatory Cells to Promote Progression of Hepatocellular Carcinoma and Resistance to Sorafenib. Gastroenterology 2016;150:1646-58.e17. 10.1053/j.gastro.2016.02.040 [DOI] [PubMed] [Google Scholar]
- 18.Wilson CL, Jurk D, Fullard N, et al. NF kappaB1 is a suppressor of neutrophil-driven hepatocellular carcinoma. Nat Commun 2015;6:6818. 10.1038/ncomms7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang R, Tang J, Chen Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun 2017;8:15129. 10.1038/ncomms15129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiedemann GM, Knott MM, Vetter VK, et al. Cancer cell-derived IL-1alpha induces CCL22 and the recruitment of regulatory T cells. Oncoimmunology 2016;5:e1175794. 10.1080/2162402X.2016.1175794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Wang B, Wu J, et al. Association of preoperative EpCAM Circulating Tumor Cells and peripheral Treg cell levels with early recurrence of hepatocellular carcinoma following radical hepatic resection. BMC Cancer 2016;16:506. 10.1186/s12885-016-2526-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li G, Liu D, Cooper TK, et al. Successful chemoimmunotherapy against hepatocellular cancer in a novel murine model. J Hepatol 2017;66:75-85. 10.1016/j.jhep.2016.07.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, Ma XL, Tian L, et al. Serum IgG4:IgG Ratio Predicts Recurrence of Patients with Hepatocellular Carcinoma after Curative Resection. J Cancer 2017;8:1338-46. 10.7150/jca.18030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadalayil L, Benini R, Pallan L, et al. A simple prognostic scoring system for patients receiving transarterial embolisation for hepatocellular cancer. Ann Oncol 2013;24:2565-70. 10.1093/annonc/mdt247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology 2011;53:1020-22. 10.1002/hep.24199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Németh J, Stein I, Haag D, et al. S100A8 and S100A9 are novel nuclear factor kappa B target genes during malignant progression of murine and human liver carcinogenesis. Hepatology 2009;50:1251-62. 10.1002/hep.23099 [DOI] [PubMed] [Google Scholar]
- 27.Gebhardt C, Riehl A, Durchdewald M, et al. RAGE signaling sustains inflammation and promotes tumor development. J Exp Med 2008;205:275-85. 10.1084/jem.20070679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D, Zhang Y, Zhang H, et al. CADM2, as a new target of miR-10b, promotes tumor metastasis through FAK/AKT pathway in hepatocellular carcinoma. J Exp Clin Cancer Res 2018;37:46. 10.1186/s13046-018-0699-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Zaidi S, Rao S, et al. Analysis of Genomes and Transcriptomes of Hepatocellular Carcinomas Identifies Mutations and Gene Expression Changes in the Transforming Growth Factor-beta Pathway. Gastroenterology 2018;154:195-210. 10.1053/j.gastro.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma XL, Jiang M, Zhao Y, et al. Application of Serum Annexin A3 in Diagnosis, Outcome Prediction and Therapeutic Response Evaluation for Patients with Hepatocellular Carcinoma. Ann Surg Oncol 2018;25:1686-94. 10.1245/s10434-018-6402-0 [DOI] [PubMed] [Google Scholar]
- 31.Senturk S, Mumcuoglu M, Gursoy-Yuzugullu O, et al. Transforming growth factor-beta induces senescence in hepatocellular carcinoma cells and inhibits tumor growth. Hepatology 2010;52:966-74. 10.1002/hep.23769 [DOI] [PubMed] [Google Scholar]
- 32.Sandbothe M, Buurman R, Reich N, et al. The microRNA-449 family inhibits TGF-β-mediated liver cancer cell migration by targeting SOX4. J Hepatol 2017;66:1012-21. 10.1016/j.jhep.2017.01.004 [DOI] [PubMed] [Google Scholar]
- 33.Yan W, Liu X, Ma H, et al. Tim-3 fosters HCC development by enhancing TGF-β-mediated alternative activation of macrophages. Gut 2015;64:1593-604. 10.1136/gutjnl-2014-307671 [DOI] [PubMed] [Google Scholar]
- 34.Chen CL, Tsukamoto H, Liu JC, et al. Reciprocal regulation by TLR4 and TGF-β in tumor-initiating stem-like cells. J Clin Invest 2013;123:2832-49. 10.1172/JCI65859 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]