Abstract
c-Myc (hereafter, Myc) is a cancer driver whose abundance is regulated by the SCFFbw7 ubiquitin ligase and proteasomal degradation. Fbw7 binds to a phosphorylated Myc degron centered at threonine 58 (T58), and mutations of Fbw7 or T58 impair Myc degradation in cancers. Here, we identify a second Fbw7 phosphodegron at Myc T244 that is required for Myc ubiquitylation and acts in concert with T58 to engage Fbw7. While Ras-dependent Myc serine 62 phosphorylation (pS62) is thought to stabilize Myc by preventing Fbw7 binding, we find instead that pS62 greatly enhances Fbw7 binding and is an integral part of a high-affinity degron. Crystallographic studies revealed that both degrons bind Fbw7 in their diphosphorylated forms and that the T244 degron is recognized via a unique mode involving Fbw7 arginine 689 (R689), a mutational hotspot in cancers. These insights have important implications for Myc-associated tumorigenesis and therapeutic strategies targeting Myc stability.
Two phosphorylation-dependent signals cooperatively regulate Myc degradation by the SCF-Fbw7 ubiquitin ligase.
INTRODUCTION
The Myc oncoprotein is a basic helix-loop-helix transcription factor (TF) that dimerizes with Max and controls both specific target genes and global transcriptional amplification (1, 2). Myc has far-reaching biologic activities in cellular processes such as proliferation, differentiation, metabolism, stem cell biology, and apoptosis (2, 3). A network of Myc-binding proteins regulates Myc’s various functions, and normal cells use many mechanisms to tightly control Myc activity and abundance (4). Myc is a potent oncoprotein that drives numerous hallmarks of tumorigenesis. Tumors disrupt Myc regulation in several ways, including genomic alterations (e.g., translocations and amplification), mutations of Myc itself, and post-translational controls such as Myc degradation (2, 5, 6). Murine and human cell models have shown that Myc-associated cancers remain highly dependent on Myc activity, which has led to numerous therapeutic strategies that seek to antagonize Myc function (7–9).
Phosphorylation-dependent Myc degradation driven by the SCFFbw7 ubiquitin ligase is commonly targeted by mutations in cancers. Fbw7 is a substrate receptor of an SCF (Skp1/Cul1/F-box protein) ubiquitin ligase that binds to phosphorylated substrates via its β-propeller WD40 domain and to the rest of the SCF complex through its F-box motif (10). Fbw7 thus brings substrates into proximity with ubiquitin-conjugating enzymes that attach polyubiquitin chains to substrates to signal their proteasomal degradation. Fbw7 targets ~30 known substrates for degradation, most of which are TFs and key oncoproteins (e.g., Myc, Notch, and Jun) (11, 12). Fbw7 is one of the most mutated tumor suppressor genes and exhibits an unusual mutational spectrum. Most common are heterozygous missense mutations, targeting one of three arginine residues (termed Fbw7ARG) that directly interact with phosphorylated substrates (11, 13).
Fbw7 forms homodimers with important functional consequences. First, dimers can potentially interact with two substrate degrons simultaneously, which enables high-avidity binding between Fbw7 and substrates with low-affinity suboptimal degrons (14, 15). Next, Fbw7 dimerization can expand the range of substrate lysines that are accessible for ubiquitin conjugation (16, 17). Last, dimerization may explain the predominance of heterozygous Fbw7ARG mutations in cancer, which are thought to dominantly inhibit wild-type (WT) Fbw7 by forming impaired Fbw7wt/Fbw7ARG heterodimers (10, 13, 18).
Fbw7 substrates contain a consensus degron motif that must be phosphorylated in its central position to bind Fbw7. Most Fbw7 substrate degrons are also phosphorylated in their +4 position, which is sometimes replaced by a negatively charged amino acid (10). The Myc threonine 58 (T58) region is a consensus Fbw7 degron in which T58 and serine 62 (S62) are the central and +4 residues, respectively (19, 20). T58 phosphorylation is catalyzed by glycogen synthase kinase 3 (GSK3) and requires a priming phosphorylation at S62 (21–23). Previous studies found that oncogenic Ras signaling appeared to increase S62 phosphorylation and Myc stability, which led to the hypothesis that opposing degron phosphorylations regulate Myc turnover: S62 phosphorylation stabilizes Myc by preventing Fbw7 binding, whereas T58 phosphorylation triggers Fbw7 binding and Myc degradation (24–27). The notion that a “stabilizing” S62 phosphorylation in Myc is required to prime a “destabilizing” T58 phosphorylation is paradoxical and has been reconciled by a model in which S62 must be dephosphorylated before Fbw7 binding to T58 monophosphorylated Myc (28, 29). However, this model differs markedly from how phosphorylation regulates other Fbw7 substrates, in which +4 phosphorylation increases degron affinity (16, 30, 31).
Here, we dissected the Fbw7-Myc interaction with an array of methodologic approaches. We identify a second diphosphorylated degron in Myc, which synergizes with the T58 phosphodegron to bind Fbw7, and we elucidate a critical positive role of S62 phosphorylation in forming a high-affinity complex with the F-box protein. We show that the two Myc degrons are simultaneously recognized by dimeric Fbw7, which drives Myc turnover. Our data provide mechanistic insights into the complex interactions between the Myc oncoprotein and the Fbw7 tumor suppressor that have important implications for both tumorigenesis and cancer therapy.
RESULTS
Identification of a second Fbw7 phosphodegron in Myc
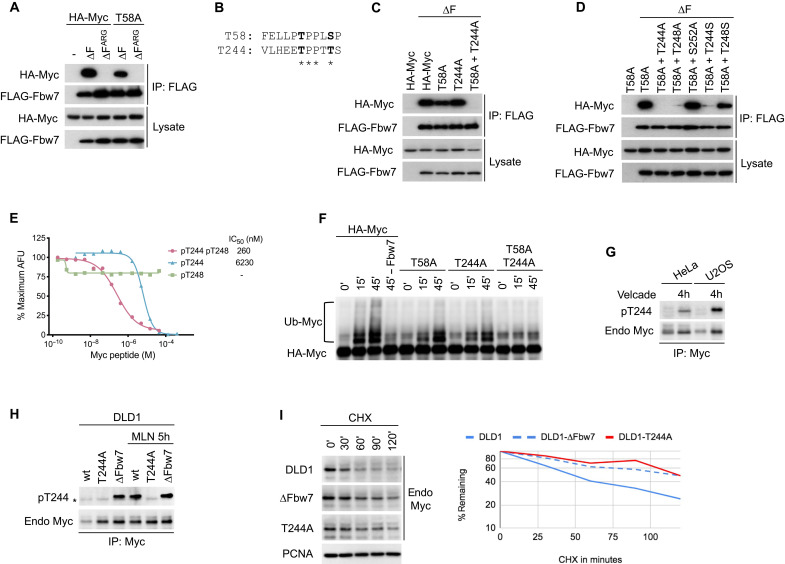
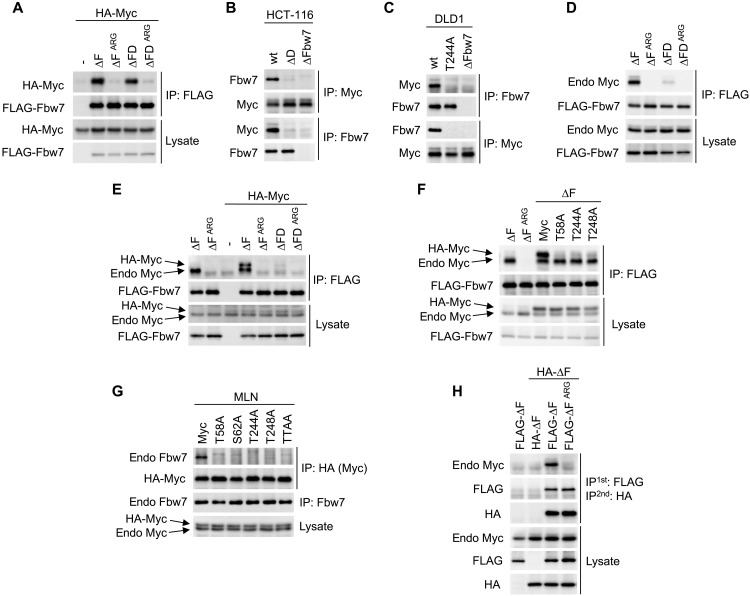
We and others previously identified the Myc T58 phosphorylation site as an essential part of an Fbw7 phosphodegron that regulates Myc protein stability (19, 20). Although T58 phosphorylation is required for Fbw7 recognition, Myc containing an alanine mutation at T58 (T58A) still binds to coexpressed Fbw7 lacking the F-box (ΔF to uncouple binding from turnover). This interaction was dependent on phosphorylation, because it was lost with a cancer-associated arginine mutation (∆FARG) (Fig. 1A), which compromises the Fbw7 arginine triad responsible for binding degron phosphates. This unexpected result strongly suggested the presence of a second phosphorylation-dependent Myc motif that directs Fbw7 binding. Myc contains a potential second phosphodegron at amino acid T244 near a cancer-associated minor hotspot in lymphomas (32), and this region is also mutated in some transforming v-Myc oncogenes found in avian retroviruses (Fig. 1B). These findings prompted us to test whether this motif constitutes a second Fbw7 binding element. Figure 1C shows that, in cotransfection assays, Myc binds to Fbw7 when either T58 or T244 are mutated, but that binding is lost only when both sites are disabled, suggesting that either can mediate Fbw7 binding. As expected for canonical Fbw7 degrons, binding via the T244 degron required T248 (the +4 position) and was impaired by mutating the central threonine to serine, which reduces binding affinity (Fig. 1D) (33). These data suggested that both T244 and T248 are phosphorylated in vivo, because binding of a phosphodegron typically depends on phosphorylation of both sites.
Fig. 1. Identification of the T244 degron.
(A) HEK293 cells were cotransfected with HA-Myc and FLAG-Fbw7, and lysates were immunoprecipitated (IP) with FLAG antibody and blotted with HA antibody. F-box deletion (∆F) uncouples binding from turnover. (B) Alignment of the T58 and T244 degrons. (C and D) Assays as in (A). To isolate the T244 degron, all mutants were made in a T58A mutant background in (D). S252 was tested for its potential role as a priming site for degron phosphorylations. (E) Quantitative peptide competition analysis of Myc peptides with a cyclin E pT380/pS384 degron peptide for Fbw7 binding as measured in arbitrary fluorescent units (AFU). (F) Myc or mutants were immunoprecipitated from transfected HEK293 cell lysates, and washed beads were subjected to in vitro ubiquitylation. Fbw7 was omitted in lane 4 (−Fbw7) as negative control. Reactions were analyzed by Western blotting. (G) HeLa and U2OS cell lines were treated with proteasome inhibitor Velcade for 4 hours (lanes 2 and 4), and lysates were immunoprecipitated against Myc and blotted as indicated. While Myc levels increase upon proteasome inhibition, pT244 appears to be increased relative to total Myc levels. (H) Isogenic DLD1 WT cells or their homozygously mutated cell lines (T244A or Fbw7 deletion) were treated with MLN4924 (right lanes only), and lysates were immunoprecipitated against Myc and blotted as indicated. Asterisk marks an antibody cross-reaction of the IP antibody heavy chain. (I) DLD1 cells or isogenic derivatives homozygously deleted for Fbw7 or T244A Myc mutated were treated with cycloheximide (CHX) and blotted for Myc steady-state abundance. Proliferating cell nuclear antigen (PCNA) serves as loading control. Bands were quantitated and plotted on the right.
To validate this idea, we quantified the in vitro binding affinity between Fbw7 and the T244 degron in its three phosphorylation states using the amplified luminescent proximity homogeneous assay (ALPHA) (Fig. 1E). In this assay, untagged T244 degron phosphopeptides were used to compete with a biotinylated cyclin E pT380/pS384 degron peptide for Fbw7 binding. The diphosphorylated T244 degron (pT244/pT248) interacts with Fbw7 with an affinity of about 260 nM, whereas the T244 monophosphorylated peptide (pT244) had a 24-fold reduction in its affinity toward Fbw7. We did not detect any competition with a T248 monophosphorylated peptide (pT248). These data directly support the notion that phosphorylation at both sites is required for high-affinity Fbw7 binding.
We next tested whether the putative T244/T248 degron directs Myc ubiquitylation. Figure 1F shows that, compared with WT Myc, mutation of either T58 or T244 compromised in vitro Myc ubiquitylation by SCFFbw7 and completely abolished ubiquitylation when both sites were mutated. Therefore, although each degron may alternatively bind to Fbw7 upon coexpression, as shown in Figure 1C, the efficiency of in vitro Myc ubiquitylation is maximized when both degrons are present.
To study the role of endogenous Myc T244 phosphorylation, we developed an anti–phospho-T244 Myc mouse monoclonal antibody, which readily demonstrated T244 phosphorylation on ectopic Myc (fig. S1). Endogenous Myc T244 phosphorylation was harder to detect but was specifically enriched (compared to total Myc abundance) upon proteasome inhibition, which is consistent with a role in Myc turnover (Fig. 1G). We next generated endogenous Myc T244A homozygous knockin DLD1 colorectal carcinoma cells, in which T244 was mutated to alanine (T244A) and thus cannot be phosphorylated (fig. S2). Myc abundance was elevated in T244A cells (compared to isogenic WT cells) and mirrored Myc abundance in isogenic Fbw7-deleted cells (Fig. 1H). Endogenous Myc T244 phosphorylation was undetectable in WT cells but strongly increased in Fbw7 knockout cells and upon inhibition of Fbw7 function by a pharmacologic Cullin neddylation inhibitor, MLN4924. Thus, like rate-limiting degron phosphorylations in other Fbw7 substrates, the amount of T244-phosphorylated Myc was dependent upon Fbw7 and proteasome function. Myc half-life was extended in T244A cells to a similar degree as seen in Fbw7 knockout DLD1 cells, as shown by its slowed decay upon cycloheximide (CHX) treatment (Fig. 1I). In summary, these data identify a second Fbw7 phosphodegron in Myc at T244/T248 that regulates Fbw7 binding and Myc stability.
Structural analysis of the T244 degron
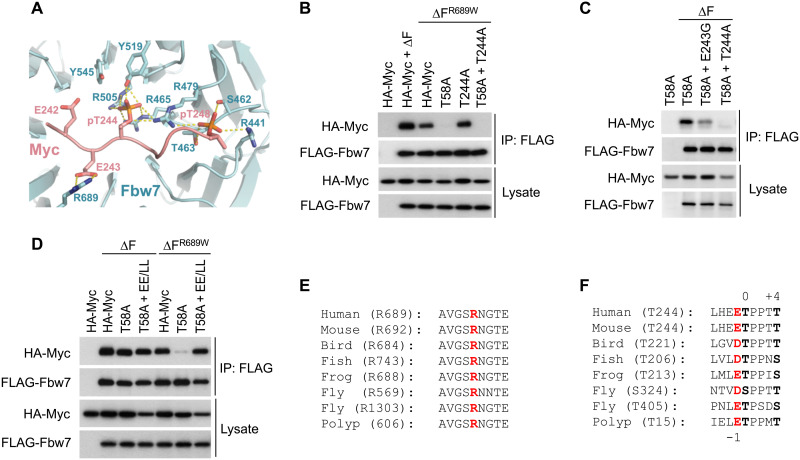
To further verify that the T244 degron acts as a canonical phosphodegron for Fbw7 binding, we determined the crystal structure of Fbw7 in complex with the diphosphorylated degron peptide at 2.55 Å resolution (Fig. 2A and table S1). The Myc T244 degron is anchored to the top surface of the Fbw7 WD40 domain. Its core region (residues 244 to 248) adopts the same conformation as the cyclin E T380 degron, where the two phosphorylated residues make a network of hydrogen bonds and electrostatic contacts with a host of Fbw7 amino acids, including three hallmark cancer hotspot arginine residues (R465, R479, R505, and Y519 for Myc pT244; and S462, T463, R441, and R479 for Myc pT248). Unlike the cyclin E T380 degron, however, the Myc T244 degron can only be traced up to the −3 position upstream of the central threonine. Such a binding mode could be associated with two glutamate residues (E242 and E243), which are immediately upstream of T244 and are distinct from the hydrophobic residues that are favored in these positions for other degrons. In the crystal structure, the side chain of E243 at the −1 position makes electrostatic interactions with the guanidinium group of Fbw7 R689, whereas E242 at the −2 position is in close vicinity to Fbw7 tyrosine 545 (Y545). Together, they deviate the N-terminal region of the Myc T244 degron from the hydrophobic interactions cyclin E’s T380 degron makes with Fbw7.
Fig. 2. Structure analysis of the T244 degron.
(A) Close-up view of the interface formed between Fbw7 and the Myc T244 degron. Fbw7 is colored in cyan with its Myc-interacting residues shown in sticks. The Myc T244 degron is colored in salmon. Hydrogen bonds and electrostatic interactions are indicated by yellow dashed lines. (B to D) HEK293 cells were transfected as indicated, immunoprecipitated against FLAG-Fbw7 (∆F), and blotted for associated Myc with HA antibody. (E) Alignment of the Fbw7 region centered at R689 (red). Bird, chicken; fish, zebrafish; frog, Xenopus; worm, Caenorhabditis elegans; fly, fruitfly; and polyp, Hydra vulgaris. Polyp Fbw7 was assembled from predicted partial transcripts. (F) Alignment of the T244 degron. The central 0 and +4 position phosphorylations are in black and bold, and the conserved negative charge in −1 is in red. Fruitflies lack the T58 degron entirely and contain two versions of the T244 degron instead, while hydra’s Fbw7 degrons are reversed such that the T244-type degron is located at the N terminus.
Consistent with the crystal structure, and in contrast to the T58 degron, Fbw7 binding to the T244 degron exhibited strong R689 dependence as evidenced by the inability of T58A Myc (which uses the T244 degron) to bind to an Fbw7 R689W mutant (Fig. 2B). R689 is a mutational hotspot in cancer and is most commonly converted to either tryptophan (W) or glutamine (Q) (fig. S3A). Fbw7 binding via the Myc T244 degron was also dependent on E243, as a cancer-derived E243G mutant was severely compromised in Fbw7 binding (Fig. 2C). The residual binding of the E243G mutant is likely due to additional water-mediated backbone contacts made by R689, which may explain the more severe effect upon R689 mutation. Converting the Myc T244 degron glutamates in the −1 and −2 positions (E242 and E243) to hydrophobic leucine residues, as found in cyclin E and other substrates, restored binding to Fbw7 R689W (Fig. 2D), likely due to compensatory interactions of these residues with Fbw7’s hydrophobic groove, as described for cyclin E. Both Fbw7 R689 and Myc E243 are highly conserved throughout evolution (Fig. 2, E and F). These findings expand the Fbw7 degron consensus motif and suggest a critical role for R689 in substrate binding. We also determined how additional tumor-derived T244 degron mutations affect Fbw7 binding and Myc T244 phosphorylation. While not as crucial as the core degron phosphorylation sites (T244 and T248), other proximal mutations also decreased T244 phosphorylation and Fbw7 binding, suggesting that they result in intermediate disruption of Fbw7-mediated Myc turnover in cancers (fig. S3, B and C).
S62 phosphorylation stimulates Myc-Fbw7 binding
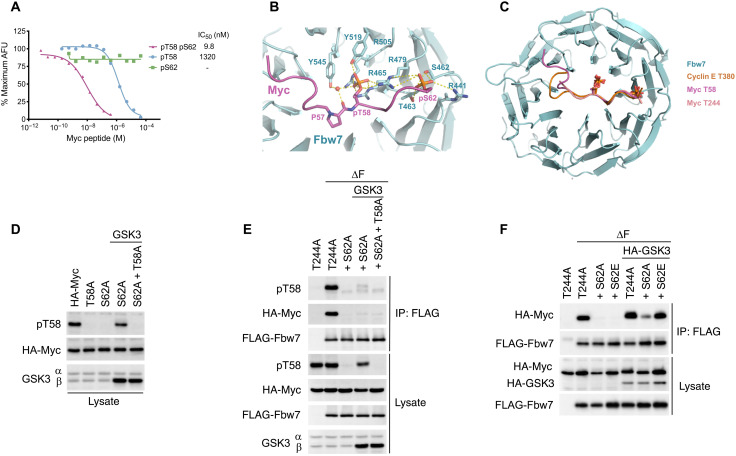
Because the T244 degron and other Fbw7 degrons use two phosphates to bind Fbw7, we reexamined how phosphorylation regulates the Myc T58 degron interaction with Fbw7. We used the ALPHA-based competition assay to determine the binding affinity of T58 degron peptides in different phosphorylation states with Fbw7. The affinity of the diphosphorylated pT58/pS62 Myc peptide for Fbw7 was about 150-fold higher than monophosphorylated pT58 Myc (9.8 versus 1320 nM) (Fig. 3A). Thus, S62 phosphorylation greatly increases, rather than decreases, the affinity of Myc for Fbw7. Accordingly, an S62A mutation prolonged Myc’s half-life in proliferating cells (fig. S4), although it is not possible to determine a role of pS62 independent of its priming function for T58 phosphorylation.
Fig. 3. S62 phosphorylation increases the affinity for Fbw7.
(A) Quantitative peptide competition assay of T58 degron phosphopeptides with a cyclin E pT380/pS384 peptide for Fbw7 binding. IC50, median inhibitory concentration. (B) Close-up view of the interface formed between Fbw7 and the Myc T58 degron. Fbw7 is colored in cyan, with its Myc-interacting residues shown in sticks. The Myc T58 degron is colored in magenta. Hydrogen bonds and electrostatic interactions are indicated by yellow dashed lines. Water is shown in red sphere. (C) Superimposition of the Myc T58, Myc T244, and cyclin E T380 degrons in complex with Fbw7 WD40 domain. (D) Enforced GSK3β expression omits the need for pS62 priming. HEK293 cells were transfected with Myc (or phosphosite mutants) and GSK3β, and lysates were blotted as indicated. This experiment also indicates that S62A mutation preserves the epitope of the phospho-T58 antibody. (E) Lysates of transfected HEK293 cells were immunoprecipitated with FLAG antibody against Fbw7 and blotted for Myc and pT58 Myc. All Myc constructs contained a T244A mutation to isolate the T58 degron (see the main text). (F) Assay as in (E), except that HA-tagged GSK3β was used and detected along with Myc in the HA blot.
To further elucidate the role of S62 phosphorylation in Fbw7 binding, we determined the crystal structure of a pT58/pS62 Myc peptide bound to Fbw7. The core region (residues 58 to 62) aligns perfectly with the corresponding region of the Myc T244 degron and the cyclin E T380 degron (Fig. 3, B and C). Notably, pS62 forms hydrogen bonds with the side chains of Fbw7 S462, T463, R441, and R479 in the exact same manner as the +4 phosphorylations of the Myc T244 degron (pT248) and the cyclin E T380 degron (pS384). This is consistent with the enhanced Fbw7 binding affinity by the phosphorylation of S62 as measured in our peptide competition assay. The upstream region (residues 53 to 57) of the T58 degron packs against the hydrophobic groove of Fbw7 analogous to the cyclin E T380 degron, but the conformation of this region varies considerably. Binding of the T58 upstream region is accompanied by water-mediated hydrogen bonds formed between the backbone carbonyl group of proline 57 (P57) and the hydroxyl group of Fbw7 Y545. Together, the extensive contacts between the Myc T58 degron and Fbw7 are in accordance with their high binding affinity in the single-digit nanomolar range and hinges on phosphorylation of both T58 and S62.
We also determined the role of S62 phosphorylation in Fbw7-Myc binding in vivo. T58 monophosphorylated Myc was proposed to be the preferred Myc species for Fbw7 binding in cells (26), but the dependence of T58 phosphorylation on prior S62 priming phosphorylation makes it difficult to produce the T58 monophosphorylated Myc needed to test this model that is not also S62 phosphorylated. Enforced GSK3β expression overcame this limitation, because it was able to phosphorylate T58 in an S62A mutant and to a similar extent as seen with WT Myc (Fig. 3D). We generated T58 monophosphorylated Myc S62A and also isolated Fbw7 binding via the T58 degron by using Myc T244A. As shown in Fig. 3E, binding of T58 monophosphorylated Myc to Fbw7 is severely compromised compared with WT Myc. In contrast, a Myc S62E mutation that mimics S62 phosphorylation restored Fbw7 binding (Fig. 3F). In agreement with our in vitro and structural results, S62 phosphorylation increased binding of Myc to Fbw7 also in a cellular context.
S62 is phosphorylated in Fbw7 complexes
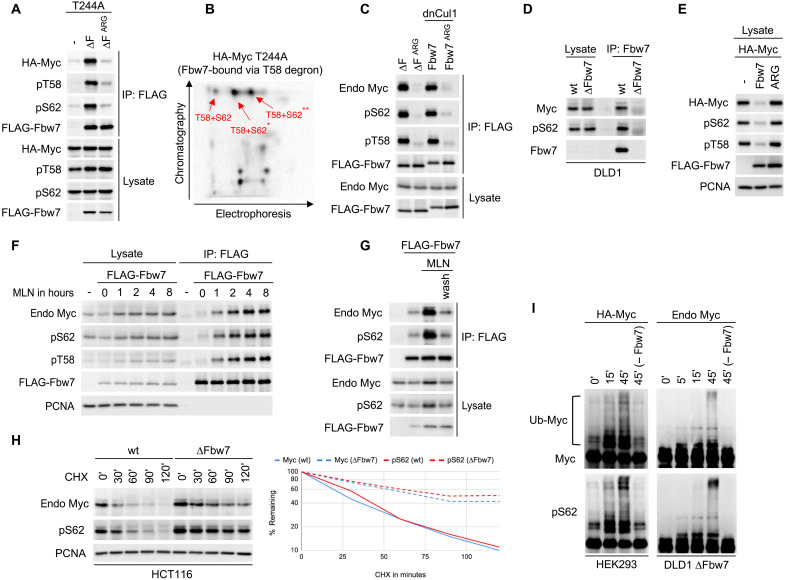
To determine whether pS62 must be dephosphorylated for Fbw7 binding (26), we assessed Myc’s phosphorylation status in Fbw7 complexes. We functionally isolated the T58 degron by using a T244A mutant and disabled Myc turnover with Fbw7 (∆F) (Fig. 4A). Both pT58 and pS62 were detected in Myc T244A bound to Fbw7 ΔF, indicating that pS62 Myc is not excluded from Fbw7 complexes mediated via the T58 degron. The specificity of the commercial phospho-antibodies was confirmed with transfected mutant HA (hemagglutinin)-Myc versions of the respective phosphorylation sites (fig. S5). We next used two-dimensional (2D) phosphopeptide mapping and phospho–amino acid analyses to show that T58 and S62 were both phosphorylated on the same Myc T244A molecule bound to Fbw7 (Fig. 4B and fig. S6). This analysis only detected diphosphorylated pT58/pS62 Myc bound to Fbw7. We did not detect T58 monophosphorylated Myc in Fbw7 complexes or otherwise. Endogenous S62-phosphorylated Myc also bound exogenous Fbw7 when its turnover was prevented by either an Fbw7 F-box deletion or by coexpression of dominant-negative Cul1 (dnCul1) (Fig. 4C). Last, endogenous Myc S62 phosphorylation was readily detected in endogenous Fbw7 immunoprecipitates from DLD1 cells treated with MLN4924 (to allow stable Fbw7-Myc binding) (Fig. 4D). Therefore, S62 phosphorylation does not prevent Myc from binding to Fbw7 and does not need to be dephosphorylated for this interaction.
Fig. 4. S62 is phosphorylated in cellular Fbw7 complexes.
(A) HEK293 cells were transfected with Myc T244A (to isolate the T58 degron) and Fbw7 (∆F) and lysates analyzed as indicated. (B) HEK293 cells transfected as in (A) were radiolabeled, and Fbw7-associated Myc was subjected to 2D phospho-peptide analysis. Only pT58/pS62 diphosphorylated peptides were identified in Fbw7 complexes (for details, see fig. S6). (C) Coimmunoprecipitation of endogenous (endo) Myc with transfected Fbw7 from HEK293 cell lysates. The interaction was stabilized either by F-box deletion (∆F) or with dominant-negative Cul1 (dnCul1). (D) Coimmunoprecipitation of endogenous Myc with endogenous Fbw7 from DLD1 cells treated with MLN4924 (4 hours). Isogenic DLD1 ∆Fbw7 cells serve as “no bait” control. (E) HEK293 cells were cotransfected and blotted as indicated (PCNA: loading control). (F) HEK293 cells were transfected with Fbw7, treated with MLN4924, and analyzed as indicated (PCNA: loading control). (G) Assay similar to (F). Samples in lanes 3 and 4 were treated with MLN4924 (4 hours). The last sample (lane 4) was washed and incubated in fresh media for one additional hour. (H) Isogenic HCT116 cells (WT or ∆Fbw7) were treated with CHX for the indicated times, and lysates were blotted for endogenous Myc or pS62 (PCNA: loading control). Quantification on the right. (I) Myc immunoprecipitated from transfected HEK293 cells (left) or endogenous Myc from DLD1 ∆Fbw7 cells (right) was subjected to in vitro ubiquitylation (Ub) for the indicated times and blotted for Myc or pS62. Fbw7 was omitted in the last lane (−Fbw7) as a control.
We also examined how Fbw7 function regulates pS62 Myc abundance. Analogously to total Myc, pS62 Myc was eliminated by coexpressed Fbw7 (Fig. 4E). Conversely, pS62 Myc accumulated in Fbw7 complexes with similar kinetics as pT58 Myc in MLN4924-treated cells to inhibit the ubiquitin ligase (Fig. 4F and fig. S7) and was rapidly lost from Fbw7 complexes upon reactivation of the ligase via drug washout (Fig. 4G). pS62 Myc was also stabilized to the same extent as total Myc in Fbw7-deleted cells (∆Fbw7), as shown by its slowed decay after CHX treatment, compared with WT cells (Fig. 4H). Last, in vitro Myc ubiquitylation by recombinant SCFFbw7 revealed S62 phosphorylation in high–molecular weight Myc-ubiquitin conjugates (Fig. 4I). Collectively, these data show that S62 phosphorylation does not prevent Fbw7 binding or Myc turnover in cells or inhibit Fbw7-mediated Myc ubiquitylation. Rather, S62 phosphorylation increases Myc’s affinity for Fbw7 and binds to the +4 phosphate-binding pocket analogously to the high-affinity cyclin E T380/S384 degron. The binding of the Myc T58 degron to Fbw7 thus exhibits the same dependence on two degron phosphates as found in other substrates.
The T58 and T244 degrons cooperatively drive Fbw7 binding
Fbw7 is a dimeric protein, and the presence of two Myc degrons suggested that they may potentially cooperate to recruit Fbw7 dimers (14). We thus tested this idea and found that Myc degron cooperation and a binding requirement for Fbw7 dimers depended on the abundance of Myc and whether we studied exogenous or endogenous Myc. In the case of overexpressed Myc, each degron independently mediated stable Fbw7 binding (Fig. 1C), and therefore binding did not require Fbw7 dimerization, as shown by similar amounts of Myc binding to either Fbw7 dimers (∆F) or monomers (∆FD) (Fig. 5A). In contrast, the endogenous Myc-Fbw7 interaction requires Fbw7 dimers. We used HCT116 cells that we previously engineered to only express endogenous Fbw7 monomers due to homozygous deletion of the Fbw7 dimerization domain (ΔD) (14). Endogenous Myc binding to endogenous Fbw7 was severely impaired in Fbw7ΔD cells (Fig. 5B), supporting the idea that, in a physiologic context, endogenous Myc uses endogenous Fbw7 dimers, likely to engage two degrons. Notably, endogenous Myc from DLD1-T244A cells did not bind to endogenous Fbw7 (Fig. 5C), explaining its prolonged half-life (Fig. 1I). Therefore, Myc regulation by Fbw7 appears to require both of its degrons and Fbw7 dimers. These data also suggest that the Myc T58 degron alone is insufficient to drive stable Fbw7 binding.
Fig. 5. Myc degrons cooperate to recruit Fbw7 dimers.
(A) HEK293 cells were cotransfected with FLAG-Fbw7 and HA-Myc and analyzed as indicated. ∆FD has a deletion in the Fbw7 dimerization domain in addition to the F-box (∆F). (B) HCT116 or their isogenic gene-targeted cell lines were treated with MLN4924 (4 hours) and lysates were analyzed by reciprocal immunoprecipitations. (C) DLD1 cells or their isogenic gene-targeted derivatives were treated with MLN4924 (4 hours) and lysates were analyzed by reciprocal immunoprecipitations. (D) HEK293 cells were transfected with the indicated Fbw7 constructs and analyzed for associated endogenous Myc. (E and F) Assay as in (A), except cells were cotransfected with 50 ng (E) or 100 ng (F) of Myc DNA per 6-cm dish [as compared with 1.5 μg in (A)]. Exogenous HA-Myc migrates just above endogenous Myc. Myc was blotted with anti-Myc antibody to detect both endogenous and exogenous proteins. (G) HA-Myc and its phosphorylation site mutants were transfected at physiologic levels (300 ng of DNA per 10-cm dish), and cells were treated with MLN4924 (4 hours) and analyzed as indicated. (H) HEK293 cells were cotransfected with differentially tagged Fbw7 (∆F), and lysates were first immunoprecipitated anti-FLAG, eluted with FLAG peptide and subjected to a second immunoprecipitation anti-HA to specifically isolate heterodimers, and analyzed by Western blotting.
We hypothesized that the different binding requirements for endogenous versus overexpressed proteins might be explained by the high concentration of overexpressed Myc and/or Fbw7, which may overcome normal affinity requirements and allow either degron to bind to Fbw7 monomers independently. Endogenous Myc bound poorly to ectopic Fbw7 monomers (ΔFD) compared with Fbw7 dimers (ΔF), indicating that low Myc levels are critical for Fbw7 dimer–dependent interactions (Fig. 5D). We thus reduced the amount of exogenous Myc expression to physiologic levels, which faithfully recapitulated the dimer-dependent Fbw7 binding seen with endogenous Myc (Fig. 5E and fig. S8). These “dimer-dependent” transfection conditions enabled us to directly test whether the T58 and T244 degrons cooperatively recruit Fbw7 dimers. As shown in Fig. 5 (F, for exogenous Fbw7, and G, for endogenous Fbw7), mutation of either degron abolished the Fbw7 interaction, demonstrating that the T58 and T244 degrons are simultaneously required for Fbw7 binding, as are dimers.
Heterozygous Fbw7ARG mutations are thought to act as dominant negatives that poison WT Fbw7 through dimerization (10). However, the oncogenic consequences of these missense mutations on Fbw7 function remain poorly understood. We used sequential immunoprecipitation of differentially tagged Fbw7 constructs to generate Fbw7wt/Fbw7ARG heterodimers, consisting of one WT protomer and one Fbw7ARG mutant protomer, to prove that Fbw7 binding to endogenous Myc requires two functional protomers in the Fbw7 dimer. HA-tagged Fbw7∆F was coexpressed with either FLAG-Fbw7ΔF or FLAG-Fbw7∆FARG in human embryonic kidney (HEK)293 cells, sequentially immunoprecipitated to specifically pull down Fbw7wt/Fbw7ARG heterodimers (see legend) and blotted for associated endogenous Myc (Fig. 5H). As expected, Fbw7wt/Fbw7wt dimers bound to endogenous Myc (lane 3). However, Fbw7wt/Fbw7ARG heterodimers failed to bind Myc (lane 4). We further examined the importance of Fbw7 dimerization on endogenous Myc stability by measuring Myc turnover in HCT116 cells with engineered homozygous ΔD or heterozygous Fbw7+/ARG mutations, which either eliminate or reduce the amount WT Fbw7 dimers, respectively. Myc was stabilized in both backgrounds, although somewhat less so in Fbw7+/ARG cells, because of residual WT Fbw7 homodimers produced by the remaining Fbw7 WT allele (presumably ~25%) (fig. S9). Collectively, these data strongly support our model that the two Myc degrons cooperatively engage the two promoters of an Fbw7 dimer (Fig. 6).
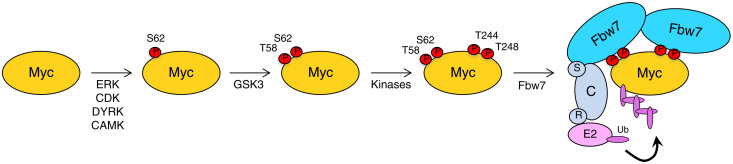
Fig. 6. Model for cooperative binding of both Myc degrons to Fbw7 dimers.
Myc is first phosphorylated on S62 by various kinases, followed by GSK3-mediated T58 phosphorylation. So far, unidentified kinases phosphorylate T244 and T248 and the two degrons recruit Fbw7 dimers whose SCF complex proteins (S, Skp1; C, Cul1; and R, Rbx1) use ubiquitin-conjugating enzymes (E2) to polyubiquitylate Myc, followed by its rapid degradation by the proteasome. This allows for additional kinase signaling input to regulate Myc’s destruction by Fbw7.
DISCUSSION
While several Myc ubiquitylation pathways are known (26), phosphorylation-dependent Myc degradation involving the T58 degron and Fbw7 is the most studied, largely because of its critical role in tumorigenesis. This pathway first came to light with the realization that v-Myc genes in avian retroviruses and translocated Myc alleles in Burkitt lymphomas share T58 mutations that deregulate Myc activity and impair its turnover (34–37). The identification of Fbw7 as a tumor suppressor that targets T58-phosphorylated Myc demonstrated that this pathway is also disrupted by Fbw7 mutations in tumors (11).
Ras-driven mitogenic pathways stabilize Myc, and this has been linked to increased S62 phosphorylation (24, 25). Moreover, studies in transgenic mouse and cell models have found different phenotypes associated with T58A and S62A mutations, lending additional support to the idea that these phosphorylations have distinct functions (22, 28, 38–40). However, the precise role of S62 has remained confusing in that the proposed stabilizing function of S62 phosphorylation seems inconsistent with its priming function for the destabilizing T58 phosphorylation by GSK3. Resolution of this conundrum is relevant to understanding normal and oncogenic Myc regulation, for instance, via oncogenic Ras signaling (41). It is also critical for the ongoing development of therapeutic approaches seeking to provoke Myc degradation in tumors via altered phosphorylation. Here, we describe new functions for Myc regulatory phosphorylations, including the following: (i) the identification of the T244 degron as a signal that is essential for Fbw7-mediated Myc turnover, (ii) the finding that S62 phosphorylation promotes Fbw7-mediated Myc ubiquitylation and degradation and greatly increases the affinity of the T58 degron for Fbw7, and (iii) that cooperative interactions of both Myc degrons with dimeric Fbw7 is required for Fbw7-mediated Myc ubiquitylation. Our study uncovers the complex circuitry underlying phosphorylation-induced degradation of Myc and demonstrates that S62 phosphorylation plays an integral role in driving this degradation.
How do we reconcile these findings with previous studies indicating that pS62 stabilizes Myc and must be dephosphorylated for Fbw7 binding? While most previous studies correlated S62 phosphorylation with Myc stability (27, 28), we directly determined how pS62 affects Fbw7 binding. The previously reported stabilizing aspects of pS62 could thus reflect pathways associated with S62 phosphorylation that do not involve Fbw7 binding, such as nuclear pore association (42). The finding that oncogenic Ras stabilizes Myc, in part, through an Fbw7-independent pathway supports this possibility (41). T58 degron phosphorylations also affect Myc in other ways, such as altered transcriptional activity, which may contribute to the reported unique S62A phenotypes, compared with T58A (43–46). The many experimental systems used to study Myc stability could also lead to discordant results regarding the functions of these phosphorylations. For example, we show here that specific Myc-Fbw7 interactions depend highly upon their relative abundances. In summary, our data do not exclude Fbw7-independent roles for S62 phosphorylation in Myc stability. Instead, they demonstrate that, rather than oppose Fbw7 binding, S62 phosphorylation forms an integral part of the Fbw7 recognition signal.
The essential role of the T244 degron in Fbw7-mediated Myc ubiquitylation adds new complexity to the control of Myc stability by yet unidentified signaling pathways. While it shares features of canonical Fbw7 degrons such as the central and +4 phosphates, its affinity is reduced, and its interaction mode with Fbw7 differs from other reported structures. The crystal structure shows that, while the T244 core degron interacts with Fbw7 almost identically to both the Myc T58 degron and cyclin E T380 degrons, the upstream interactions differ significantly in each structure. For the T244 degron, we demonstrate a crucial role for a negative charge in the −1 position and Fbw7 R689. While not as common as mutations of the arginine residues of the central phosphate-binding triad (Fbw7ARG), R689 is a mutational hotspot in cancers that we speculate may be selected by the stabilization of substrates with degrons that resemble the T244 degron in their −1 upstream position.
The essential role of the T244 degron is compatible with other studies of Myc degradation that have focused solely on the T58 degron. Moreover, its role was only revealed by studying endogenous levels of Myc and Fbw7, and this may partly explain why it has gone largely unnoticed to date. The large and acidic nature of the tryptic peptide containing T244 and T248 likely prevented detection of these phosphorylations in 2D phospho-peptide analysis and in published proteomic surveys. One other report also found that T244 phosphorylation may contribute to Myc stability in tumors and that this region (especially P245) represents a minor mutational hotspot in Burkitt’s and other lymphomas (32). This study noticed that P245 mutation phenocopied T58 mutations, and we provide the mechanism for this observation here with our model of degron cooperation.
The use of Fbw7 dimerization mutants allowed us to ascertain that Fbw7 dimers are required for Myc binding via the cooperative interactions of the two degrons. We isolated Fbw7 dimers to demonstrate the importance of a fully functional Fbw7 dimer in Myc binding, which revealed that Fbw7 heterodimers consisting of Fbw7wt and Fbw7ARG protomers cannot bind Myc. This finding has important implications for tumorigenesis involving heterozygous Fbw7ARG mutations, because their oncogenic phenotypes are thought to be different from Fbw7 null mutations and may involve substrate-specific activities (47). Fbw7+/ARG tumors presumably express a mixture of Fbw7wt/Fbw7wt, Fbw7wt/Fbw7ARG, and Fbw7ARG/Fbw7ARG dimers. The finding that the Fbw7wt/Fbw7ARG heterodimers cannot bind to Myc shows one way in which these mutations dominantly inhibit Fbw7 and suggests that Myc may be a critical target of Fbw7ARG mutations in cancers.
Loss of degron cooperation with Fbw7 dimers likely represents the underlying reason for the frequent cancer-associated selection of heterozygous Fbw7ARG mutations. Two cooperating phosphodegrons allow for additional control of substrate degradation by the signaling pathways that mediate phosphorylation of each degron but consequently rely on fully functional Fbw7 dimers for turnover. While many substrates may contain a single degron with sufficient affinity for Fbw7, cooperating degrons likely have suboptimal affinity and synergistically recruit the ligase through their combined avidity. Other substrates have been noted to contain secondary degrons, but their role in cooperatively binding to Fbw7 dimers is not yet known. Substrates may also provide two cooperating degrons via homo- or heterodimerization. For instance, sterol regulatory element–binding protein (SREBP) dimers contain a degron in each of its protomers (14). Other dimeric ubiquitin ligases also use two degrons to bind to substrates. For example, the binding of Nrf2 to Keap1 uses both a high- and a low-affinity degron simultaneously (48).
It is perhaps unexpected that the high-affinity T58 degron is insufficient to recruit Fbw7 and relies on the cooperation with the T244 degron. We thus assume that, at physiologic levels, the T58 degron somehow cannot use all of the contacts it makes with Fbw7 in vitro, perhaps due to a binding partner of Myc’s transcriptional activation domain that overlaps with a portion of the Fbw7 degron and thereby renders the degron ineffective and reliant on the cooperation with the T244 degron. This idea might explain the increased affinity of the T58 degron upon overexpression if a degron-masking binding partner is out-titrated by transfection or perhaps by cancer-associated overexpression or amplification. If correct, there might be physiologic conditions in which the T58 degron may function independent of T244 in the absence of this putative binding partner. Perhaps this is the case for N-Myc and L-Myc regulation by Fbw7, as neither contains a recognizable second Fbw7 degron. In case of N-Myc, the partial occupancy of the T58 degron by Aurora kinase A leads to N-Myc stabilization (49, 50). The possibility that the T58 degron may suffice for Fbw7 binding in some conditions, perhaps in tumors with high Myc expression, might explain the observation that the T244 degron is mutated less frequently than the T58 degron in cancers. That is, autonomous T58 degron function might remove the selective pressure for T244 mutations. Some degron mutations may also have consequences other than just Fbw7 binding, whereas others may be deleterious, which may also affect the frequency of degron mutations.
Myc degradation has also been linked to its function as a TF, and an engineered stable version of Myc that lacks all lysine residues (and cannot be ubiquitylated) is unable to stimulate transcription (51, 52). These data are also consistent with findings that T58A Myc has decreased transactivation potential and fails to induce Bim (44, 46). Fbw7-mediated Myc degradation may thus be a crucial activator of Myc-induced transcription. Because some Myc turnover must be maintained for Myc-induced tumorigenesis, this model may help to explain why heterozygous Fbw7ARG mutation is favored over complete loss of Fbw7, as cells still contain some WT Fbw7 dimers.
While the therapeutic potential of Myc inhibition in cancer is clear, TFs are notoriously difficult pharmacologic targets, and Myc is often considered “undruggable” (9). Some approaches have targeted Myc’s functions as a TF by preventing its association with Max (53). Others have targeted Myc abundance, rather than activity. For example, bromodomain inhibitors that down-regulate Myc transcription are in clinical trials (54–56). Another emerging strategy attempts to prevent Myc stabilization in Ras-associated cancers by inhibiting kinases that phosphorylate and stabilize Myc, including S62 kinases (e.g., ERK5 and CDK9) (41, 57). While the mechanism through which these kinase inhibitors destabilize Myc is thought to involve pS62 and Fbw7 binding, our data question the presumed role of S62 phosphorylation in Myc stabilization and show that this is not likely to involve Fbw7. Moreover, our data suggest that inhibiting S62 phosphorylation could even be detrimental in some cases. A deeper understanding of how phosphorylations control Myc stability will thus help to advance these promising approaches.
MATERIALS AND METHODS
Cell cultures
HEK293, DLD1, HCT116, U2OS, C33A, HeLa, and RPE-1 (Retinal Pigmented Epithelial-1) (hTERT) cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and penicillin-streptomycin in standard incubator conditions (humidified at 37°C with 5% CO2).
Cell line manipulation and generation
All transfections were done in HEK293 cells using calcium phosphate precipitation overnight. Cells were washed and incubated for an additional 24 hours before harvest. For CHX treatments, identically seeded cell dishes were grown to about 75% confluence and incubated in CHX (50 μg/ml) for the indicated time points. Cell treatments with Velcade (500 nM) or MLN4924 (500 nM) were typically done for 4 hours, unless indicated otherwise. DLD1-T244A homozygous knockin cells were generated by adeno-associated virus gene targeting as previously described (58). Homozygous deletions of Fbw7 in DLD1 and HCT116 cells as well as homozygous HCT116-∆D and heterozygous HCT116+/ARG cells were described previously (14, 58, 59).
Plasmids and antibodies
Myc and all mutants thereof are in pCS2+ vector containing a single HA tag at the N terminus. Fbw7 and all mutants thereof are in p3xFLAG-CMV24 (Sigma-Aldrich), except HA-Fbw7, which was cloned into pCS2+ with a single N-terminal HA tag. All mutations were generated by the QuikChange method (Stratagene) and verified by sequencing. Fbw7 ∆F has the entire F-box deleted, while Fbw7 ∆D has a deletion of five critical amino acids in the dimerization domain (14). The following antibodies were used: FLAG: M2 (Sigma-Aldrich), HA: Y11 (Santa Cruz Biotechnology), Myc: D3N8F (Cell Signaling Technology), pT58: EPR17923 (Abcam ab185655), pS62: ERP17924 (Abcam ab185656), pT244: in-house mouse monoclonal, Fbw7: A301-720A and A301-721A (Bethyl Laboratories), proliferating cell nuclear antigen (PCNA): PC-10 (Santa Cruz Biotechnology), and GSK3: 368662 (Calbiochem).
Immunoprecipitation
Cells were harvested on ice in radioimmunoprecipitation assay (RIPA) lysis buffer containing protease and phosphatase inhibitors and sonicated for 10 s, and lysates were cleared by centrifugation. Supernatants were equalized for protein concentration after Bradford analysis and added to protein A/G Sepharose beads precoupled with the respective antibody. Samples were rotated at 4°C for 90 min, and beads were washed and denatured in 2× Laemmli sample buffer by heating for 5 min. Samples were then analyzed by immunoblotting.
Immunoblotting
Cells were lysed on ice in RIPA buffer containing protease and phosphatase inhibitors and sonicated and cleared by centrifugation. Protein concentration was determined by Bradford analysis, and supernatants were equalized. Lysates were then used for immunoprecipitation and/or direct denaturation in 2× or 4× Laemmli sample buffer, followed by 5 min of heating at 95°C. Samples were run on SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gels for separation and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore) by semi-dry electroblotting. Membranes were blocked for 10 min in 5% milk and probed with primary antibodies for 1 hour to overnight. Blots were washed, incubated with horseradish peroxidase–conjugated secondary antibody, washed again, and analyzed by enhanced chemiluminescence either on film (Kodak) or by an imager (ChemiDoc Touch Imaging System). Signal quantifications were performed by digital band analysis using Image Lab software.
In vitro ubiquitylation
Overexpressed or endogenous Myc was immunoprecipitated from cells to obtain phosphorylated Myc species. Washed beads were then subjected to enzymatic reactions containing E1 (50 ng), E2 (hCDC34 and UBCH5, both at 500 ng), Fbw7 copurified with Skp1 (40 ng), neddylated Cul1 copurified with Rbx1 (50 ng), ubiquitin (5 μg), 5 mM adenosine 5′-triphosphate, and 2 mM dithiothreitol (DTT) in buffer containing 50 mM tris (pH 8), 50 mM NaCl, and 5 mM MgCl2. Reactions were incubated at 30°C for the indicated times while shaking in the thermal mixer at 1500 revolutions per minute (Boekel Scientific), terminated with 4× Laemmli sample buffer, heated for 5 min, and analyzed by immunoblotting.
2D phospho-peptide mapping and phospho–amino acid analysis
Phospho-peptide analysis was essentially performed as in (30). Briefly, transfected HEK293 cells were incubated with orthophosphate for 2 hours (1 mCi/ml), washed, and lysed. Cleared supernatants were immunoprecipitated for Myc (with HA antibody) or Fbw7 (with FLAG antibody), separated by SDS-PAGE, transferred to PVDF membranes, and exposed on film. Radioactive markers were used to determine the precise locations of Myc on the membrane, which was then excised and trypsinated overnight. Peptides were separated on thin layer chromatography (TLC) plates by electrophoresis (pH 1.9) and chromatography (isobutyric buffer). Plates were exposed using radioactive markers to precisely locate spots for elution of phosphopeptides for phospho–amino acid analysis by acid hydrolysis and 2D separation by electrophoresis along with a mix of cold phospho–amino acid standards (pS, pT, and pY). Phospho–amino acids were visualized with ninhydrin spray, and TLC plates were exposed on a PhosphorImager.
Protein purification and crystallization
Glutathione S-transferase (GST)–tagged Fbw7 (residues 263 to 707) and a truncated version of Skp1 were coexpressed in BL21 (DE3) Escherichia coli (16). The bacteria were lysed using a microfluidizer in buffer containing 20 mM tris-HCl (pH 8.0), 200 mM NaCl, 5 mM DTT, leupeptin (1 μg/ml), pepstatin (1 μg/ml), and 100 μM phenylmethylsulfonyl fluoride. The Fbw7-Skp1 complex was isolated from the soluble cell lysate with Pierce Glutathione Agarose (Thermo Fisher Scientific). For the peptide competition assay, the complex was further purified by a HiTrap Q HP anion exchange column and a Superdex 200 increase gel filtration column. For crystallization, the GST tag was removed by TEV protease. The complex was further purified with anion exchange and gel filtration chromatography and concentrated to 13 mg/ml. The crystals of the Fbw7-Skp1 complex were grown at 25°C using hanging drop vapor diffusion. One microliter of the Fbw7-Skp1 complex was mixed with 0.5 μl of reservoir solution containing 0.1 M bicine (pH 6.5) and 1.6 M Li2SO4. Four days later, crystals were harvested and soaked with either 1 mM Myc T58 degron peptide or 1 mM Myc T244 degron peptide in cryoprotectant [0.1 M bicine (pH 6.5) and 2.2 M Li2SO4] for 1 hour before flash freezing in liquid nitrogen. The Myc T58 peptide sequence is EDIWKKFELLP(pT)PPL(pS)PSRR. The Myc T244 peptide sequence is PEPLVLHEE(pT)PPT(pT)SSDSEEEQEDEEIDVV.
Data collection and structure determination
X-ray diffraction data were collected at Advanced Light Source Beamline 8.2.1. The dataset was integrated and scaled using the HKL2000 package (60). The structure was solved by molecular replacement and Phaser from the Phenix suite of programs. The models were built and refined with COOT and Phenix.
Quantitative peptide competition assay
Synthetic biotinylated cyclin E peptide was captured by streptavidin-coated AlphaScreen donor beads, while GST-tagged Fbw7 was captured by anti-GST AlphaLISA acceptor beads. The donor and acceptor beads were brought into proximity by the interactions between Fbw7 and cyclin E peptides. Upon excitation at 680 nm, the donor beads generate singlet oxygen, which will interact with the thioxene derivatives on the acceptor beads to emit light at 615 nm. Nonbiotinylated Myc T58 or Myc T244 degron peptides were added in the reaction to compete with the biotinylated cyclin E peptide to bind to Fbw7. The reaction volume is 100 μl with 0.3 nM GST-Fbw7, 3 nM biotinylated cyclin E peptide, donor beads (5 μg/ml), acceptor beads (5 μg/ml), and the indicated concentrations of Myc peptides. The assay buffer is composed of 20 mM tris-HCl (pH 8.0), 200 mM NaCl, 0.02% Tween 20, and bovine serum albumin (50 ng/ml).
Acknowledgments
N.Z. is a Howard Hughes Medical Institute Investigator.
Funding: This work was supported by National Institutes of Health grant R01 CA215647-05 (B.E.C.) and NIH/NCI Cancer Center Support Grant P30 CA015704 (B.E.C.).
Author contributions: Conceptualization: B.E.C., M.W., and N.Z. Methodology: M.W., B.W., and D.-V.R. Investigation: M.W., B.W., D.-V.R., J.S., and Y.H. Visualization: M.W., B.E.C., N.Z., and B.W. Supervision: B.E.C. and N.Z. Writing—original draft: M.W., B.E.C., and N.Z. Validation: M.W., B.E.C., and N.Z.
Competing interests: B.E.C. and N.Z. are equity holders and paid consultants for Coho Therapeutics. B.E.C. receives research funding from Coho Therapeutics that was not used to fund or support any aspect of this work. N.Z. is a member of the scientific advisory boards of Kymera Therapeutics and Seed Therapeutics. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Structural coordinates have been deposited in the Protein Data Bank with accession codes 7T1Z (Myc N-terminal degron in complex with FBW7) and 7T1Y (Myc C-terminal degron in complex with FBW7).
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Table S1
REFERENCES AND NOTES
- 1.Baluapuri A., Wolf E., Eilers M., Target gene-independent functions of MYC oncoproteins. Nat. Rev. Mol. Cell Biol. 21, 255–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dang C. V., MYC on the path to cancer. Cell 149, 22–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabay M., Li Y., Felsher D. W., MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 4, a014241 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conacci-Sorrell M., McFerrin L., Eisenman R. N., An overview of MYC and its interactome. Cold Spring Harb. Perspect. Med. 4, a014357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalkat M., De Melo J., Hickman K. A., Lourenco C., Redel C., Resetca D., Tamachi A., Tu W. B., Penn L. Z., MYC deregulation in primary human cancers. Genes 8, 151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaub F. X., Dhankani V., Berger A. C., Trivedi M., Richardson A. B., Shaw R., Zhao W., Zhang X., Ventura A., Liu Y., Ayer D. E., Hurlin P. J., Cherniack A. D., Eisenman R. N., Bernard B., Grandori C.; Cancer Genome Atlas Network , Pan-cancer alterations of the MYC oncogene and its proximal network across the cancer genome atlas. Cell Syst. 6, 282–300.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen-Petersen B. L., Sears R. C., Mission possible: Advances in MYC therapeutic targeting in cancer. BioDrugs 33, 539–553 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madden S. K., de Araujo A. D., Gerhardt M., Fairlie D. P., Mason J. M., Taking the Myc out of cancer: Toward therapeutic strategies to directly inhibit c-Myc. Mol. Cancer 20, 3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitfield J. R., Beaulieu M. E., Soucek L., Strategies to inhibit Myc and their clinical applicability. Front. Cell Dev. Biol. 5, 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welcker M., Clurman B. E., FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8, 83–93 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Davis R. J., Welcker M., Clurman B. E., Tumor suppression by the Fbw7 ubiquitin ligase: Mechanisms and opportunities. Cancer Cell 26, 455–464 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yumimoto K., Nakayama K. I., Recent insight into the role of FBXW7 as a tumor suppressor. Semin. Cancer Biol. 67, 1–15 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Akhoondi S., Sun D., von der Lehr N., Apostolidou S., Klotz K., Maljukova A., Cepeda D., Fiegl H., Dafou D., Marth C., Mueller-Holzner E., Corcoran M., Dagnell M., Nejad S. Z., Nayer B. N., Zali M. R., Hansson J., Egyhazi S., Petersson F., Sangfelt P., Nordgren H., Grander D., Reed S. I., Widschwendter M., Sangfelt O., Spruck C., FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 67, 9006–9012 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Welcker M., Larimore E. A., Swanger J., Bengoechea-Alonso M. T., Grim J. E., Ericsson J., Zheng N., Clurman B. E., Fbw7 dimerization determines the specificity and robustness of substrate degradation. Genes Dev. 27, 2531–2536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welcker M., Clurman B. E., Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2, 7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hao B., Oehlmann S., Sowa M. E., Harper J. W., Pavletich N. P., Structure of a Fbw7-Skp1-cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26, 131–143 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Tang X., Orlicky S., Lin Z., Willems A., Neculai D., Ceccarelli D., Mercurio F., Shilton B. H., Sicheri F., Tyers M., Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell 129, 1165–1176 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Davis H., Tomlinson I., CDC4/FBXW7 and the “just enough” model of tumourigenesis. J. Pathol. 227, 131–135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yada M., Hatakeyama S., Kamura T., Nishiyama M., Tsunematsu R., Imaki H., Ishida N., Okumura F., Nakayama K., Nakayama K. I., Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 23, 2116–2125 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welcker M., Orian A., Jin J., Grim J. E., Harper J. W., Eisenman R. N., Clurman B. E., The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 9085–9090 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henriksson M., Bakardjiev A., Klein G., Luscher B., Phosphorylation sites mapping in the N-terminal domain of c-myc modulate its transforming potential. Oncogene 8, 3199–3209 (1993). [PubMed] [Google Scholar]
- 22.Pulverer B. J., Fisher C., Vousden K., Littlewood T., Evan G., Woodgett J. R., Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene 9, 59–70 (1994). [PubMed] [Google Scholar]
- 23.Lutterbach B., Hann S. R., Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol. Cell. Biol. 14, 5510–5522 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sears R., Leone G., DeGregori J., Nevins J. R., Ras enhances Myc protein stability. Mol. Cell 3, 169–179 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Sears R., Nuckolls F., Haura E., Taya Y., Tamai K., Nevins J. R., Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14, 2501–2514 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farrell A. S., Sears R. C., MYC degradation. Cold Spring Harb. Perspect. Med. 4, a014365 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayes T. K., Neel N. F., Hu C., Gautam P., Chenard M., Long B., Aziz M., Kassner M., Bryant K. L., Pierobon M., Marayati R., Kher S., George S. D., Xu M., Wang-Gillam A., Samatar A. A., Maitra A., Wennerberg K., Petricoin E. F. III, Yin H. H., Nelkin B., Cox A. D., Yeh J. J., Der C. J., Long-term ERK inhibition in KRAS-mutant pancreatic cancer is associated with MYC degradation and senescence-like growth suppression. Cancer Cell 29, 75–89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeh E., Cunningham M., Arnold H., Chasse D., Monteith T., Ivaldi G., Hahn W. C., Stukenberg P. T., Shenolikar S., Uchida T., Counter C. M., Nevins J. R., Means A. R., Sears R., A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6, 308–318 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Arnold H. K., Sears R. C., Protein phosphatase 2A regulatory subunit B56alpha associates with c-myc and negatively regulates c-myc accumulation. Mol. Cell. Biol. 26, 2832–2844 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welcker M., Singer J., Loeb K. R., Grim J., Bloecher A., Gurien-West M., Clurman B. E., Roberts J. M., Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol. Cell 12, 381–392 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Sundqvist A., Bengoechea-Alonso M. T., Ye X., Lukiyanchuk V., Jin J., Harper J. W., Ericsson J., Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7). Cell Metab. 1, 379–391 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Chakraborty A. A., Scuoppo C., Dey S., Thomas L. R., Lorey S. L., Lowe S. W., Tansey W. P., A common functional consequence of tumor-derived mutations within c-MYC. Oncogene 34, 2406–2409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nash P., Tang X., Orlicky S., Chen Q., Gertler F. B., Mendenhall M. D., Sicheri F., Pawson T., Tyers M., Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414, 514–521 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Salghetti S. E., Kim S. Y., Tansey W. P., Destruction of Myc by ubiquitin-mediated proteolysis: Cancer-associated and transforming mutations stabilize Myc. EMBO J. 18, 717–726 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bahram F., von der Lehr N., Cetinkaya C., Larsson L. G., c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood 95, 2104–2110 (2000). [PubMed] [Google Scholar]
- 36.Gregory M. A., Hann S. R., c-Myc proteolysis by the ubiquitin-proteasome pathway: Stabilization of c-Myc in Burkitt’s lymphoma cells. Mol. Cell. Biol. 20, 2423–2435 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatia K., Huppi K., Spangler G., Siwarski D., Iyer R., Magrath I., Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nat. Genet. 5, 56–61 (1993). [DOI] [PubMed] [Google Scholar]
- 38.Wang X., Cunningham M., Zhang X., Tokarz S., Laraway B., Troxell M., Sears R. C., Phosphorylation regulates c-Myc’s oncogenic activity in the mammary gland. Cancer Res. 71, 925–936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Escamilla-Powers J. R., Sears R. C., A conserved pathway that controls c-Myc protein stability through opposing phosphorylation events occurs in yeast. J. Biol. Chem. 282, 5432–5442 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benassi B., Fanciulli M., Fiorentino F., Porrello A., Chiorino G., Loda M., Zupi G., Biroccio A., c-Myc phosphorylation is required for cellular response to oxidative stress. Mol. Cell 21, 509–519 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Vaseva A. V., Blake D. R., Gilbert T. S. K., Ng S., Hostetter G., Azam S. H., Ozkan-Dagliyan I., Gautam P., Bryant K. L., Pearce K. H., Herring L. E., Han H., Graves L. M., Witkiewicz A. K., Knudsen E. S., Pecot C. V., Rashid N., Houghton P. J., Wennerberg K., Cox A. D., Der C. J., KRAS suppression-induced degradation of MYC is antagonized by a MEK5-ERK5 compensatory mechanism. Cancer Cell 34, 807–822.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su Y., Pelz C., Huang T., Torkenczy K., Wang X., Cherry A., Daniel C. J., Liang J., Nan X., Dai M. S., Adey A., Impey S., Sears R. C., Post-translational modification localizes MYC to the nuclear pore basket to regulate a subset of target genes involved in cellular responses to environmental signals. Genes Dev. 32, 1398–1419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Albert T., Urlbauer B., Kohlhuber F., Hammersen B., Eick D., Ongoing mutations in the N-terminal domain of c-Myc affect transactivation in Burkitt’s lymphoma cell lines. Oncogene 9, 759–763 (1994). [PubMed] [Google Scholar]
- 44.Gupta S., Seth A., Davis R. J., Transactivation of gene expression by Myc is inhibited by mutation at the phosphorylation sites Thr-58 and Ser-62. Proc. Natl. Acad. Sci. U.S.A. 90, 3216–3220 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang D. W., Claassen G. F., Hann S. R., Cole M. D., The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol. Cell. Biol. 20, 4309–4319 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemann M. T., Bric A., Teruya-Feldstein J., Herbst A., Nilsson J. A., Cordon-Cardo C., Cleveland J. L., Tansey W. P., Lowe S. W., Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature 436, 807–811 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.King B., Trimarchi T., Reavie L., Xu L., Mullenders J., Ntziachristos P., Aranda-Orgilles B., Perez-Garcia A., Shi J., Vakoc C., Sandy P., Shen S. S., Ferrando A., Aifantis I., The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 153, 1552–1566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tong K. I., Kobayashi A., Katsuoka F., Yamamoto M., Two-site substrate recognition model for the Keap1-Nrf2 system: A hinge and latch mechanism. Biol. Chem. 387, 1311–1320 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Richards M. W., Burgess S. G., Poon E., Carstensen A., Eilers M., Chesler L., Bayliss R., Structural basis of N-Myc binding by Aurora-A and its destabilization by kinase inhibitors. Proc. Natl. Acad. Sci. U.S.A. 113, 13726–13731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otto T., Horn S., Brockmann M., Eilers U., Schuttrumpf L., Popov N., Kenney A. M., Schulte J. H., Beijersbergen R., Christiansen H., Berwanger B., Eilers M., Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell 15, 67–78 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Jaenicke L. A., von Eyss B., Carstensen A., Wolf E., Xu W., Greifenberg A. K., Geyer M., Eilers M., Popov N., Ubiquitin-dependent turnover of MYC antagonizes MYC/PAF1C complex accumulation to drive transcriptional elongation. Mol. Cell 61, 54–67 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Endres T., Solvie D., Heidelberger J. B., Andrioletti V., Baluapuri A., Ade C. P., Muhar M., Eilers U., Vos S. M., Cramer P., Zuber J., Beli P., Popov N., Wolf E., Gallant P., Eilers M., Ubiquitylation of MYC couples transcription elongation with double-strand break repair at active promoters. Mol. Cell 81, 830–844.e13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han H., Jain A. D., Truica M. I., Izquierdo-Ferrer J., Anker J. F., Lysy B., Sagar V., Luan Y., Chalmers Z. R., Unno K., Mok H., Vatapalli R., Yoo Y. A., Rodriguez Y., Kandela I., Parker J. B., Chakravarti D., Mishra R. K., Schiltz G. E., Abdulkadir S. A., Small-molecule MYC inhibitors suppress tumor growth and enhance immunotherapy. Cancer Cell 36, 483–497.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Letson C., Padron E., Non-canonical transcriptional consequences of BET inhibition in cancer. Pharmacol. Res. 150, 104508 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Bechter O., Schoffski P., Make your best BET: The emerging role of BET inhibitor treatment in malignant tumors. Pharmacol. Ther. 208, 107479 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Delmore J. E., Issa G. C., Lemieux M. E., Rahl P. B., Shi J., Jacobs H. M., Kastritis E., Gilpatrick T., Paranal R. M., Qi J., Chesi M., Schinzel A. C., McKeown M. R., Heffernan T. P., Vakoc C. R., Bergsagel P. L., Ghobrial I. M., Richardson P. G., Young R. A., Hahn W. C., Anderson K. C., Kung A. L., Bradner J. E., Mitsiades C. S., BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blake D. R., Vaseva A. V., Hodge R. G., Kline M. P., Gilbert T. S. K., Tyagi V., Huang D., Whiten G. C., Larson J. E., Wang X., Pearce K. H., Herring L. E., Graves L. M., Frye S. V., Emanuele M. J., Cox A. D., Der C. J., Application of a MYC degradation screen identifies sensitivity to CDK9 inhibitors in KRAS-mutant pancreatic cancer. Sci. Signal. 12, eaav7259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grim J. E., Gustafson M. P., Hirata R. K., Hagar A. C., Swanger J., Welcker M., Hwang H. C., Ericsson J., Russell D. W., Clurman B. E., Isoform- and cell cycle-dependent substrate degradation by the Fbw7 ubiquitin ligase. J. Cell Biol. 181, 913–920 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rajagopalan H., Jallepalli P. V., Rago C., Velculescu V. E., Kinzler K. W., Vogelstein B., Lengauer C., Inactivation of hCDC4 can cause chromosomal instability. Nature 428, 77–81 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Otwinowski Z., Minor W., Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Table S1