Abstract
Background
Breast invasive carcinoma (BRCA) has a poor prognosis. Numerous studies have shown that SET domain bifurcated histone lysine methyltransferase 1 (SETDB1) is involved in the initiation and progression of many cancers. This study aims to reveal the potential mechanism of SETDB1 in the development and progression of BRCA.
Methods
The ONCOMINE database, TIMER database, UALCAN database and GEPIA database were used to analyze the expression of SETDB1 in human cancers. We evaluated the expression level of SETDB1 in cell lines by quantitative real-time polymerase chain reaction (qPCR), and the survival analysis of SETDB1 was performed on PrognoScan and Kaplan-Meier plotter websites. The upstream regulator was obtained from starBase database.
Results
We confirmed that SETDB1 messenger RNA (mRNA) level showed high expression in breast cell lines, and we also found that SETDB1 showed high expression in many types of cancers. Moreover, SETDB1 overexpression was positively correlated with poor prognosis in BRCA. Furthermore, we first predicted miR-29a-3p was a potential upstream regulator of SETDB1 in BRCA. Our findings indicated that SETDB1 might play a carcinogenic role by increasing the infiltration of immune cell and influencing immune checkpoint expression.
Conclusions
This study suggested that miR-29a-3p can mediate the expression of SETDB1 with poor prognosis and tumor immune infiltration in BRCA.
Keywords: Breast invasive carcinoma (BRCA), SET domain bifurcated histone lysine methyltransferase 1 (SETDB1), miR-29a-3p, immune infiltration, prognosis
Introduction
Breast invasive carcinoma (BRCA) is a phenomenon in which breast epithelial cells become uncontrollable under the action of various carcinogens. Breast cancer is a local disease, but it can metastases to lymph nodes and distant organs (1). The incidence of breast cancer in women worldwide is 24.2%, ranking first among women’s cancers, 52.9% of which occur in developing countries (2,3). Many risk factors associated with the occurrence and progression of BRCA have been reported, such as genetic history, overnutrition, and obesity (4). Despite the significant advances in diagnosis, treatment and prognosis, the prognosis for people with BRCA remains unsatisfactory, with 0.5 million deaths worldwide every year (3). Thus, it is urgent to seek effective therapeutic targets for BRCA.
SETDB1 (SET domain bifurcated histone lysine methyltransferase 1) is a protein lysine methyltransferase located on human chromosome 1q21.3 (5). SETDB1 can be expressed in the liver, brain, heart, ovary, and many other tissues. Previous studies showed that SETDB1 acts as a critical factor in the development of primordial germ cells (6,7). Accumulating evidence has shown that SETDB1 is involved in many human cancers (8-10). Loss of SETDB1 histone methyltransferase activity can influence alternative splicing in kidney renal clear cell carcinoma (KIRC) and could potentially target therapy (11). The relationship between SETDB1 and tumor immune invasion in BRCA has rarely been reported.
MicroRNAs (miRNAs) are a major class of small noncoding RNAs that exist widely in eukaryotes. miRNA specifically binds to target messenger RNA (mRNA) to inhibit post-transcriptional gene expression and plays an important role in regulating gene expression, cell cycle, and developmental sequence of organisms. Accumulating evidences has shown that miRNAs regulate tumor progression and metastasis by interacting with target genes in the cells (12-14).
We assessed the expression analysis and the prognostic values of SETDB1 in human cancers. And then, we predicted that miR-29a-3p was associated with the SETDB1. Finally, we analyzed the correlation between SETDB1 expression with immune cell infiltration and immune checkpoints in BRCA. Those findings indicate that miR-29a-3p could mediate the expression of SETDB1 correlates with poor prognosis and immune infiltration in BRCA. We present the following article in accordance with the REMARK checklist (available at https://dx.doi.org/10.21037/tcr-21-1527).
Methods
The expression of SETDB1 in human cancer
The ONCOMINE database (www.oncomine.org) was used to analyze the expression of SETDB1 in various cancer types.
Cell line and RNA extraction
We obtained the human breast cancer cell lines (MDA-MB-231, MCF-7) and a human mammary epithelial cell line (MCF-10A) from ATCC Cell Lines (Manassas, VA, USA). The cells were cultured with DMEM (Meilunbio, Dalian, China) with 10% fetal bovine serum (Gibco, USA) and 1% concentration of penicillin/streptomycin (Meilunbio, Dalian, China) at 37 °C with 5% CO2. Total RNA was obtained from the cell lines using TRIzol reagent (Invitrogen, USA) according to the instructions. The absorbance ratio of A260/A280 and A260/A230 was determined by an ND1000 spectrophotometer for RNA quantification. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
qPCR
qPCR was detected with a CFX96 real-time PCR system (Bio-Rad, USA). One µg RNA of each sample was transcribed into cDNA using HiScript III RT SuperMix for qPCR kit (Vazyme, China) according to the kit instruction after RNA extraction. According to the manufacturer’s protocols, the qPCR was using ChamQ Universal SYBR qPCR Master Mix (Vazyme, China). The expression level of SETDB1 mRNA was calculated using the 2−ΔΔCt method and normalized to GAPDH. The primer sequences of SETDB1 were as follows: forward primer: 5'-GACTCTCTGAGACAACTTCCAAGGA-3' and reverse primer: 5'-CAGGGATTGAGGGAGGAACA-3'. The primer sequences of GAPDH were as follows: forward primer: 5'-TGCACCACCAACTGCTTAGC-3' and reverse primer: 5'-GGCATGCACTGTGGTCATGAG-3'.
Survival analysis
We evaluated the relationship between SETDB1 expression and survival in different cancer types with PrognoScan (http://dna00.bio.kyutech.ac.jp/PrognoScan/index.html) and Kaplan-Meier plotter (http://www.kmplot.com/analysis/) (15,16). The relationship between SETDB1 expression level and prognosis was searched in PrognoScan, including overall survival (OS), disease-free survival (DFS), disease-specific survival (DSS), event-free survival (EFS) and relapse-free survival (RFS). We analyzed the correlation of SETDB1 expression between OS and RFS in pan-cancer using a Kaplan-Meier plotter.
Candidate miRNA prediction
starBase (http://starbase.sysu.edu.cn/) is a platform to perform the interaction of RNA and explore survival and differential expression analysis (17). We predicted the correlation analysis of candidate miRNA and SETDB1 and performed the expression of miR-29a-3p in BRCA.
GEPIA database analysis
GEPIA is a research database for analyzing cancer and routine based on TCGA and GTEx data (http://gepia.cancer-pku.cn/) (18). We used GEPIA to determine the expression of SETDB1 with immune checkpoints in BRCA.
TIMER database analysis
TIMER (Tumor Immune Estimation Resource) (https://cistrome.shinyapps.io/timer/) is an online research tool for tumor immunology (19). We used TIMER to assess the correlation of SETDB1 expression between the level of immune cell infiltration and immune checkpoints expression in BRCA.
UALCAN database analysis
UALCAN is an online website for analyzing cancer data (http://ualcan.path.uab.edu/index.html) (20). We used UALCAN to assess the gene and protein level of SETDB1 in BRCA.
Statistical analysis
The results are shown as mean ± SEM. T-test was used to analyze the differences between the two groups. The threshold was set as P value <0.05 and log-rank P value <0.05.
Results
The expression of SETDB1 in human cancer and cell lines
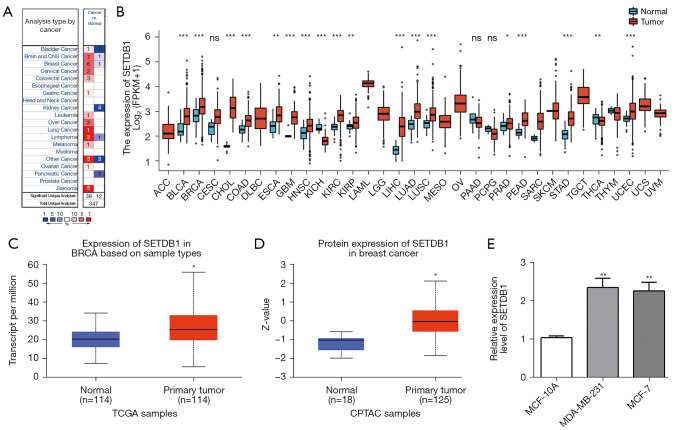
We analyzed the expression of SETDB1 in human cancer from the ONCOMINE database. SETDB1 was significantly upregulated in the brain, breast, cervical, colorectal, gastric, leukemia, liver, lung, lymphoma, myeloma, and sarcoma. Meanwhile, lower expression of SETDB1 was found in the kidney and pancreatic (Figure 1A).
Figure 1.
The expression of SETDB1 in human cancers and cell lines. (A) The expression of SETDB1 in different cancers based on the ONCMINE database; (B) SETDB1 expression level in different cancer types in TIMER; (C) SETDB1 expression level in BRCA in the UALCAN database; (D) the protein expression of SETDB1 in BRCA in the UALCAN database; (E) the expression of SETDB1 level in MDA-MB-231, MCF-7, and MCF-10A by using qPCR. *, P<0.05; **, P<0.01; ***, P<0.001. SETDB1, SET domain bifurcated histone lysine methyltransferase 1; ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous carcinoma; KICH, kidney Chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
Next, we evaluate SETDB1 expression in pan-cancer using the TIMER database. As shown in Figure 1B, SETDB1 expression in BLCA, BRCA, CHOL, COAD, ESCA, GBM, HNSC, KIRC, KIRP, LIHC, LUAD, LUSC, PRAD, READ, STAD, THCA, and UCEC was increased, and we found SETDB1 was only downregulated in KICH.
Similarly, the gene level and protein level of SETDN1 were also higher than the normal tissues (Figure 1C,1D). We also assessed the expression of SETDB1 in cell lines. As shown in Figure 1E, the relative expression of SETDB1 was higher in breast cancer cell lines than in normal cells.
The prognostic values of SETDB1 in human cancer
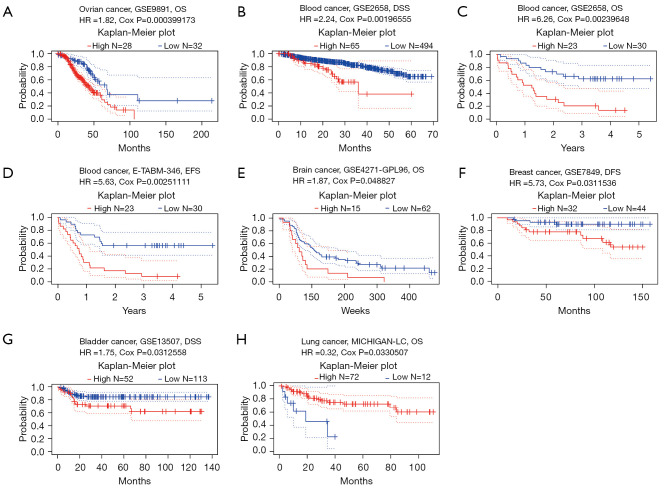
We assessed the prognostic value of SETDB1 for human cancer in PrognoScan, as shown in Figure 2. SETDB1 played a detrimental role in ovarian cancer (OS: HR =1.82, Cox P=0.000399173), blood cancer (DSS: HR =2.24, Cox P=0.00196555; OS: HR =6.26, Cox P=0.00239648; EFS: HR =5.63, Cox P=0.00251111), brain cancer (OS: HR =1.87, Cox P=0.048827), breast cancer (DFS: HR =5.73, Cox P=0.311536) and lung cancer (OS: HR =0.32, Cox P=0.0330507). However, the SETDB1 have a protective role in bladder cancer (DSS: HR =1.75, Cox P=0.0312558).
Figure 2.
Survival analysis of SETDB1 in different cancer types in PrognoScan. (A) OS (n=278) in ovarian cancer cohort GSE9891; (B-D) DSS (n=559), OS (n=53) EFS (n=53) and in blood cancer cohort GSE2658 and E-TABM-346; (E) OS (n=77) in brain cancer cohort GSE4271-GPL96; (F) DFS (n=76) in breast cancer cohort GSE7849; (G) DSS (n=165) in bladder cancer cohort GSE13507; (H) OS (n=86) in lung cancer cohort MICHIGAN-LC. SETDB1, SET domain bifurcated histone lysine methyltransferase 1; EFS, event-free survival; DSS, disease-specific survival; DFS, disease-free survival; OS, overall survival.
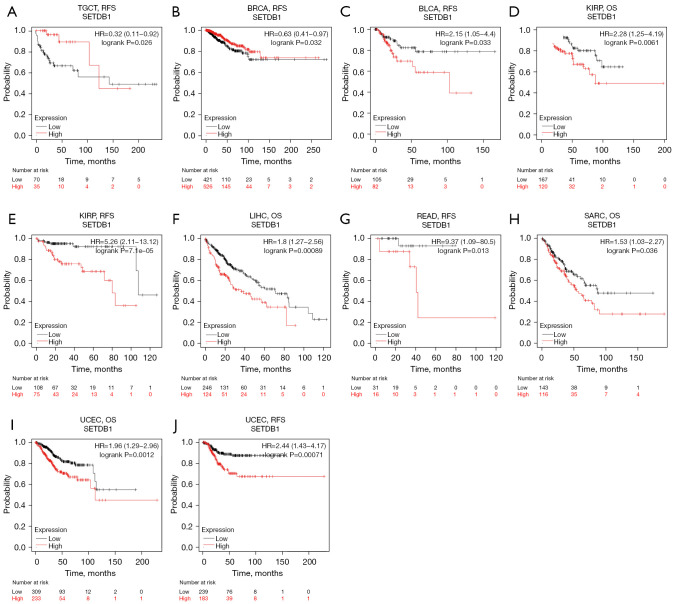
We then further evaluated SETDB1-related survival (OS and RFS) from the Kaplan-Meier plotter (Figure 3). SETDB1 was found to have detrimental prognostic factors in BLCA (RFS: HR =2.15, 95% CI: 1.05 to 4.4, longrank P=0.033), ESCA (OS: HR =0.64, 95% CI: 0.48 to 0.85, logrank P=0.029), KIRP (OS: HR =2.28, 95% CI: 1.25 to 4.19, logrank P=0.0061; RFS: HR =5.26, 95% CI: 2.11 to 13.12, logrank P=0.000071), LIHC (OS: HR =1.8, 95% CI: 1.27 to 2.56, logrank P=0.00089), READ (RFS: HR =9.37, 95% CI: 1.09 to 80.5, logrank P=0.013), SARC (OS: HR =1.53, 95% CI: 1.03 to 2.27, logrank P=0.036), UCEC (OS: HR =1.96, 95% CI: 1.29 to 2.96, logrank P=0.0012; RFS: HR =2.44, 95% CI: 1.43 to 4.17, logrank P=0.00071). Meanwhile, we found that SETDB1 has a protective factor in TGCT (RFS: HR =0.32, 95% CI: 0.11 to 0.92, logrank P=0.026) and BRCA (RFS: HR =0.63, 95% CI: 0.41 to 0.97, logrank P=0.032).
Figure 3.
Survival analysis of SETDB1 in different cancers in Kaplan-Meier plotter. RFS of (A) TGCT, (B) BRCA, (C) BLCA, (E) KIRP, (G) READ and (J) UCEC. OS of (D) KIRP, (F) LIHC, (H) SARC and (I) UCEC. SETDB1, SET domain bifurcated histone lysine methyltransferase 1; RFS, relapse-free survival; OS, overall survival; TGCT, testicular germ cell tumors; BRCA, breast invasive carcinoma; BLCA, bladder urothelial carcinoma; KIRP, kidney renal papillary cell carcinoma; READ, rectum adenocarcinoma; UCEC, uterine corpus endometrial carcinoma; LIHC, liver hepatocellular carcinoma; SARC, sarcoma.
By combining the survival significance analysis of the two databases, we found that the expression of SETDB1 was associated with poor BRCA prognosis, and SETDB1 may be a biomarker for poor BRCA prognosis.
Prediction and analysis of upstream miRNAs of SETDB1
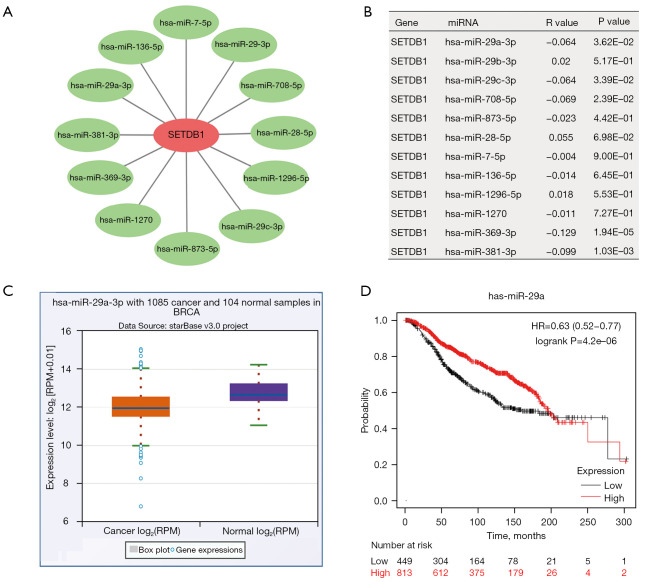
There are many kinds of gene expression regulation. Non-coding RNA plays a vital role in the regulation of gene expression. We used the software to predict the upstream miRNA regulating SETDB1 and obtained 12 candidate miRNAs (P<0.05). We then used Cytoscape software to establish a network of miRNA-related SETDB1 (Figure 4A,4B). miRNA has a negative correlation with the target gene. Among these miRNAs, the expression and prognostic of miR-29a-3p and miR-381-3p in BRCA were determined. However, miR-381-3p has been reported in a previous study. Interestingly, we found that miR-29a-3p expression was down in BRCA, and its high expression was positively correlated to prognosis (Figure 4C,4D). Those findings may suggest that miR-29a-3p may be a new potential regulator of SETDB1 in BRCA.
Figure 4.
miR-29a-3p as a potential upstream regulator of SETDB1 in BRCA. (A) Network diagram of miRNAs and SETDB1; (B) the correlation of miRNAs and SETDB1 in BRCA; (C) the expression of miR-29a-3p in BRCA. P<0.05; (D) the prognostic value of miR-29a in BRCA. SETDB1, SET domain bifurcated histone lysine methyltransferase 1; BRCA, breast invasive carcinoma.
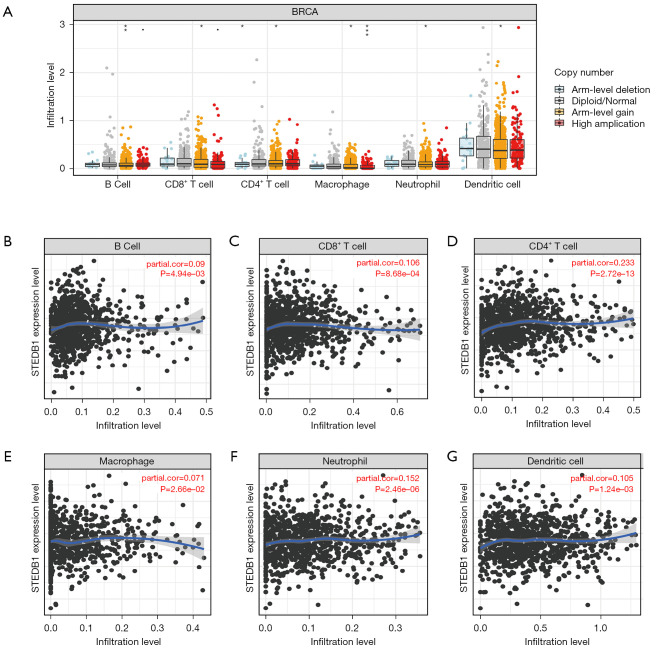
Expression correlation of SETDB1 and immune cell infiltration in BRCA
Immune cell infiltration in the tumor microenvironment affects the prognosis of tumor therapy. To explore SETDB1 expression in immune cell infiltration, we analyzed the relationships between SETDB1 and infiltrating immune cells by using TIMER and the GEPIA database. As shown in Figure 5A, we found significant changes in immune cell infiltration levels with different copy numbers of SETDB1 in BRCA. We then analyzed the relationship between SETDB1 expression and the level of immune cell infiltration. SETDB1 expression was significantly positively correlated with infiltrating levels of B cell (r=0.09, P=4.94e-03), CD8+ T cell (r=0.106, P=8.68e-04), CD4+ T cells (r=0.233, P=2.72e-13), macrophage (r=0.071, P=2.66e-02), neutrophil (r=0.152, P=2.46e-06), and dendritic cell (r=0.105, P=1.24e-03) in BRCA (Figure 5B-5G).
Figure 5.
The correlation of immune cell infiltration with SETDB1 expression in BRCA. (A) Infiltration level of different immune cells under different copy numbers of SETDB1 in BRCA. The relationship of SETDB1 expression level with B cell (B), CD8+ T cell (C), CD4+ T cell (D), macrophage (E), neutrophil (F), or dendritic cell (G) infiltration level in BRCA. *, P<0.05; **, P<0.01; ***, P<0.001. SETDB1, SET domain bifurcated histone lysine methyltransferase 1; BRCA, breast invasive carcinoma.
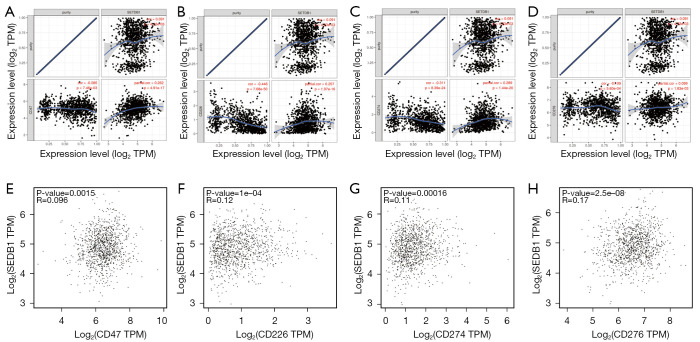
Correlation between SETDB1 and immune checkpoints in BRCA
The abnormal expression and function of immune checkpoint molecules are one of the crucial causes of many diseases. We assessed the relationship of SETDB1 with immune checkpoints in BRCA. As shown in Figure 6A-6D, SETDB1 expression was significantly positively correlated with CD47 (r=0.262, P=4.91e-17), CD226 (r=0.257, P=1.97e-16), CD274 (r=0.289, P=1.44e-20) and CD276 (r=0.099, P=1.83e-03) in BRCA. Moreover, we found a positive correlation of SETDB1 with CD47 (R=0.096, P=0.0015), CD226 (R=0.12, P=1e-04), CD274 (R=0.11, P=1.6e-04) and CD276 (R=0.17, P=2.5e-08) in BRCA according to the GEPIA database (Figure 6E-6H).
Figure 6.
Correlation of SETDB1 expression level with CD47, CD226, CD274, and CD276 in BRCA. Spearman correlation of SETDB1 with expression of (A) CD47, (B) CD226, (C) CD274, and (D) CD276 in BRCA by using TIMER. The correlation of SETDB1 expression with (E) CD47, (F) CD226, (G) CD274, and (H) CD276 in BRCA determined by the GEPIA database. SETDB1, SET domain bifurcated histone lysine methyltransferase 1; BRCA, breast invasive carcinoma.
Conclusions
Currently, BRCA is still the first hazard to women’s health. Clarifying the molecular mechanism of breast cancer cells might provide critical ways for finding effective therapeutic targets for prognosis. Histone lysine methylation can activate or inhibit gene transcription and is related to tumor transformation (21). SETDB1 is a member of the H3K9 methyltransferase. SETDB1 has played a role as an oncogene in many human cancers (22-24). Fei et al. (25) found SETDB1 was overexpressed in LIHC, and a previous study reported that silencing SETDB1 expression inhibited the growth of tumor cells (26).
In the current study, we performed the expression analysis of SETDB1 in human cancers using the TIMER and ONCOMINE databases collected from TCGA data. Survival analysis for SETDB1 indicated that BRCA patients with high expression of SETDB1 had a poor prognosis.
Noncoding RNAs (ncRNAs) are RNAs transcribed from the genome. Although the majority of ncRNAs do not encode any proteins, but they play a vital role in regulating gene expression (27,28). Numerous studies have found that ncRNAs are associated with the emergence and development of many diseases (29-31). We used the starBase database to predict potential miRNAs that could bind to SETDB1. Finally, we obtained 12 miRNAs that have the potential to regulate the expression of SETDB1. However, among those miRNAs, we found 10 miRNAs have significant changes (P<0.05), and then, using the starBase database, we found only miR-29a-3p and miR-381-3p were found downregulated in BRCA. Nevertheless, the previous study has shown that miR-381-3p/SETDB1 axis played a critical role in the metastasis of BRCA (32). In this study, we found that miR-29a-3p also has the potential to bind SETDB1, and its expression was correlated with the BRCA patent prognosis.
Immune cells play an essential role in the human body, including lymphocytes, T cells, macrophages, neutrophils and dendritic cells. Some studies have shown that the infiltration of immune cells in tumors is correlated to the metastasis, treatment, and prognosis of tumors (33,34). In this study, we found that SETDB1 was positively related to many immune cells in BRCA, including macrophage, B cell, CD8+ T cell, neutrophil, CD4+ T cell and dendritic cells. We also found that high expression of SETDB1 was associated with immune checkpoints in BRCA, such as CD47, CD226, CD274 and CD276. These findings demonstrated that tumor immune infiltration might account for SETDB1-mediated BRCA, and targeting SETDB1 might increase the efficacy of immunotherapy in BRCA.
Our studies showed that SETDB1 was highly expressed in BRCA and breast cancer cell lines and positively correlated with poor prognosis in BRCA. Furthermore, we first predicted miR-29a-3p was a potential upstream regulator of SETDB1 in BRCA. Our findings also suggested that SETDB1 may play a carcinogenic role by increasing tumor immune cell infiltration and influencing immune checkpoint expression. Finally, miR-29a-3p/SETDB1 may be a novel therapeutic target for the treatment of BRCA. However, further research is needed to confirm this novel finding.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81800264).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-1527
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-1527). The authors have no conflicts of interest to declare.
References
- 1.Gradishar WJ, Anderson BO, Abraham J, et al. Breast Cancer, Version 3.2020, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2020;18:452-78. 10.6004/jnccn.2020.0016 [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Siegel RL, Ward EM, et al. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol Biomarkers Prev 2016;25:16-27. 10.1158/1055-9965.EPI-15-0578 [DOI] [PubMed] [Google Scholar]
- 4.Ginsburg O, Bray F, Coleman MP, et al. The global burden of women's cancers: a grand challenge in global health. Lancet 2017;389:847-60. 10.1016/S0140-6736(16)31392-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukuda K, Shinkai Y. SETDB1-Mediated Silencing of Retroelements. Viruses 2020;12:596. 10.3390/v12060596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackburn ML, Chansky HA, Zielinska-Kwiatkowska A, et al. Genomic structure and expression of the mouse ESET gene encoding an ERG-associated histone methyltransferase with a SET domain. Biochim Biophys Acta 2003;1629:8-14. 10.1016/S0167-4781(03)00155-6 [DOI] [PubMed] [Google Scholar]
- 7.Liu S, Brind'Amour J, Karimi MM, et al. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev 2014;28:2041-55. 10.1101/gad.244848.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong CM, Wei L, Law CT, et al. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology 2016;63:474-87. 10.1002/hep.28304 [DOI] [PubMed] [Google Scholar]
- 9.Wu PC, Lu JW, Yang JY, et al. H3K9 histone methyltransferase, KMT1E/SETDB1, cooperates with the SMAD2/3 pathway to suppress lung cancer metastasis. Cancer Res 2014;74:7333-43. 10.1158/0008-5472.CAN-13-3572 [DOI] [PubMed] [Google Scholar]
- 10.Kostaki M, Manona AD, Stavraka I, et al. High-frequency p16(INK) (4A) promoter methylation is associated with histone methyltransferase SETDB1 expression in sporadic cutaneous melanoma. Exp Dermatol 2014;23:332-8. 10.1111/exd.12398 [DOI] [PubMed] [Google Scholar]
- 11.Ho TH, Park IY, Zhao H, et al. High-resolution profiling of histone h3 lysine 36 trimethylation in metastatic renal cell carcinoma. Oncogene 2016;35:1565-74. 10.1038/onc.2015.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 13.Qadir MI, Faheem A. miRNA: A Diagnostic and Therapeutic Tool for Pancreatic Cancer. Crit Rev Eukaryot Gene Expr 2017;27:197-204. 10.1615/CritRevEukaryotGeneExpr.2017019494 [DOI] [PubMed] [Google Scholar]
- 14.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol 2009;4:199-227. 10.1146/annurev.pathol.4.110807.092222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizuno H, Kitada K, Nakai K, et al. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics 2009;2:18. 10.1186/1755-8794-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lou W, Chen J, Ding B, et al. Identification of invasion-metastasis-associated microRNAs in hepatocellular carcinoma based on bioinformatic analysis and experimental validation. J Transl Med 2018;16:266. 10.1186/s12967-018-1639-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howe FS, Fischl H, Murray SC, et al. Is H3K4me3 instructive for transcription activation? Bioessays 2017;39:1-12. 10.1002/bies.201600095 [DOI] [PubMed] [Google Scholar]
- 22.Spyropoulou A, Gargalionis A, Dalagiorgou G, et al. Role of histone lysine methyltransferases SUV39H1 and SETDB1 in gliomagenesis: modulation of cell proliferation, migration, and colony formation. Neuromolecular Med 2014;16:70-82. 10.1007/s12017-013-8254-x [DOI] [PubMed] [Google Scholar]
- 23.Kim HA, Koo BK, Cho JH, et al. Notch1 counteracts WNT/β-catenin signaling through chromatin modification in colorectal cancer. J Clin Invest 2012;122:3248-59. 10.1172/JCI61216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Cai K, Wang J, et al. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells 2014;32:2858-68. 10.1002/stem.1795 [DOI] [PubMed] [Google Scholar]
- 25.Fei Q, Shang K, Zhang J, et al. Histone methyltransferase SETDB1 regulates liver cancer cell growth through methylation of p53. Nat Commun 2015;6:8651. 10.1038/ncomms9651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez-Paredes M, Martinez de Paz A, Simó-Riudalbas L, et al. Gene amplification of the histone methyltransferase SETDB1 contributes to human lung tumorigenesis. Oncogene 2014;33:2807-13. 10.1038/onc.2013.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J, Chen X, Shan C, et al. Autophagy in cardiovascular diseases: role of noncoding RNAs. Mol Ther Nucleic Acids 2021;23:101-18. 10.1016/j.omtn.2020.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet 2006;15 Spec No 1:R17-29. 10.1093/hmg/ddl046 [DOI] [PubMed] [Google Scholar]
- 29.Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int J Mol Sci 2018;19:1310. 10.3390/ijms19051310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brandenburger T, Salgado Somoza A, Devaux Y, et al. Noncoding RNAs in acute kidney injury. Kidney Int 2018;94:870-81. 10.1016/j.kint.2018.06.033 [DOI] [PubMed] [Google Scholar]
- 31.Jaé N, Dimmeler S. Noncoding RNAs in Vascular Diseases. Circ Res 2020;126:1127-45. 10.1161/CIRCRESAHA.119.315938 [DOI] [PubMed] [Google Scholar]
- 32.Wu M, Fan B, Guo Q, et al. Knockdown of SETDB1 inhibits breast cancer progression by miR-381-3p-related regulation. Biol Res 2018;51:39. 10.1186/s40659-018-0189-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokratous G, Polyzoidis S, Ashkan K. Immune infiltration of tumor microenvironment following immunotherapy for glioblastoma multiforme. Hum Vaccin Immunother 2017;13:2575-82. 10.1080/21645515.2017.1303582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Willigen WW, Bloemendal M, Gerritsen WR, et al. Dendritic Cell Cancer Therapy: Vaccinating the Right Patient at the Right Time. Front Immunol 2018;9:2265. 10.3389/fimmu.2018.02265 [DOI] [PMC free article] [PubMed] [Google Scholar]