Abstract
Although COVID-19 emerged as a major concern to public health around the world, no licensed medication has been found as of yet to efficiently stop the virus spread and treat the infection. The SARS-CoV-2 entry into the host cell is driven by the direct interaction of the S1 domain with the ACE-2 receptor followed by conformational changes in the S2 domain, as a result of which fusion peptide is inserted into the target cell membrane, and the fusion process is mediated by the specific interactions between the heptad repeats 1 and 2 (HR1 and HR2) that form the six-helical bundle. Since blocking this interaction between HRs stops virus fusion and prevents its subsequent replication, the HRs inhibitors can be used as anti-COVID drugs. The initial drug selection is based on existing molecular databases to screen for molecules that may have a therapeutic effect on coronavirus. Based on these premises, we chose two approved drugs to investigate their interactions with the HRs (based on docking methods). To this end, molecular dynamics simulations and molecular docking were carried out to investigate the changes in the structure of the SARS-CoV-2 spike protein. Our results revealed, cefpiramide has the highest affinity to S protein, thereby revealing its potential to become an anti-COVID-19 clinical medicine. Therefore, this study offers new ways to re-use existing drugs to combat SARS-CoV-2 infection.
Keywords: Coronavirus, SARS-CoV-2, COVID-19, Spike protein, Cefpiramide, Conivaptan, Inhibitor, Molecular dynamic simulation
1. Introduction
Coronaviruses (CoVs) are important zoonotic pathogens, which are well-known for their capability to infect many mammalian species [1,2]. CoVs often have effective strategies for transmission and immune evasion, particularly if epidemic and pandemic occur within dense populations of humans [3,4]. CoVs are enveloped viruses with RNA genomes [5,6]. They are classified into four genera, α-, β-, γ-, and δ-coronaviruses [7]. Among them, β-CoVs are responsible for previous Severe Acute Respiratory Syndrome (SARS) and Middle Eastern Respiratory Syndrome (MERS) epidemics [8] that were localized and have similar characteristics. The newly discovered 2019-nCoV (also known as SARS-CoV-2) is a β-CoV [9] is responsible for the newest CoV pandemic, which is mysterious, widespread, and dangerous. It has started in December 2019 in Wuhan, China and spread rapidly in more than two hundred countries, infecting almost 35 million people and causing more than a million deaths.
The entry of all CoVs into the human host cells is mediated by the homotrimeric spike proteins (S proteins or SPs) that protrude from the viral envelope forming a crown-like halo that surrounds the viral particle and that defines the name of these viruses. Sequence of the SARS-CoV-2 S protein contains 1273 residues and includes a signal peptide (residues 1–13), the S1 subunit (residues 14–685 residues), and the SP2 subunit (residues 686–1273). The S1 and S2 subunits, which are responsible for receptor binding and membrane fusion, respectively [10], are further subdivided into several functional domains and regions. In the S1 subunit, one can find an N-terminal domain (residues 14–305) and a receptor-binding domain (RBD, residues 319–541); whereas the SP2 subunit includes the fusion peptide (FP) (residues 788–806), heptapeptide repeat sequence 1 (HR1, residues 912–984), HR2 (residues 1163–1213), single-pass transmembrane (TM) domain (residues 1213–1237), and a short cytoplasm domain (residues 1237–1273). An important feature of the S2 subunit is the presence of two repeat domains, HR1 and HR2 that include a repetitive heptapeptide: HPPHCPC, where H, P, and C correspond to a hydrophobic, polar and charge residue, respectively [11], and which interact with each other, forming a six-helical bundle (6-HB), causing virus fusion to host cell [12].
The capability of virus binding to host cell receptors is determined by the spike protein. It is well documented that SARS-CoV-2 binds to the angiotensin-converting enzyme-2 (ACE-2) receptor located on the surface of some human cells [13]. Blocking the spike protein binding to human cell receptors and inhibiting the subsequent virus fusion are among the effective therapeutic strategies for fighting this deadly virus. These are also the antiviral effects of the drugs proposed in this study. Since there are no specific drugs or strategies for the SARS-CoV-2 infection treatment, finding means to efficiently control COVID-19 represents an important scientific task that drives efforts of many scientists around the globe. For example, Wu et al. examined the effects of multiple drugs on the intracellular phase of the virus and on the interactions of SARS-CoV-2 proteins with the intracellular proteins and constructed a database of anti-viral drugs for SARS-CoV-2 [14]. In the same line, we investigated the effects of drugs on the extracellular phase of the viral life and on the SARS-CoV-2 S2. Functions of many proteins encoded by the CoV genome and associated with SARS-CoV-2 pathology remain a mystery. We are presenting here a computational analysis of one of these mysterious proteins, spike protein, which in addition to playing a role in the host cell entry might function as a potential modulator of host immunity to delay or attenuate the immune response against the viruses.
In this study, we investigated the structure of the spike protein of SARS-CoV-2 after binding to various drugs (Table S1) and identified drugs that potentially interact with spike protein with high-affinity. We further discussed potential candidates for the treatment of SARS-CoV-2 infection and COVID-19, the changes they induce in the spike structure, and strategies to inhibit virus entry. In this study, we investigated the structural changes induced in the fusion core of the S protein by all the drugs listed in Table S1, and the best drug is suggested for the potential utilization in the treatment of COVID-19.
2. Materials and methods
2.1. Virtual selection of ligand for binding to SARS-CoV-2 spike protein
With comprehensive studies, the most efficient compounds were selected as virtual ligands. The approved drugs were selected. Various software was used to investigate the changes in S2 conformation. The chemical structure of selected drugs were downloaded from the chem-spider (http://www.chemspider.com) site. The three-dimensional structure of the 6-HB post-fusion core containing HR1 and HR2 domains in the SARS-CoV-2 S protein S2 subunit (PDB ID 6LXT) was obtained from Protein Data Bank (PDB) [15].
2.2. Methods
2.2.1. Molecular docking
The molecular docking assays for the Spike protein in the presence of all drugs was carried out using Auto dock 4.2.6 software. The structure of the S2 and all drugs were obtained from PDB (RCSB), chem-spider, and drug bank site, respectively. They were saved in PDB format after optimizing by chimera 1.13.1 software [16]. For determining the best drugs that bind to Spike protein, for molecular dynamic simulation study, the initial docking was performed. All receptor and ligand files were prepared in PDBQT format using Auto-Dock Tools. All possible binding sites were explored through blind docking as there was no assumption regarding the relative affinity of the receptors various binding sites. Blind docking was done with a grid box that covering all dimensions of the receptor. A standard docking protocol with the addition of Gasteiger atomic charges and assignment of default atom-types was implemented. In the molecular docking assays, the best conformation for all drugs were selected. After computing the binding free energy and binding mode between all drugs and spike protein-2 using Auto-Dock 4.2.1 suites, the two lowest docking energy of all drugs were analyzed in MD simulation. The grid box size for the both Conivaptan and Cefpiramide-spike protein-2 complex were set 80 Å × 80 Å × 80 Å. The required hydrogen atoms were attached to the spike protein-2. The grid spacing was 1 Å. For determining the type of interaction, R-studio discovery (version V 16.1) and chimera software were applied. The choice of the best binding mode in each binding site was based on the higher affinity of the obtained structures, and then the molecular dynamic simulation was performed.
2.2.2. Molecular dynamic simulation
The GROMACS 2018.1 package conducted 30-ns MD simulation on the elected complex of protein-drugs obtained from docking studies [17]. Molecular dynamics simulations analysis of spike protein-2 and both conivaptan and cefpiramide were performed. The G43a1 force-field parameters were utilized for spike protein, counter ions, and water. The G43a1force field, with the intermolecular potential denoted as a sum of Lennard-Jones (LJ) force and pairwise Coulomb interaction, was applied to provide the topology and interaction parameters. The prodrug server was employed as an automated interface to allocate the parameters [18]. The two complexes were immersed in a defined triclinic box of the SPC water model with 10 Å padding from the walls [19]. The box of complex (2.4 × 2.4 × 6.82 Å3) was filled with the appropriate amount of water molecules and five Na+ for neutralization. The 9054 and 5765 water molecules with the SPC model were added to the protein-conivaptan and protein-cefpiramide boxes, respectively. The system was minimized energy with a maximum of 100,000 steps by the steepest descent method to remove inappropriate contacts. Succeeding, the position restrained simulation with confining the position of heavy atoms of the system, at 300 K has been performed for 500 ps (NVT condition). Then, another 10000-ps simulation has been performed to equilibrate the system at 1 bar pressure while keeping the temperature of the system constant at 300 K (NPT condition). Also, the leap-frog algorithm with a time step of 1 fs was utilized to support the periodic boundary condition and integration of motion equations. The Berendsen thermostat and the Parrinello–Rahman barostat were applied under NVT and NPT conditions, respectively. Then were used to analyze the GROMACS simulation results [20].
3. Results and discussion
3.1. SARS-CoV-2 molecular docking studies
In this study, we utilzied in silico methods, such as molecular docking and Molecular Dynamic (MD) simulation to work with SARS-CoV-2 spike protein, which is located on the surface of the virus and is responsible for the two processes (receptor binding fusion to the host cell) assocate with the viral cell entry. It is likely to be cleaved into S1 and S2 subuits after interaction with the host receptor. Molecular docking represents a broadly used approah to determine the binding site of the selected drugs and was successfully used to characterize the interaction between drug candidates and the spike protein of the coronavirus [21]. In this study, we selected two drugs, conivaptan and cefpiramide, from the list of the 80 approved drugs in Table S1, based on their lowest binding energy to the SARS-CoV-2 S2 protein as evaluated by the Auto Dock 4.2.6 software. In docking experiments, conivaptan and cefpiramide were efficiently bound to the heptapeptide repeat domain of the S2 protein leading to the potential inhibition of the SARS-CoV-2 activation. The conivaptan hydroxyl and amine groups form two hydrogen bonds to the Gln A:920 and Asp A:1192 of S2 (Fig. 1 ). Also, there are six Van der Waals interactions between the conivaptan rings and Glu A:1202, Gln A:913, Gln A:1201, Leu A:1197, Ser A:1196, and Phe A:927 of the S2 protein. conivaptan is an inhibitor that bind to the top of the S2 at the position of the HR2 domain (Fig. 2 ). For the conivaptan-spike protein complex, the value of binding Gibbs Free Energy (∆G˚) has been measured about -6.64 kJ/mol (Table 1 ). The negative free energy values showed that the formation of the conivaptan-spike protein complex was spontaneous. The docking results indicated that conivaptan could bind to S2 with high affinity. This binding caused conformational alterations in the S2 protein which potentially can lead the inhibition of the fusion of SARS-CoV-2 with human cell.
Fig. 1.
Two-dimensional diagram of interaction of conivaptan with SARS-CoV-2 spike protein-2.
Fig. 2.
conivaptan docked in the binding pocket of S2 protein of SARS-CoV-2 using AutoDock software. The image was made using a chimera.
Table 1.
Results of the conivaptan docking to the SARS-CoV-2 S2 protein.
| Lowest Binding energy (kcal/mol) | Inhibition Constant (298.15K), µM | Final intermolecular energy (kcal/mol) | VdW+H-bond + dissolve energy (kcal/mol) | Electrostatic Energy (kcal/mol) | Final total internal energy (kcal/mol) |
|---|---|---|---|---|---|
| -6.64 | 13.57 | -7.83 | -7.77 | -0.07 | -0.6 |
Among the 80 drugs anayzed in our study, cefpiramide was ranked second (after conivaptan) based on the significant binding affinity (- 6.04 kcal/mol) to S2 and the presece of several binding modes. Many S2 residues can be involved in cefpiramide-S2 interactions and several pockets capable of interaction with the cefpiramide were found in the HR1 domain. Based on the consensus binding affinity and meaningful interactions with the significant S2 residues, cefpiramide is being recommended as another possible binding partner of the S2 protein. The chosen bound conformation of cefpiramide with S2 is represented in Figs. 3 and 4 , and in Tables 1 and 2 . Many notable interactions, such as hydrogen bonds (dark green), Van der Waals interactions (light green), and alkyl interactions (purple) can be found between the surrounding S2 residues and cefpiramide (see details in Fig. 3). This analysis revealed that the cefpiramide occupies the internal binding pockets (HR1 domain) of S2, shows significant binding affinity and forms several close interactions with the residues Ser A:937, Lys A:933, Ala A:930, Ser A:929, Gln A:926, and Phe A:927. Based on these data it seems that both conivaptan and cefpiramide are capable of inhibiting the coronavirus binding to the host cell.
Fig. 3.
Two-dimensional diagram of cefpiramide interactions with SARS-CoV-2 spike protein-2.
Fig. 4.
cefpiramide docked in the binding pocket of S2 protein of SARS-CoV-2 using AutoDock software. The image was made using a chimera.
Table 2.
Results of the cefpiramide docking to the SARS-CoV-2 spike protein-2.
| Lowest Binding energy (kcal/mol) | Inhibition Constant (298.15K), µM | Final Intermolecular energy (kcal/mol) | vdW+H-bond + dissolve energy (kcal/mol) | Electrostatic Energy (kcal/mol) | Final Total Internal Energy (kcal/mol) |
|---|---|---|---|---|---|
| -6.04 | 37.35 | -8.13 | -6.34 | -1.79 | -1.05 |
3.2. Molecular dynamic (MD) simulations of SARS-CoV-2
The study of the structural properties, flexibility, compaction, and conformational changes of the SARS-CoV-2 S2 subunit in the absence and presence of the approved drugs, conivaptan and cefpiramide, were surveyed using Molecular Dynamic (MD) simulations. These target drugs were selected from the set of 80 approved drugs based on the results of their molecular docking to the SARS-CoV-2 protein S2, and the best-docked complexes were taken for the MD simulations.
3.2.1. Analysis of the SARS-CoV-2 spike protein-2 conformation
In the MD simulations, evaluation of the Root Mean Square Deviations (RMSDs) is the most accurate method for investigation of the average distances between the backbone atoms of proteins and a ligand or other proteins [22]. Also, the RMSD can be used for monitoring the equilibrium processes of the system and the stability of protein structure upon the binding of a small ligand [23]. In general, RMSD in MD simulation represents a means to assess conformational deviation from the initial protein structure [24]. Furthermore, the RMSD could be used to compare the stability of protein structure considering the absence and presence of a ligand. As illustrated by Fig. 5 , the RMSD values of protein and its complexes experienced increase during 1-29 ns of simulation and subsequently reached the plateau, indicating that the systems achieved the steady state. The SARS-CoV-2 S2 alone and, S2-conivaptan and S2-cefpiramide complexes reached their equilibrium confirmations after 29, 80, and 80 ns, respectively. According to the estimated RMSD values, binding of conivaptan and cefpiramide led to the higher fluctuations in the SARS-CoV-2 S2 protein. It is well documented that the decreased values of the protein RMSD upon complex formation represent stabilization to the target structure [25]. Since the S2 RMSD values were increased in the presence of the conivaptan and cefpiramide, we can suggest that the resulting protein-drug complexes had higher structural flexibility and lower conformational stability than the protein alone. This indicated binding-induced destabilization of the protein structure reflected in the noticeable deviations in the residues positions from their states in the unbound protein.
Fig. 5.
Time dependence of rmsd. Cα rmsd values for unliganded SARS-CoV-2 spike protein-2 and SARS-CoV-2 spike protein-2-complex during 10000 ps MD simulation.
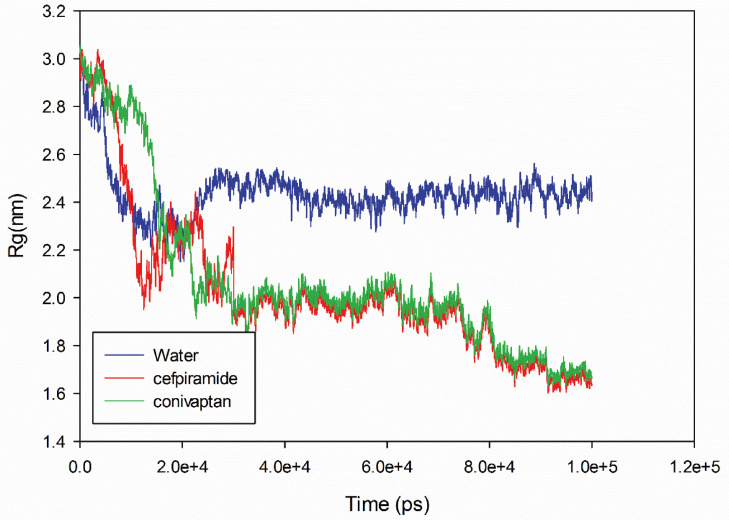
When a biomolecule, such as protein, undergoes changes in compaction, these changes can be estimated. To gain the corresponding information on the structural compactness of protein conformation during MD simulation, the protein gyration radius (RG) was investigated. In general, the radius of gyration of a molecule is the root-mean-square distance of all atoms from its center of gravity [26]. Therefore, radius of gyration provides a means to estimate the correlations between protein dimension, protein compaction, and protein folding [27]. Fig. 6 shows that the time course of changes in the RG value of the SARS-CoV-2 S2 protein and its complexes with conivaptan and cefpiramide show a certain downward trend. It is also seen that binding of cefpiramide caused larger changes in compaction than binding of conivaptan. obviously, the RG changes in S2 complexed with conivaptan and cefpiramide were easily distinguishable from the RG changes in S2 alone. The changes in cefpiramide are larger than conivaptan. This might be because of the differences between the conivaptan and cefpiramide binding to S2, where conivaptan binds to the interior of the HR2 helix and fusion core region, whereas cefpiramide interacts with the HR1 head region. There is a strong interaction between HR1 and HR2 domains within the fusion core region. conivaptan binds to the fusion core region disrupting its structure. cefpiramide binds to HR1 domain and affects the interaction between the two HR domains.
Fig. 6.
Time evolution of the radius of gyration (Rg) during 10000 ps of MD simulation of unliganded SARS-CoV-2 spike protein-2 and SARS-CoV-2 spike protein-2-complex.
3.2.2. Flexibility analysis of the SARS-CoV-2 spike protein-2
When a dynamic system fluctuates in some portions, the extent of fluctuations can be computed. One of the widely used methods to study the protein fluctuations is the roots mean square fluctuation (RMSF), which is a key measure of the flexibility of the residue over the whole simulation time [28]. The greater the amino acid RMSF value, the more flexibility it shows in the process of binding. Therefore, the regions, where conivaptan and cefpiramide have largest impacts on protein residues, were evaluated by RMSF analysis (Fig. 7 ). This analysis revealed that binding of both conivaptan and cefpiramide caused the increase in the average RMSF values of S2 reflecting the increase in the local flexibility of the protein.
Fig. 7.
The RMSF values of unliganded SARS-CoV-2 spike protein-2 and SARS-CoV-2 spike protein-2-complex were plotted against residue numbers.
Based on these results, we conclude that the structural dynamics and flexibility of S2 protein increases in the presence of these drugs. Furthermore, in the presence of cefpiramide, S2 protein was more flexible, which could be related to the mode of the drug coordination. The higher backbone RMSF values with higher standard deviation (Fig. 6 and Table 3 ) represent higher thermal instability, a conclusion supported by subsequent thermodynamics analysis (see below).
Table 3.
The average and standard deviations of RMSD and RMSF of SARS-CoV-2 spike protein-2 with conivaptan and cefpiramide.
| complex | RMSF (nm) | RMSD (nm) | RG (nm) |
|---|---|---|---|
| SARS-CoV-2 spike protein | 0.41 ± 0.17 | 1.10 ± 0.05 | 2.43 ± 0.04 |
| Conivaptan-spike protein complex | 0.42 ± 0.21 | 2.16 ± 0.08 | 1.72 ± 0.02 |
| Cefpiramide-spike protein complex | 0.57 ± 0.27 | 2.14 ± 0.08 | 1.69 ± 0.02 |
3.2.3. Effects of drug binding on the secondary structure of SARS-CoV-2 spike protein-2
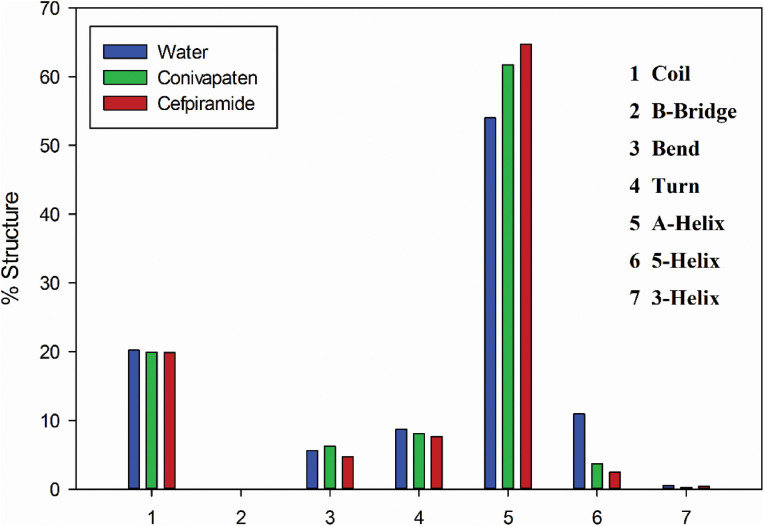
During the simulation process, the conformational changes in the SARS-CoV-2 spike protein-2 can be further clarified by calculating the secondary structure content in this protein alone and in it complexes with conivaptan and cefpiramide. As shown in Fig. 8 and Table 4 , the structure of the SARS-CoV-2 protein S2 was characterized by the presence of 62.6%, 23.4%, 10.12%, and 0% of the α-helical, random coil, β-turn, and β-sheet structure, respectively. Therefore, the dominant type of secondary structure in the SARS-CoV-2 spike protein-2 is α-helix. This analysis also showed that the α-helical content of the S2 protein increased to 74.93% and 71.58% as a result of protein interaction with conivaptan and Cefpiramide, respectively. The random coil content decreased slightly from 23.01% and 23.08 %, respectively, whereas the β-turn and β-sheet contents were stable at around 0%, which indicated that conivaptan and cefpiramide binding influenced the secondary structure of the SARS-CoV-2 spike protein-2. Hence, the drug binding-induced increase in the α-helical structure of S2 was accompanied by some decrease in its random coil and β-turn content. However, these changes in the secondary structure of SARS-CoV-2 spike protein-2 during its binding of conivaptan and cefpiramide were not considerable, which is consistent with the results of RG, RMSD, and H-bond analyses. On the other hand, although the drug-induced changes in the secondary structure content of the protein were rather minor, they do not exclude a possibility that the conformational changes induced by cefpiramide and conivaptan in the S2 protein can cause alterations in the SARS-CoV-2 binding to the human cells.
Fig. 8.
Secondary structure content of the free SARS-CoV-2 spike protein-2 and S2 in the complexes with cefpiramide and conivaptan.
Table 4.
Secondary structure content of spike protein-2 in various forms.
| Spike Protein-2 | A-Helix (%) | 5-Helix | 3-Helix | β-Bridge (%) | β-Turn (%) | Random Coil (%) | Bend (%) |
|---|---|---|---|---|---|---|---|
| S2 alone | 62.06 | 12.69 | 0.61 | 0.010 | 10.12 | 23.44 | 6.49 |
| S2-cefpiramid complex | 71.58 | 4.3 | 0.31 | 0.080 | 9.36 | 23.08 | 7.26 |
| S2-conivaptan complex | 74.92 | 2.87 | 0.51 | 0.110 | 8.86 | 23.01 | 5.44 |
3.2.3. Solvent accessible surface area and H-bonding analyses of SARS-CoV-2 protein S2
Next, changes in the Solvent Accessible Surface Area (SASA) upon binding of drugs to the SARS-CoV-2 spike protein-2 were analyzed, and the corresponding results are presented in Fig. 9 . The unbound form of the S2 protein was characterized by the SASA of 66.59 nm2, whereas SASA decreased slightly after the protein complexation with drugs. Combined with the RG data, these observations suggested some drug-induced compaction of S2 protein. We speculate that the attraction between the hydrophilic residues of the S2 protein and drugs resulted in the observed SASA changes.
Fig. 9.
Solvent Accessible Surface Area of the free SARS-CoV-2 spike protein-2 and S2 in the complexes with cefpiramide and conivaptan.
Since hydrophobic interactions and hydrogen bonds (H-bonds) are important for protein structure, the H-bonding of three structures were analyzed, and the corresponding results are reported in Table 5 . The binding of cefpiramide and conivaptan resulted in some decrease in the number of the internal H-bonds in S2 protein (Table 5), indicating that the conformation of SARS-CoV-2 spike protein-2 changed after drugs binding.
Table 5.
The average and standard deviations of SASA and H-bonds of SARS-CoV-2 spike protein-2 with conivaptan and cefpiramide.
| Complex | SASA | Internal hydrogen bonds |
|---|---|---|
| CoV-2 spike protein-2 | 64.71 ± 0.97 | 91.13 ± 5.05 |
| Conivaptan-spike protein-2 complex | 66.07 ± 1.06 | 89.32 ± 4.94 |
| Cefpiramide-spike protein-2 complex | 66.37 ± 1.06 | 90.32 ± 4.9 |
3.2.4. Evaluation of the accessible surface areas of all the residues in the SARS-CoV-2 protein S2 in the presence of conivaptan and cefpiramide
The accessible surface area (ASA) is the surface area of a biomolecule that is accessible to a solvent [29] that can be applied to forecast the secondary structure of protein [30,31]. According to the results of the analysis of ASA of all the S2 amino acids (see Table S2), binding of conivaptan and cefpiramide drugs causes considerable changes in ASA of several residues due to the hydrogen bond with the drugs. Also, computed ASA of some residue indicated that the ASA of conivaptan and cefpiramide become smaller after interaction S2 with conivaptan and cefpiramide.
3.2.5. Complexation energy analysis of SARS-CoV-2 spike protein-2
Proteins are stable due to cooperative intramolecular forces that maintain the native state. Transitions to non-native states are prevented by free energy barriers that typically cannot be overcome by the natural conformational fluctuations that occur in free proteins. Interaction of a protein interacts with the ligand can increase these structural fluctuations. In the cases where the magnitude of the fluctuations is sufficiently high, the extra kinetic energy imparted on the molecule becomes great enough for free energy barriers to be overcome, allowing the transitions to non-native conformations. At this point, the protein experience conformational alterations and starts to unfold. Base on the information shown in Table 6 , cefpiramide has strong interaction with spike protein-2 and therefore can be considered as a potential medicine for COVID-19. Approximately 300 K temperature controlled systems. The temperature of the box was kept constant by the thermostat Nose-Hoover.
Table 6.
The average and standard deviations of temperature (K) of kinetic (EKCMT) and potential (EPTOT) and total (ETOT) energies (kJ/mol).
| complex | Temperature (K) | EKCMT (kJ/mol) | EPTOT (kJ/mol) | ETOT (kJ/mol) |
|---|---|---|---|---|
| SARS-CoV-2 spike protein | 299.7 ± 2.9 | 69767 ± 674 | -385390 ± 5890 | -315623 ± 5812 |
| Conivaptan-spike protein complex | 300.2 ± 2.9 | 71892 ± 699 | -395118 ± 5456 | -323225 ± 5547 |
| Cefpiramide-spike protein complex | 300.1 ± 2.4 | 71874 ± 699 | -323242 ± 5546 | -323242 ± 5546 |
The S2 subunits of SARS-CoV-2 and SARS-CoV are highly conserved according to the sequence alignment, with the 92.6% and 100% sequence identity in the HR1 and HR2 domains, respectively. Based on this reason, HR2 was selected for this investigation. The S1 section of the spike protein directly interacts with the receptor on the cell surface, whereas the fusion process is mediated by heptad repeats (HRs). The Receptor Binding Domain (RBD) of S1 binds to ACE2 receptor in human cell surface. HR2 and HR1 interact with each other in an antiparallel manner. Affecting this interaction is able to stop virus fusion and prevent virus replication thereafter. Therefore, it is expected that after cefpiramide and conivaptan binding to HR domains, virus fusion would be altered that would prevent virus replication thereafter. The interaction between HR1 and HR2 shows that there were several hydrogen bonds between the HR1 amino acids Asn925, Gln935, Gln949, Asn953, and Asn960 and the HR2 amino acids Ala1174, Val1177, Ile1179, Gln1180, Ala1190, Asn1194, and Ile1198 [32]. After cefpiramide and conivaptan binding, some of the hydrogen bonds between HR1 and HR2 are destroyed. In this way, these drugs may inhibit the virus fusion to ACE2 receptors.
3.2.6. MM-PBSA calculations
The Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA), developed by Koll-man and Case, is a method for precisely determining the extent of ligand-protein interaction [33,34].To the calculation of binding free energy of docked system (drugs-spike protein complex) by MMPBSA, the molecular mechanics energies (EMM) and continuum solvent Poisson–Boltzmann (PB) model for polar salvation and solvent-accessible surface area (SA) for nonpolar salvation were considered. The following equation was used to calculate the system's overall free energy [35]:
The snapshots were drawn from each 10000 Ps of MD tracks then the average was taken for the analysis of the binding-free energy. The binding-free energies of Conivaptan and Cefpiramide-with spike protein were computed and depicted in Table 7 .
Table 7.
The binding-free energies of Conivaptan and Cefpiramide with spike proteina.
| MMPBSA | ΔEVdW (kJ mol–1) | ΔEElec (kJ mol–1) | ΔGPB (kJ mol–1) |
|---|---|---|---|
| Water | -1306.472626 ± 115.5779565 | -13873.70166 ± 616.9440591 | -25561.7 ± 335.575 |
| Conivaptan | -1276.04 ± 112.3876 | -13099 ± 772.1717 | -26461.3 ± 501.8864 |
| Cefpiramide | -1266.04 ± 102.3876 | -12089 ± 672.0317 | -23481.3 ± 401.7864 |
ΔEVdW:Van der Walls interactions, ΔEElec: electrostatic energy, and ΔGPB: polar binding.
4. Conclusions
The fusion process between the SARS-CoV-2 and host cells is mediated by the heptad repeats (HRs) located within the S2 subunit of the SARS-CoV-2 spike protein. Altering HR interaction can stop virus fusion and prevent virus replication. Since it has been reported that the HR inhibitors can be used as anti-COVID drugs, we selected two approved drugs to investigate their binding to HRs. The results of this study provide a suitable opportunity for a clinical assessment of these drugs. If the clinical results are consistent with the results of this article, these two drugs may be involved in the treatment of the pandemic severe acute respiratory syndrome coronavirus 2. In this study, molecular docking and molecular dynamics (MD) simulations were carried out to investigate the changes in the SARS-CoV-2 protein S structure. Through these MD simulations and molecular docking studies, we explain how the structure and conformation of the SARS-CoV-2 Spike protein-2 were affected by conivaptan and cefpiramide. The conivaptan binding site was located at the top of S2 crowns at the position of HR2 according to the molecular docking results. This drug is anticipated to bind to the Gln920 and Asp1192 amino acids of the spike protein, which play a key role in ACE-2 attachment. The cefipiramid docking results revealed that the binding site was located on the Ser937, Lys933, Ala930, Ser929, Gln926, and Phe927 on the HR1 domain. Drug binding caused a conformational hindrance. Furthermore, the RMSD of the S2 protein in the presence of conivaptan and cefpiramide was increased and protein structure was destabilized. In addition to the fact that cefpiramide had larger number of hydrogen bonds with S2, its binding energy to S2 compared to conivaptan was slightly lower. Therefore, cefpiramide is expected to have higher binding affinity to the S2 and show a stronger inhibitory effect on the virus fusion. Our results suggest that cefpiramide has the potential to become an anti-COVID-19 clinical medicine. Our findings could provide new target drugs for more comprehensive in vitro and in vivo studies, as well as define new strategies for the treatment of SARS-CoV-2 infection. Furthermore, it offers new ways to re-use existing drugs to fight with the SARS-CoV-2 infection.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2022.132488.
Appendix. Supplementary materials
References
- 1.Forni D., Cagliani R., Clerici M., Sironi M. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye Z.W., Yuan S., Yuen K.S., Fung S.Y., Chan C.P., Jin D.Y. Int. J. Biol. Sci. 2020;16:1686. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y., Peng F., Wang R., Guan K., Jiang T., Xu G., Sun J., Chang C. J. Autoimmun. 2020 [Google Scholar]
- 4.Bowdle A., Munoz-Price L.S. Anesthesiol. J. Am. Soc. Anesthesiol. 2020;132:1292–1295. doi: 10.1097/ALN.0000000000003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pillay T.S. J. Clin. Pathol. 2020 [Google Scholar]
- 6.Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceraolo C., Giorgi F.M. J. Med. Virol. 2020;92:522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y., Guo Y., Pan Y., Zhao Z.J. Biochem. Biophys. Res. Commun. 2020 [Google Scholar]
- 10.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chambers P., Pringle C.R., Easton A.J. J. Gen. Virol. 1990;71(Pt 12):3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 12.Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S. Cell. Mol. Immunol. 2020:1–3. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker H., Parkkila S. BioRxiv. 2020 [Google Scholar]
- 14.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X. Acta Pharma. Sin. B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canrong W., Yang Y., Yang L., Zhang P., Wang Y., Wang Q. ChinArxiv. 2020 [Internet] [Google Scholar]
- 16.Scudellari M. Nature. 2020;581:252–255. doi: 10.1038/d41586-020-01444-z. [DOI] [PubMed] [Google Scholar]
- 17.Al-Khafaji K., AL-DuhaidahawiL D., Tok T.T. J. Biomol. Struct. Dyn. 2020:1–11. [Google Scholar]
- 18.A. Parvez, S. Yadav, V. Pandey, X. Qi, (2020).
- 19.Zhang X., Yan J., Wang H., Wang Y., Wang J., Zhao D. J. Biomol. Struct. Dyn. 2020:1–14. [Google Scholar]
- 20.Nath P., Sharma K., Kumar K., Goyal A. Int. J. Biol. Macromol. 2020;148:364–377. doi: 10.1016/j.ijbiomac.2020.01.116. [DOI] [PubMed] [Google Scholar]
- 21.Yu R., Chen L., Lan R., Shen R., Li P. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.S. Raschka, in, 2014.
- 23.Li D., Zhang Y., Zhao R.N., Fan S., Han J.G. J. Mol. Model. 2014;20:2122. doi: 10.1007/s00894-014-2122-y. [DOI] [PubMed] [Google Scholar]
- 24.Monhemi H., Housaindokht M.R., Nakhaei Pour A. J. Phys. Chem. B. 2015;119:10406–10416. doi: 10.1021/acs.jpcb.5b03970. [DOI] [PubMed] [Google Scholar]
- 25.Sadeghi-Kaji S., Shareghi B., Saboury A.A., Farhadian S. J. Biomol. Struct. Dyn. 2019 doi: 10.1080/07391102.2019.1568306. [DOI] [PubMed] [Google Scholar]
- 26.O. Kratky, P. Laggner, (2003).
- 27.Mendichi R., Šoltés L., Giacometti Schieroni A. Biomacromolecules. 2003;4:1805–1810. doi: 10.1021/bm0342178. [DOI] [PubMed] [Google Scholar]
- 28.Ceruso M.A., Amadei A., Nola A.D. Protein Sci. 1999;8:147–160. doi: 10.1110/ps.8.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee B., Richards F. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 30.Momen-Roknabadi A., Sadeghi M., Pezeshk H., Marashi S.A. BMC Bioinf. 2008;9:357. doi: 10.1186/1471-2105-9-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adamczak R., Porollo A., Meller J. Proteins Struct. Funct. Bioinf. 2005;59:467–475. doi: 10.1002/prot.20441. [DOI] [PubMed] [Google Scholar]
- 32.Sun H., Li Y., Liu P., Qiao C., Wang X., Wu L., Liu K., Hu Y., Su C., Tan S. Emerg. Microb. Infect. 2020:1–19. doi: 10.1080/22221751.2020.1770631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Shabib N.A., Khan J.M., Malik A., Alsenaidy M.A., Rehman M.T., AlAjmi M.F., Alsenaidy A.M., Husain F.M., Khan R.H. J. Mol. Liq. 2018;269:511–520. [Google Scholar]
- 34.Mohseni S.S., Nasri F., Davari K., Mirzaie S., Moradzadegan A., Abdi F., Farzaneh F. Comput. Biol. Chem. 2018;77:279–290. doi: 10.1016/j.compbiolchem.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Alavi F.S., Ghadari R., Zahedi M. J. Iran. Chem. Soc. 2017;14:19–35. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.