Abstract
Background
Gastric cancer (GC) is one of the most common malignant diseases worldwide, the incidence and mortality for GC is still high, thus it is urgently important to identify the effective and reliable biomarkers to evaluate GC and the underlying molecular events.
Methods
The study integrated four Gene Expression Omnibus (GEO) profile datasets and The Cancer Genome Atlas (TCGA) dataset to screen differentially expressed genes (DEGs), screened key genes by performing the Kaplan-Meier analysis, univariate and multivariate-cox analysis. Further analysis were performed to evaluate and validate the prognostic value of the key genes based on TCGA database and online websites. In addition, mechanism analysis of the key genes was performed thought biological processes and KEGG pathway analysis.
Results
In the study, 192 DEGs (92 up-regulated and 100 down-regulated) were identified from the GEO and TCGA datasets. Next, gene ontology (GO) for DEGs focused primarily on cell adhesion, extracellular region and extracellular matrix structural constituent. Then four significant key genes were screened by performed the Kaplan-Meier analysis, univariate and multivariate-cox analysis. By using Kaplan-Meier plotter and OncoLnc, the expression level was associated with a worse prognosis. In addition, the area under curve (AUC) for time-dependent receiver operating characteristic (ROC) indicated a moderate diagnostic value. Furthermore, the expression of collagen triple helix repeat containing 1 (CTHRC1), serpin family E member 1 (SERPINE1), Versican (VCAN) was associated with tumor size, Uroplakin 1B (UPK1B) expression was associated with distant metastasis. Finally, multiple biological processes and signaling pathway associated with key genes revealed the underlying mechanism in GC.
Conclusions
Taken together, CTHRC1, SERPINE1, VCAN, UPK1B were novel potential prognostic molecular markers for GC, which acted as oncogene to promote the development of GC.
Keywords: Gastric cancer (GC), data mining, prognostic markers
Introduction
The incidence and mortality for gastric cancer (GC) have been appreciably declining for several decades. However, GC is still the fourth most common cancer and the second leading cause of cancer deaths worldwide (1-3). In China alone, there were about 679/100,000 of new GC cases and 798/100,000 of death GC cases and accounting for the third of malignant tumor incidence and mortality in 2015 (4). The pathogenesis of GC is multifactorial, including genetic susceptibility and environmental factors, cell cycle, DNA repair, metabolism, cell-to-cell and cell-to-matrix interactions, apoptosis, angiogenesis, and immune surveillance contribute to cancer development (5). However, although there have been extensive previous studies on the molecular mechanism of GC formation and progression, the molecular mechanism of GC is not yet clear. Due to high morbidity and mortality in GC, it is urgently important to reveal the causes and the underlying molecular mechanisms. Thus, identifying novel diagnostic and prognostic biomarkers remains critical importance for stomach cancer.
In this work, we have downloaded four original microarray datasets GSE79973 (6), GSE26899 (7), GSE54129 (8), GSE63089 (9), from NCBI-Gene Expression Omnibus database (NCBI-GEO), there are total of 262 GC cases and 88 normal cases available. differentially expressed genes (DEGs) between cancer tissues and normal tissues were obtained from GEO and TCGA gene expression profile, respectively. Then, we overlapped the four GEO and TCGA gene expression profiles and identified 204 overlapped genes, DAVID was used to perform GO enrichment analysis and KEGG enrichment analysis on the overlapped genes. Next, some analysis has been performed to screen the key genes, including: the Kaplan-Meier analysis, univariate and multivariate-cox analysis for overall survival (OS). In addition, to evaluate and validate the prognostic value of the key genes, we performed the correlation analysis between TMN and expression of key genes based on TCGA data, the Kaplan-Meier analysis based on the online website including Kaplan-Meier plotter and OncoLnc, ROC analysis for OS and DFS, univariate and multivariate-cox analysis for DFS. Furthermore, the co-expressed genes associated with GC were identified by using Coexpedia, the biological processes and KEGG-signaling pathway were predicted via using R software. Finally, Gene set enrichment analysis (GSEA) was performed to further investigate pathways of four key genes that may be associated with GC.
Methods
Identification and processing of microarray data
We used the “GC” OR “gastric carcinoma” keyword to search gene expression profiles from GEO database (http://www.ncbi.nlm.nih.gov/geo/), and four qualified gene expression profiles (GSE54129, GSE79973, GSE63089, GSE26899) were identified with platform and series matrix file(s) being downloaded as TXT files, type of data were RMA signal intensity and standardized, and log2 transformed. The dataset information was presented in Table 1.
Table 1. Details for GEO gastric cancer data.
| Reference | Sample | GEO | Platform | Normal | Tumor |
|---|---|---|---|---|---|
| He et al. | Gastric cancer | GSE79973 | GPL570 | 10 | 10 |
| Oh et al. | Gastric cancer | GSE63089 | GPL5175 | 45 | 45 |
| Hippo et al. | Gastric cancer | GSE54129 | GPL570 | 21 | 111 |
| Siegel et al. | Gastric cancer | GSE26899 | GPL6947 | 12 | 96 |
GEO, Gene Expression Omnibus.
Identification of DEGs and overlapped genes
R annotation package was performed to convert the probe into gene symbol. Next, SVA package was used for background correction, merge package was applied to combine the four gene expression data according to the gene symbol. Then, gene differential expression analysis between normal cases and tumor cases was performed by using limma package in the Bioconductor package from GEO and TCGA gene expression profile, with corrected P value <0.05 and absolute log fold change (FC) >1 being considered as the cutoff criterion. Finally, overlapped genes were identified from four GEO and TCGA gene expression profiles.
Overlapped genes enrichment analysis
The DAVID database (https://david.ncifcrf.gov/) is an essential foundation for the success of any high-throughput gene function analysis. We used DAVID to perform GO annotations analysis on overlapped genes.
Identification and validation of clinically relevant hub genes
The Kaplan-Meier analysis was performed to screen the survival-related genes, univariate and multivariate-cox analysis for OS was conducted to identify the key genes from the survival-related genes. To evaluate and validate the prognostic value of the key genes, we performed the Kaplan-Meier analysis based on the online website including Kaplan-Meier plotter (http://kmplot.com/) and OncoLnc (http://www.oncolnc.org/). The Kaplan-Meier analysis for disease free survival (DFS) based on TCGA dataset, univariate and multivariate-cox analysis for DFS by mining TCGA dataset, the receiver operating characteristic (ROC) analysis for OS and DFS, the correlation analysis between TMN and expression of key genes based on TCGA data. The gene expression level ≤ median was regarded as low expression, otherwise was regarded as high expression.
Biological processes and signaling pathway analysis for the co-expressed genes associated with GC
To explore the potential mechanisms for the key genes, we identified the co-expressed genes associated with key genes by using Coexpedia (http://www.coexpedia.org/), biological processes and KEGG-signaling pathway for the co-expressed genes associated with GC were predicted by R software.
Gene set enrichment analysis
To further investigate pathways of four key genes that may be associated with GC, GSEA was performed using the JAVA program (http://www.broadinstitute.org/gsea) with TCGA dataset. Expression of each key gene was set to annotate phenotypes, 1,000 times were performed for gene set permutations. The nominal P value <0.05 was used to sort the pathways enriched in each phenotype.
Results
The DEG of GEO gene expression profiles
We performed background correction on the GEO expression profiles. The result was shown in Figure 1. Then, we analyzed the DEGs of integrated GEO and TCGA gene expression profiles by using the limma package (FDR <0.05, absolute log FC >1), 219 up-regulated genes and 179 down-regulated genes were obtained from GSE26899, GSE54129, GSE63089 and GSE79973, 1,110 up-regulated genes and 1566 down-regulated genes were obtained from TCGA dataset. After using Venny, 92 up-regulated genes and 100 down-regulated genes were overlapped across four GEO and TCGA datasets (Figure 2).
Figure 1.
Standardization of gene expression. (A) The standardization of GSE26899 data, (B) the standardization of GSE54129 data, and (C) the standardization of GSE63089 data. (D) the standardization of GSE79973 data. The green bar represents the data before normalization, and the red bar represents the normalized data.
Figure 2.
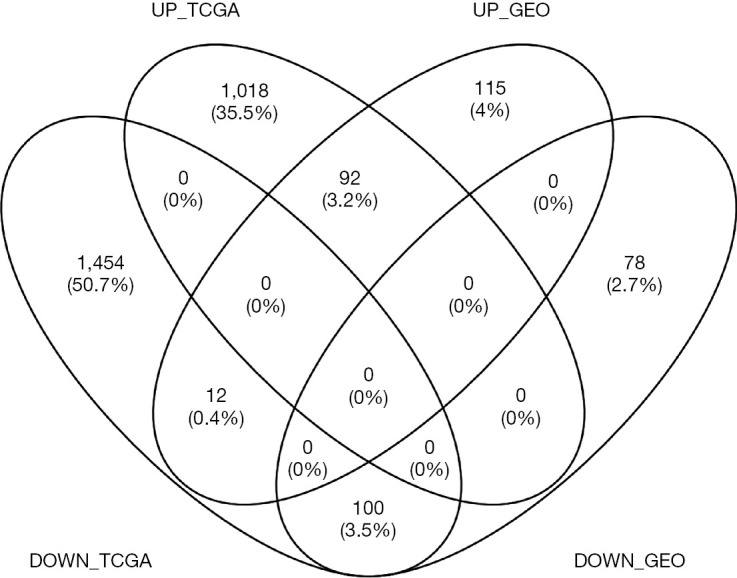
Venn plot of the DEGs between the integrated four GEO datasets and the TCGA dataset. DEGs, differentially expressed genes; GEO, Gene Expression Omnibus; TCGA, The Cancer Genome Atlas.
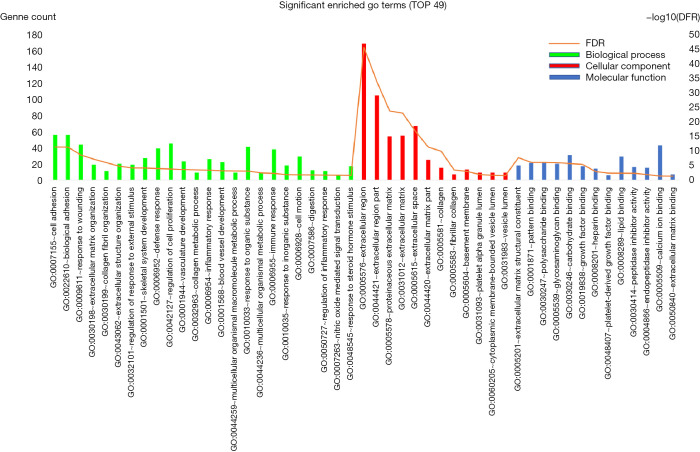
GO and KEGG enrichment analysis
Enrichment analysis of the overlapped genes was performed using the DAVID online site (corrected P value<0.05). The enrichment analysis was divided into three functional groups, including biological processes, cell composition and molecular function, biological processes. In the biological processes group, the differential genes were mainly enriched in cell adhesion and biological adhesion. In the cell composition, the differential genes were mainly enriched in the extracellular region and the extracellular region part. In the molecular function, the differential genes were mainly enriched in the extracellular matrix structural constituent and pattern binding (Figure 3).
Figure 3.
GO enrichment analysis of overlapped genes into three functional groups: molecular function, biological processes, and cell composition. GO, gene ontology.
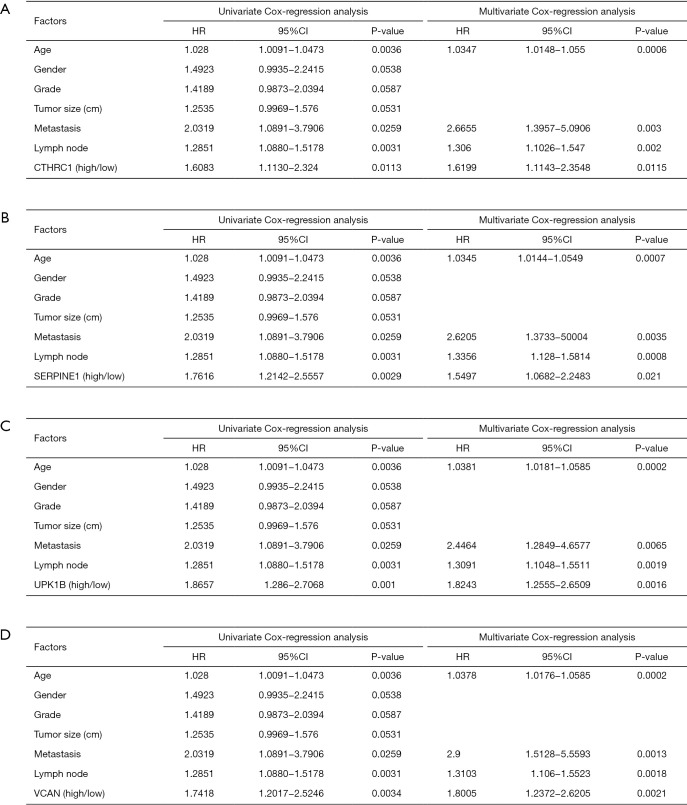
Identification of four key genes from overlapped genes
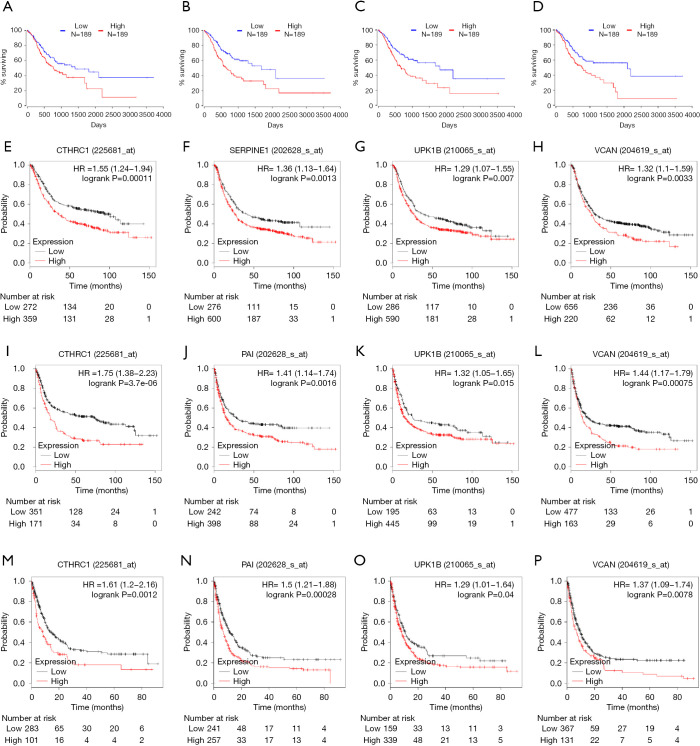
Twenty-three survival-related genes were identified by performing the Kaplan-Meier analysis, and high expression level was associated with a poorer OS (Table S1).Then, we identified four significant key genes by conducting univariate and multivariate-Cox analysis for OS, including CTHRC1, SERPINE1, UPK1B, VCAN, with HR >1 (P<0.05) (Figure 4).
Table S1. Twenty-three survival related genes were identified by performing the Kaplan-Meier analysis (P<0.05).
| Gene | P value |
|---|---|
| SERPINE1 | 0.000219 |
| UPK1B | 0.001473 |
| ANGPT2 | 0.005681 |
| AADAC | 0.006003 |
| PDGFRB | 0.01205 |
| TNFRSF11B | 0.012199 |
| OLFML2B | 0.012447 |
| LOX | 0.013269 |
| SMPD3 | 0.013456 |
| VCAN | 0.01891 |
| MAMDC2 | 0.020227 |
| ECT2 | 0.020561 |
| TUBB6 | 0.02132 |
| MFAP2 | 0.022238 |
| DPT | 0.025029 |
| COL4A1 | 0.027449 |
| COL5A2 | 0.028342 |
| CTHRC1 | 0.029945 |
| FAP | 0.040863 |
| AGT | 0.04225 |
| MAP7D2 | 0.047641 |
| MMP12 | 0.048191 |
| COL12A1 | 0.049813 |
| OSMR | 0.054358 |
| CALD1 | 0.059583 |
| INHBA | 0.067962 |
| CST2 | 0.071974 |
| CLIC6 | 0.082916 |
| COL10A1 | 0.084384 |
| S100A9 | 0.096072 |
| GUCA2B | 0.096917 |
| COL1A1 | 0.109207 |
| COL5A1 | 0.10969 |
| COL8A1 | 0.11083 |
| COL3A1 | 0.111259 |
| SYTL5 | 0.114956 |
| CDH11 | 0.124127 |
| ADH7 | 0.130015 |
| VSIG1 | 0.132332 |
| SCIN | 0.13247 |
| SPP1 | 0.13646 |
| PLLP | 0.13804 |
| PRC1 | 0.147127 |
| C6orf58 | 0.147472 |
| CIDEC | 0.151807 |
| ESM1 | 0.15414 |
| BCAT1 | 0.155191 |
| LTF | 0.157024 |
| MT1G | 0.158712 |
| PI15 | 0.158844 |
| OTC | 0.164052 |
| UGT2B15 | 0.174819 |
| TREM1 | 0.184753 |
| SOSTDC1 | 0.187739 |
| EMP3 | 0.189071 |
| PDIA2 | 0.189871 |
| COL1A2 | 0.191785 |
| OLR1 | 0.205573 |
| RNASE1 | 0.20733 |
| ASPN | 0.212696 |
| TFF2 | 0.233068 |
| SULF1 | 0.238557 |
| MT1M | 0.247873 |
| ETV4 | 0.254438 |
| KRT20 | 0.265582 |
| FBP2 | 0.265783 |
| GHRL | 0.270264 |
| ANXA10 | 0.276172 |
| MAOA | 0.276519 |
| AKR7A3 | 0.276782 |
| PBK | 0.277451 |
| SNX10 | 0.28286 |
| TNFSF4 | 0.290801 |
| KCNJ15 | 0.29299 |
| GKN1 | 0.29894 |
| SELENBP1 | 0.306575 |
| CHI3L1 | 0.311321 |
| RDH12 | 0.320753 |
| CXCL17 | 0.324846 |
| HRASLS2 | 0.327991 |
| OLFM4 | 0.32988 |
| FSCN1 | 0.358197 |
| CPXM1 | 0.364493 |
| FBXO32 | 0.36483 |
| SFRP4 | 0.369919 |
| MMP1 | 0.372373 |
| GEM | 0.383342 |
| LIFR | 0.391984 |
| IRX3 | 0.395766 |
| GKN2 | 0.399234 |
| THY1 | 0.40806 |
| CA2 | 0.41812 |
| GGT6 | 0.431151 |
| AQP9 | 0.43801 |
| CXCL5 | 0.443691 |
| VILL | 0.446059 |
| HOXC6 | 0.450283 |
| ECM1 | 0.453289 |
| APOBEC2 | 0.455273 |
| THBS2 | 0.469881 |
| CLDN2 | 0.473161 |
| RCN3 | 0.481199 |
| WNT2 | 0.489813 |
| CBR1 | 0.49536 |
| CHGA | 0.503973 |
| APOE | 0.506067 |
| CCL18 | 0.506183 |
| IGF2BP3 | 0.510406 |
| GSTA1 | 0.511807 |
| TIMP1 | 0.51665 |
| RARRES1 | 0.525777 |
| KLK6 | 0.532234 |
| SPINK7 | 0.533784 |
| MAL | 0.541585 |
| S100A8 | 0.548391 |
| SST | 0.550388 |
| CEACAM6 | 0.552092 |
| COL11A1 | 0.556812 |
| TAGLN | 0.562203 |
| LY6E | 0.563069 |
| MT1H | 0.568436 |
| KLK11 | 0.569524 |
| SSTR1 | 0.573811 |
| PMEPA1 | 0.583951 |
| MXRA5 | 0.584339 |
| CXCL9 | 0.587203 |
| TFF1 | 0.596394 |
| EPHB2 | 0.597971 |
| PLK1 | 0.600849 |
| CDH3 | 0.605368 |
| MSR1 | 0.612339 |
| F2RL2 | 0.616396 |
| C1orf116 | 0.617585 |
| S100P | 0.617652 |
| BGN | 0.624311 |
| SERPINH1 | 0.629089 |
| FPR3 | 0.634469 |
| CYP4F12 | 0.653012 |
| CAP2 | 0.654383 |
| MMP3 | 0.655905 |
| TOP2A | 0.657755 |
| ANLN | 0.658936 |
| REG3A | 0.660342 |
| KCNE2 | 0.66711 |
| PGC | 0.668703 |
| SCNN1B | 0.6804 |
| ALDH3A1 | 0.685418 |
| CCKBR | 0.685544 |
| MLLT11 | 0.687678 |
| CYP2C9 | 0.69351 |
| SULT2A1 | 0.694968 |
| ADH1C | 0.70767 |
| PSCA | 0.709882 |
| PLA2G2A | 0.712788 |
| IFITM1 | 0.71556 |
| LIPF | 0.717355 |
| LIF | 0.724972 |
| CLDN1 | 0.734878 |
| LDHD | 0.740984 |
| PIGR | 0.743844 |
| KLF4 | 0.744254 |
| SLC16A9 | 0.745095 |
| PLAU | 0.745453 |
| CAPN9 | 0.747822 |
| PBLD | 0.753745 |
| TRIP13 | 0.754745 |
| CXCL1 | 0.768736 |
| GIF | 0.780016 |
| TCN1 | 0.781361 |
| C4BPA | 0.78778 |
| TPX2 | 0.791127 |
| APOBEC1 | 0.802589 |
| GATA5 | 0.812229 |
| SLC28A2 | 0.819387 |
| SIDT2 | 0.827656 |
| ANG | 0.828256 |
| CST1 | 0.83539 |
| SCGB2A1 | 0.849791 |
| ATP4A | 0.857416 |
| MMP7 | 0.857637 |
| CXCL10 | 0.865142 |
| FMO5 | 0.885593 |
| TNFRSF17 | 0.887868 |
| SULT1B1 | 0.89062 |
| PLA2G7 | 0.892582 |
| FAM3B | 0.897619 |
| CAPN13 | 0.910593 |
| LIPG | 0.911736 |
| GAST | 0.919055 |
| CKB | 0.925075 |
| ALDOB | 0.928519 |
| AKR1C3 | 0.935679 |
| MMP9 | 0.938571 |
| ITPKA | 0.948574 |
| FCGBP | 0.952871 |
| VSIG2 | 0.954352 |
| BCAS1 | 0.957483 |
| HPGD | 0.960429 |
| PXMP2 | 0.966763 |
| CYP2C18 | 0.969566 |
| ATP4B | 0.970968 |
| CILP | 0.971054 |
| TMED6 | 0.996557 |
Figure 4.
Univariate and multivariate analysis of clinicopathologic characteristics and key genes for OS. (A) CTHRC1 (B) SERPINE1 (C) UPK1B (D) VCAN. OS, overall survival; CTHRC1, collagen triple helix repeat containing 1; SERPINE1, plasminogen activator inhibitor type 1; UPK1B, uroplakin Ib; VCAN, Verscan.
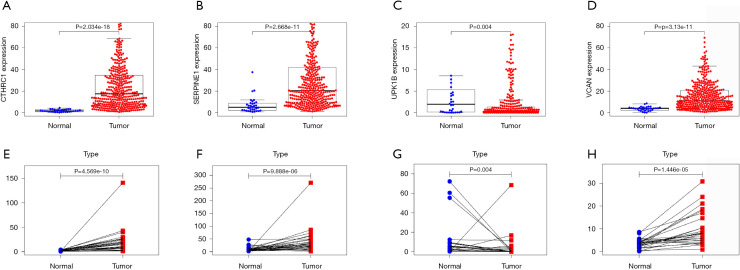
Prognostic significance for the four genes
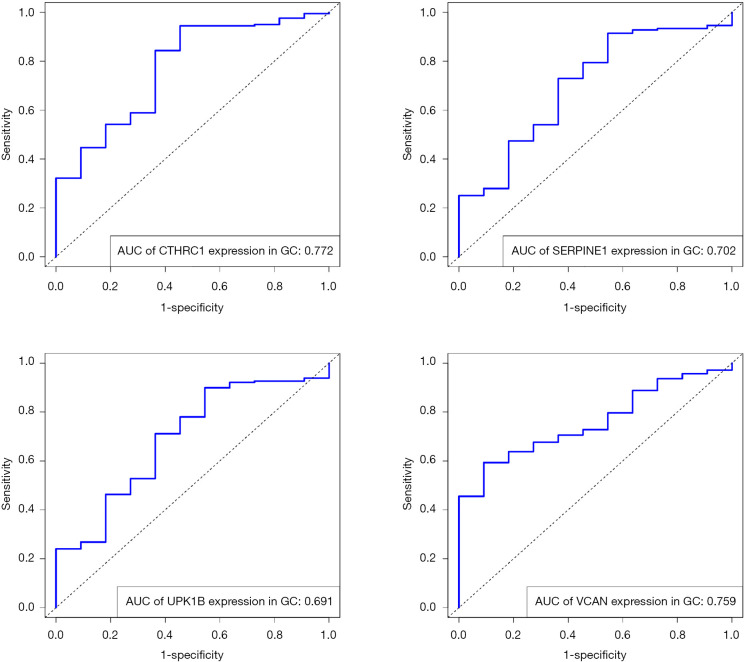
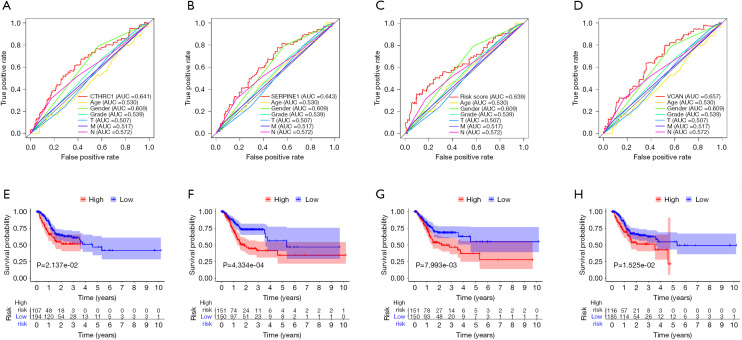
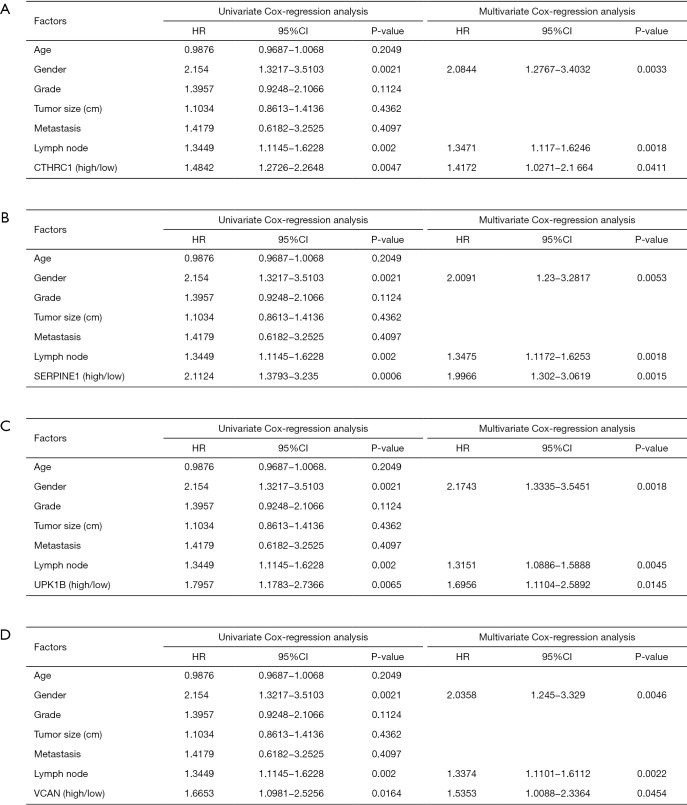
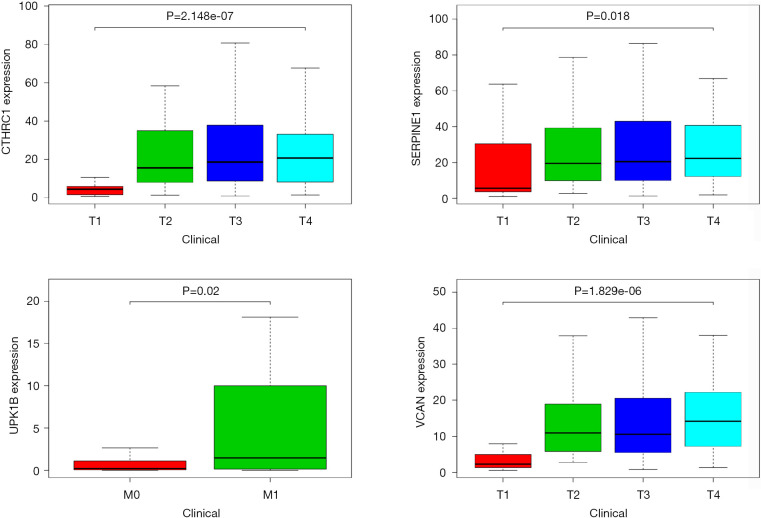
The gene expression of the cancer group was higher than the normal group from TCGA dataset for CTHRC1 (Figure 5A), SERPINE1 (Figure 5B), UPK1B (Figure 5C) and VCAN (Figure 5D). Meantime, the gene expression of the cancer group was higher than paracancerous group for CTHRC1 (Figure 5E), SERPINE1 (Figure 5F), UPK1B (Figure 5G) and VCAN (Figure 5H). By using OncoLnc, it indicated high gene expression was significantly associated with a shorter OS (Figure 6A,B,C,D). Then Kaplan Meier plotter revealed the same trend, high expression presented worse OS (Figure 6E,F,G,H), first progression (FP) (Figure 6I,J,K,L) and post progression survival (PPS) (Figure 6M,N,O,P). Next, the ROC analysis of four key genes was performed to evaluate the diagnostic value of four key genes for OS, as showed in Figure 7, all the AUC indicated a moderate diagnostic value (CTHRC1: 0.772, SERPINE1: 0.702, UPK1B: 0.691, VCAN: 0.759). Furthermore, patients with high expression level have poorer DFS than the patients with low expression level (P<0.05, Figure 8A,B,C,D). The ROC curve for DFS demonstrated that CTHRC1, SERPINE1, UPK1B and VCAN were specific and sensitive than any clinical characteristics, including age, gender, grade, tumor size, lymph node and metastasis (Figure 8E,F,G,H). In addition, univariate and multivariate-Cox analysis for DFS displayed four key genes were all powerful and independent factors for DFS (Figure 9). Finally, correlation analysis between TMN and expression of key genes was analyzed by performing Mann-Whitney-Wilcoxon Test based on TCGA data, it revealed that gene expression was associated with tumor stage, including CTHRC1, SERPINE1, VCAN. Meantime, UPK1B expression was associated with distant metastasis (Figure 10).
Figure 5.
Comparation of the key gene expression. The different expression level in normal tissue and GC (A-D), (A) CTHRC1, (B) SERPINE1, (C) UPK1B, (D) VCAN. The different expression level in paracancerous tissue and GC (E-H), (E) CTHRC1, (F) SERPINE1, (G) UPK1B, (H) VCAN. CTHRC1, collagen triple helix repeat containing 1; SERPINE1, plasminogen activator inhibitor type 1; UPK1B, uroplakin Ib; VCAN, Verscan.
Figure 6.
The performance analysis using OncoLnc (A-D) and Kaplan-Meier Plotter (E-P), (E-H) OS, (I-L) FP, (M-P) PPS. (A,E,I,M) CTHRC1, (B,F,J,N) SERPINE1, (C,G,K,O) UPK1B, (D,H,L,P) VCAN. CTHRC1, collagen triple helix repeat containing 1; SERPINE1, plasminogen activator inhibitor type 1; UPK1B, uroplakin Ib; VCAN, Verscan; GC, gastric cancer.
Figure 7.
The ROC curve for OS in GC. ROC, Time-dependent receiver operating characteristic; OS, overall survival; GC, gastric cancer.
Figure 8.
The performance analysis for DFS in GC. The ROC curve for DFS in GC (A-D), Kaplan-Meier plotter for DFS in GC (E-H). (A,E) CTHRC1, (B,F) SERPINE1, (C,G) UPK1B, (D,H) VCAN. CTHRC1, collagen triple helix repeat containing 1; SERPINE1, plasminogen activator inhibitor type 1; UPK1B, uroplakin Ib; VCAN, Verscan; GC, gastric cancer.
Figure 9.
Univariate and multivariate analysis of clinicopathologic characteristics and key genes for DFS. (A) CTHRC1, (B) SERPINE1, (C) UPK1B, (D) VCAN. DFS, disease free survival; CTHRC1, collagen triple helix repeat containing 1; SERPINE1, plasminogen activator inhibitor type 1; UPK1B, uroplakin Ib; VCAN, Verscan.
Figure 10.
Significant correlation between key gene expression and TMN in GC. T, tumor; N, lymph node; M, metastasis; GC, gastric cancer.
Biological processes and signaling pathway analysis for the co-expressed genes associated with GC
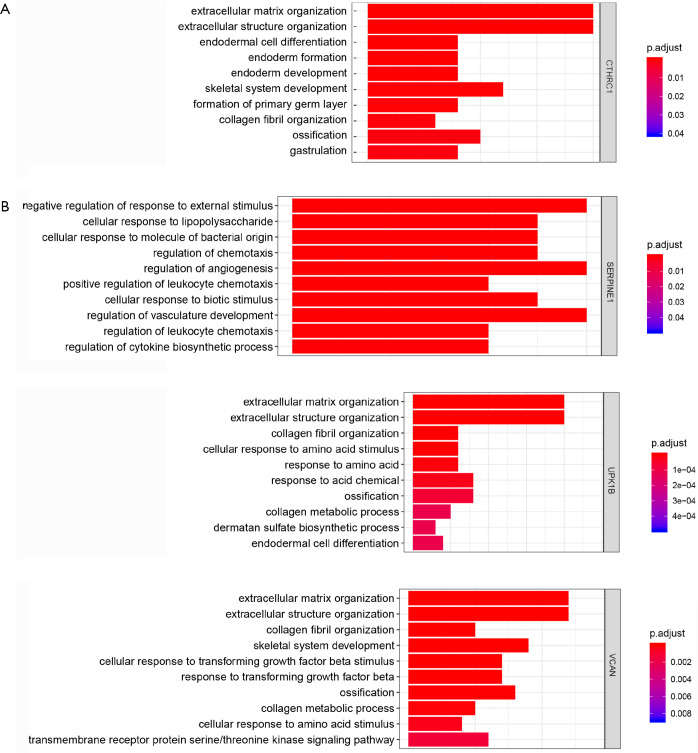
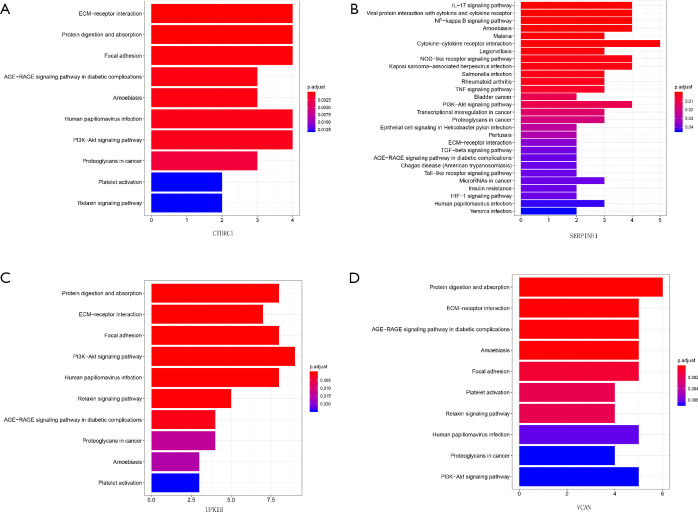
We identified the co-expressed genes associated with key genes in GC. In addition, the biological processes and signaling pathway analysis of key genes in GC were investigated. These co-expressed genes were involved in a variety of biological processes, such as endodermal cell differentiation, endoderm development, and extracellular matrix organization for CTHRC1 (Figure 11A), regulation of angiogenesis, positive regulation of leukocyte chemotaxis cellular, and regulation of vasculature development for SERPINE1 (Figure 11B), extracellular matrix organization, collagen fibril organization, collagen metabolic process, and endodermal cell differentiation for UPK1B, extracellular matrix organization, and cellular response to transforming growth factor beta stimulus for VCAN. These co-expressed genes were involved in a variety of biological processes, such as ECM-receptor interaction, AGE-RAGE signaling pathway in diabetic complications, PI3K-Akt signaling pathway for CTHRC1 (Figure 12A), such as NF-kappa B signaling pathway, PI3K-Akt signaling pathway, Toll-like receptor signaling pathway for SERPINE1 (Figure 12B), such as ECM-receptor interaction, PI3K-Akt signaling pathway, relaxin signaling pathway for UPK1B (Figure 12C), such as ECM-receptor interaction, PI3K-Akt signaling pathway for VCAN (Figure 12D).
Figure 11.
Potential biological processes for the key genes in GC. GC, gastric cancer.
Figure 12.
Potential signaling pathways for the key genes in GC. GC, gastric cancer.
GSEA identifies prognostic genes-related signaling pathway
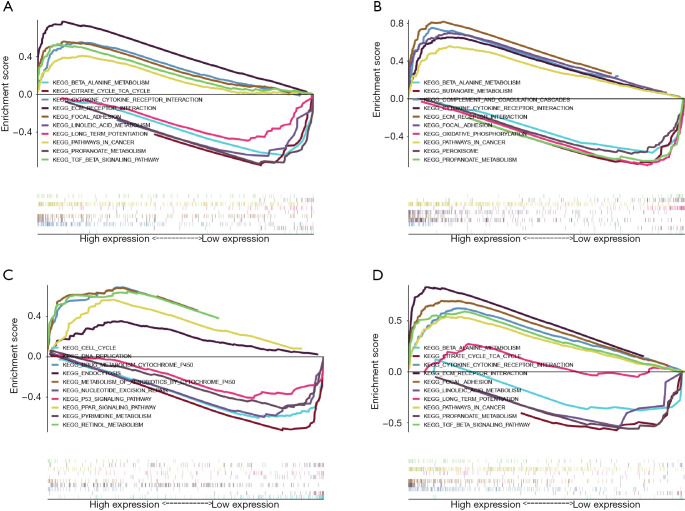
In order to further explore the mechanism of prognostic genes in patients with GC, we conducted GSEA between low and high expression group to identify the significant pathways (FDR <0.05, NOM P value <0.05). For CTHRC1, some significant pathways which were active in the high-expression group, including KEGG_ECM_RECEPTOR_INTERACTION, KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION, KEGG_TGF_BETA_SIGNALING_PATHWAY, KEGG_PATHWAYS_IN_CANCER, KEGG_FOCAL_ADHESION. Several significant pathways which were active in the low-risk group, including KEGG_PROPANOATE_METABOLISM, KEGG_CITRATE_CYCLE_TCA_CYCLE, KEGG_BETA_ALANINE_METABOLISM, KEGG_LONG_TERM_POTENTIATION, KEGG_LINOLEIC_ACID_METABOLISM (Figure 13A). The most significant pathways were presented for SERPINE1 (Figure 13B), UPK1B (Figure 13C) and VCAN (Figure 13D).
Figure 13.
Enrichment plots from GSEA for (A) CTHRC1, (B) SERPINE1, (C) UPK1B, (D) VCAN. GSEA, gene set enrichment analysis; CTHRC1, collagen triple helix repeat containing 1; SERPINE1, plasminogen activator inhibitor type 1; UPK1B, uroplakin Ib; VCAN, Verscan.
Discussion
GC is one most common malignant cancer worldwide and it is very difficult to treat the advanced-stage SC. Although the formation, progression and underlying mechanisms for GC have been revealed from some basic and clinical studies, the incidence and mortality of GC is still very high worldwide (10). Therefore, it is necessary to identify novel prognostic and therapeutic target for GC.
UPK1B is a structural protein on the surface of urothelial cells1, which was considered as the entirely specific for urothelium, recent studies have indicated that UPK1B also expressed in other tissues, including bladder, brain, eye, kidney, lung, stomach (11). UPK1B may promote the occurrence and development of cancer (12,13), UPK1B could promote the proliferation, invasion and metastasis in bladder cancer (14,15). Su et al. (16) showed abnormal expression of UPK1B in various types of cancers. However, the role of UPK1B in GC has not been reported. In this study, the different expression level between normal and cancer is of significance, high gene expression was significantly associated with a shorter OS, UPK1B is significant diagnostic factor in GC. The expression level was associated with distant metastasis, UPK1B may participate in the biological processes (extracellular matrix organization, collagen fibril organization, collagen metabolic process, endodermal cell differentiation) through ECM-receptor interaction, PI3K-Akt signaling pathway, relaxin signaling pathway to promote the metastasis in GC.
The CTHRC1 gene belongs to chromosome 8q22.3, which encoded a protein to participate in the vascularity and bone formation and so on (17). The expression level was different between normal tissue and tumor tissue for some types of tumors, including breast cancer (18), cervical cancer (19), colorectal cancer (20), liver cancer (21) and GC (22), the aberrant expression level was associated with poor OS and progression-free survival and it was the independent prognostic marker in GC in GC, which was consistent with our result. Recently, Ding et al. (23) have reported that HIF-1α/CXCR4 signaling may be involved in the migration and invasion in GC, however, the underlying molecular mechanism for CTHRC1 promoting the occurrence and development of GC is not very clear. In this study, we identify several signaling pathway which may be involved in the occurrence and development of GC.
SERPINE1 gene encodes plasminogen activator inhibitor type 1, which participated in inhibiting tissue plasminogen activator and uridylyl phosphate adenosine, the aberrant expression in many types of cancer and SERPINE1 could be an independent risk factor for various types of cancers, including head and neck cancer (24,25), esophageal cancer (26), bladder cancer (27), melanoma (28). Li et al. (29) indicated SERPINE1 is a poor prognosis for GC, and SERPINE1 could promote tumour cell proliferation, migration, and invasion by regulating EMT. However, SERPINE1 still remains largely unknown in GC. In our study, SERPINE1 was an significant diagnostic factor in GC, and we found the expression level of SERPINE1 was associated with depth of invasion, the potential signal pathways may participated in the biological process including NF-kappa B signaling pathway, PI3K-Akt signaling pathway, Toll-like receptor signaling pathway.
VCAN is a chondroitin sulfate proteoglycan, a member of the aggregating chondroitin sulfate PGs family, which is an important component of ECM (30). Verscan expression often occurs in the context of tissue remodeling, angiogenesis, including: follicular growth (31), inflammation (32), wound healing (33) or atherosclerotic lesions (34), and environmental significance around progressive tumors (35). It has been previously reported that tumor stromal cells play an important role in tumor formation and tumor progression (36), and VCAN is expressed and secreted by tumor stromal cells. Yeung et al. indicated that CAF-specific VCAN was up-regulated by TGF-β signal to promote tumorigenesis and invasion in ovarian cancer (37). The level of VCAN increased in many patients with malignant tumors includes colon cancer (38), rectal cancer (39), melanoma (40), odontogenic cancer (41), and ovarian cancer (42). In vitro and in vivo research, it has shown that VCAN can promote the proliferation, metastasis and invasion of cancer cells (43-45), with playing an important role in the formation of extracellular matrices that support tumor growth and metastasis. Shen et al. (46) reported that VCAN expression can be used as a prognostic indicator for GC patients, VCAN expression is higher in cancer tissues than in adjacent tissues, and could promote proliferation and invasion in GC cells. However, few literatures mentioned VCAN associated signaling pathways that promote the development of GC. We identified some signaling pathways that may be involved in the development of GC. This regulatory mechanism needs to be further elucidated.
Conclusions
In conclusion, by integrating four GEO and TCGA gene expression profile datasets, we identified four key genes (CTHRC1, SERPINE1, VCAN, UPK1B) which might as the novel potential prognostic molecular markers for GC. The four key genes have high prognostic performance, and could considered as independent prognostic factors for OS and DFS in GC. The four key genes act as oncogene to promote the development of GC, CTHRC1 participated in endodermal cell differentiation, extracellular matrix organization, SERPINE1 participated in regulation of angiogenesis, positive regulation of leukocyte chemotaxis cellular, regulation of vasculature development, UPK1B participated in extracellular matrix organization, collagen fibril organization, collagen metabolic process, endodermal cell differentiation, VCAN participated in extracellular matrix organization, cellular response to transforming growth factor beta stimulus. The study would provide some novel genes for the future prognosis prediction and potential molecular targeting therapy for GC.
However, further biological experiments should be performed to validate our results.
Acknowledgments
We are grateful to the reviewers for their constructive comments which led to improvements in this manuscript. In addition, we thank Bin Zhao (Official Wechat Account: Bio_Med2017) of Xiamen University for suggestions on the manuscripts.
Funding: This study was supported by Xiamen Scientific and Technological Plan (No. 3502Z20194005).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-211). The authors have no conflicts of interest to declare.
References
- 1.Bertuccio P, Chatenoud L, Levi F, et al. Recent patterns in gastric cancer: a global overview. Int J Cancer 2009;125:666-73. 10.1002/ijc.24290 [DOI] [PubMed] [Google Scholar]
- 2.Ferro A, Peleteiro B, Malvezzi M, et al. Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur J Cancer 2014;50:1330-44. 10.1016/j.ejca.2014.01.029 [DOI] [PubMed] [Google Scholar]
- 3.Peleteiro B, Severo M, La Vecchia C, et al. Model-based patterns in stomach cancer mortality worldwide. Eur J Cancer Prev 2014;23:524-31. 10.1097/CEJ.0b013e328364f2b6 [DOI] [PubMed] [Google Scholar]
- 4.Chen W. Cancer statistics: updated cancer burden in China. Chin J Cancer Res 2015;27:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Figueiredo C, Camargo MC, Leite M, et al. Pathogenesis of Gastric Cancer: Genetics and Molecular Classification. Curr Top Microbiol Immunol 2017;400:277-304. 10.1007/978-3-319-50520-6_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He J, Jin Y, Chen Y, et al. Downregulation of ALDOB is associated with poor prognosis of patients with gastric cancer. Onco Targets Ther 2016;9:6099-109. 10.2147/OTT.S110203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh SC, Sohn BH, Cheong JH, et al. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat Commun 2018;9:1777. 10.1038/s41467-018-04179-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hippo Y, Taniguchi H, Tsutsumi S, et al. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res 2002;62:233-40. [PubMed] [Google Scholar]
- 9.Zhang X, Ni Z, Duan Z, et al. Overexpression of E2F mRNAs associated with gastric cancer progression identified by the transcription factor and miRNA co-regulatory network analysis. PLoS One 2015;10:e0116979. 10.1371/journal.pone.0116979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 11.Yu J, Lin JH, Wu XR, et al. Uroplakins Ia and Ib, two major differentiation products of bladder epithelium, belong to a family of four transmembrane domain (4TM) proteins. J Cell Biol 1994;125:171-82. 10.1083/jcb.125.1.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webb GC, Finch JL, Cowled PA. Assignment of the Uroplakin 1b (Upk1b) gene to mouse chromosome 16 bands B5-C2 by in situ hybridization. Cytogenet Cell Genet 1999;84:37-8. 10.1159/000015208 [DOI] [PubMed] [Google Scholar]
- 13.Finch JL, Webb GC, Evdokiou A, et al. Chromosomal localization of the human urothelial "tetraspan" gene, UPK1B, to 3q13.3-q21 and detection of a TaqI polymorphism. Genomics 1997;40:501-3. 10.1006/geno.1996.4589 [DOI] [PubMed] [Google Scholar]
- 14.Osman I, Kang M, Lee A, et al. Detection of circulating cancer cells expressing uroplakins and epidermal growth factor receptor in bladder cancer patients. Int J Cancer 2004;111:934-9. 10.1002/ijc.20366 [DOI] [PubMed] [Google Scholar]
- 15.Olsburgh J, Harnden P, Weeks R, et al. Uroplakin gene expression in normal human tissues and locally advanced bladder cancer. J Pathol 2003;199:41-9. 10.1002/path.1252 [DOI] [PubMed] [Google Scholar]
- 16.Su J, Zhang Y, Su H, et al. A recurrence model for laryngeal cancer based on SVM and gene function clustering. Acta Otolaryngol 2017;137:557-62. 10.1080/00016489.2016.1247984 [DOI] [PubMed] [Google Scholar]
- 17.Takeshita S, Fumoto T, Matsuoka K, et al. Osteoclast-secreted CTHRC1 in the coupling of bone resorption to formation. J Clin Invest 2013;123:3914-24. 10.1172/JCI69493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai YH, Chen J, Wang XP, et al. Collagen triple helix repeat containing-1 negatively regulated by microRNA-30c promotes cell proliferation and metastasis and indicates poor prognosis in breast cancer. J Exp Clin Cancer Res 2017;36:92. 10.1186/s13046-017-0564-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li N, Chen L, Liu C, et al. Elevated CTHRC1 expression is an indicator for poor prognosis and lymph node metastasis in cervical squamous cell carcinoma. Hum Pathol 2019;85:235-41. 10.1016/j.humpath.2018.10.015 [DOI] [PubMed] [Google Scholar]
- 20.Ni S, Ren F, Xu M, et al. CTHRC1 overexpression predicts poor survival and enhances epithelial-mesenchymal transition in colorectal cancer. Cancer Med 2018;7:5643-54. 10.1002/cam4.1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H, Su L, Liu C, et al. CTHRC1 May Serve As A Prognostic Biomarker For Hepatocellular Carcinoma. Onco Targets Ther 2019;12:7823-31. 10.2147/OTT.S219429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu L, Liu L, Zhong L, et al. Cthrc1 overexpression is an independent prognostic marker in gastric cancer. Hum Pathol 2014;45:1031-8. 10.1016/j.humpath.2013.12.020 [DOI] [PubMed] [Google Scholar]
- 23.Ding X, Huang R, Zhong Y, et al. CTHRC1 promotes gastric cancer metastasis via HIF-1alpha/CXCR4 signaling pathway. Biomed Pharmacother 2020;123:109742. 10.1016/j.biopha.2019.109742 [DOI] [PubMed] [Google Scholar]
- 24.Pavon MA, Arroyo-Solera I, Cespedes MV, et al. uPA/uPAR and SERPINE1 in head and neck cancer: role in tumor resistance, metastasis, prognosis and therapy. Oncotarget 2016;7:57351-66. 10.18632/oncotarget.10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavon MA, Arroyo-Solera I, Tellez-Gabriel M, et al. Enhanced cell migration and apoptosis resistance may underlie the association between high SERPINE1 expression and poor outcome in head and neck carcinoma patients. Oncotarget 2015;6:29016-33. 10.18632/oncotarget.5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klimczak-Bitner AA, Kordek R, Bitner J, et al. Expression of MMP9, SERPINE1 and miR-134 as prognostic factors in esophageal cancer. Oncol Lett 2016;12:4133-8. 10.3892/ol.2016.5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang G, Gomes-Giacoia E, Dai Y, et al. Validation and clinicopathologic associations of a urine-based bladder cancer biomarker signature. Diagn Pathol 2014;9:200. 10.1186/s13000-014-0200-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein RM, Bernstein D, Higgins SP, et al. SERPINE1 expression discriminates site-specific metastasis in human melanoma. Exp Dermatol 2012;21:551-4. 10.1111/j.1600-0625.2012.01523.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Zhu Z, Zhao Y, et al. FN1, SPARC, and SERPINE1 are highly expressed and significantly related to a poor prognosis of gastric adenocarcinoma revealed by microarray and bioinformatics. Sci Rep 2019;9:7827. 10.1038/s41598-019-43924-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz NB, Pirok EW, 3rd, Mensch JR, Jr, et al. Domain organization, genomic structure, evolution, and regulation of expression of the aggrecan gene family. Prog Nucleic Acid Res Mol Biol 1999;62:177-225. 10.1016/S0079-6603(08)60508-5 [DOI] [PubMed] [Google Scholar]
- 31.Russell DL, Ochsner SA, Hsieh M, et al. Hormone-regulated expression and localization of versican in the rodent ovary. Endocrinology 2003;144:1020-31. 10.1210/en.2002-220434 [DOI] [PubMed] [Google Scholar]
- 32.Hakkinen L, Westermarck J, Kahari VM, et al. Human granulation-tissue fibroblasts show enhanced proteoglycan gene expression and altered response to TGF-beta 1. J Dent Res 1996;75:1767-78. 10.1177/00220345960750101001 [DOI] [PubMed] [Google Scholar]
- 33.Cattaruzza S, Schiappacassi M, Ljungberg-Rose A, et al. Distribution of PG-M/versican variants in human tissues and de novo expression of isoform V3 upon endothelial cell activation, migration, and neoangiogenesis in vitro. J Biol Chem 2002;277:47626-35. 10.1074/jbc.M206521200 [DOI] [PubMed] [Google Scholar]
- 34.Evanko SP, Raines EW, Ross R, et al. Proteoglycan distribution in lesions of atherosclerosis depends on lesion severity, structural characteristics, and the proximity of platelet-derived growth factor and transforming growth factor-beta. Am J Pathol 1998;152:533-46. [PMC free article] [PubMed] [Google Scholar]
- 35.Crowther M, Brown NJ, Bishop ET, et al. Microenvironmental influence on macrophage regulation of angiogenesis in wounds and malignant tumors. J Leukoc Biol 2001;70:478-90. [PubMed] [Google Scholar]
- 36.Calon A, Lonardo E, Berenguer-Llergo A, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet 2015;47:320-9. 10.1038/ng.3225 [DOI] [PubMed] [Google Scholar]
- 37.Yeung TL, Leung CS, Wong KK, et al. TGF-beta modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer Res 2013;73:5016-28. 10.1158/0008-5472.CAN-13-0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theocharis AD. Human colon adenocarcinoma is associated with specific post-translational modifications of versican and decorin. Biochim Biophys Acta 2002;1588:165-72. 10.1016/S0925-4439(02)00161-8 [DOI] [PubMed] [Google Scholar]
- 39.Tsara ME, Theocharis AD, Theocharis DA. Compositional and structural alterations of proteoglycans in human rectum carcinoma with special reference to versican and decorin. Anticancer Res 2002;22:2893-8. [PubMed] [Google Scholar]
- 40.Touab M, Villena J, Barranco C, et al. Versican is differentially expressed in human melanoma and may play a role in tumor development. Am J Pathol 2002;160:549-57. 10.1016/S0002-9440(10)64874-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito Y, Abiko Y, Tanaka Y, et al. Immunohistochemical localization of large chondroitin sulfate proteoglycan in odontogenic tumor. Med Electron Microsc 2002;35:173-7. 10.1007/s007950200022 [DOI] [PubMed] [Google Scholar]
- 42.Voutilainen K, Anttila M, Sillanpaa S, et al. Versican in epithelial ovarian cancer: relation to hyaluronan, clinicopathologic factors and prognosis. Int J Cancer 2003;107:359-64. 10.1002/ijc.11423 [DOI] [PubMed] [Google Scholar]
- 43.Ang LC, Zhang Y, Cao L, et al. Versican enhances locomotion of astrocytoma cells and reduces cell adhesion through its G1 domain. J Neuropathol Exp Neurol 1999;58:597-605. 10.1097/00005072-199906000-00004 [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Cao L, Yang BL, et al. The G3 domain of versican enhances cell proliferation via epidermial growth factor-like motifs. J Biol Chem 1998;273:21342-51. 10.1074/jbc.273.33.21342 [DOI] [PubMed] [Google Scholar]
- 45.Yang BL, Zhang Y, Cao L, et al. Cell adhesion and proliferation mediated through the G1 domain of versican. J Cell Biochem 1999;72:210-20. [DOI] [PubMed] [Google Scholar]
- 46.Shen XH, Lin WR, Xu MD, et al. Prognostic significance of Versican expression in gastric adenocarcinoma. Oncogenesis 2015;4:e178. 10.1038/oncsis.2015.36 [DOI] [PMC free article] [PubMed] [Google Scholar]