Abstract
Background
Whether or not EGFR mutation status detected by ddPCR in plasma predicts the effect of icotinib on patients with advanced lung adenocarcinoma was determined.
Methods
Plasma and matched tissue specimens from patients with advanced lung adenocarcinoma were collected prior to icotinib treatment. The ARMS method was used to detect EGFR mutation status in DNA extracted from tissue specimens, while the EGFR mutation status in ctDNA extracted from plasma specimens was determined by ddPCR. The therapeutic effects of icotinib were compared between patients with EGFR-activating mutations detected by ddPCR in ctDNA and ARMS in tissue DNA.
Results
EGFR mutation status was detected in 96 tissue and 100 plasma specimens. The sensitivity and positive predictive value of 19del detected in ctDNA by ddPCR was 70.97% (22/31) and 44.90% (22/49), respectively. The positive predictive value was 84.62% (22/26) and the sensitivity was 53.66% (22/41) for the L858R mutation. For the common sensitive EGFR mutations, ddPCR had a positive predictive value of 77.19% (44/57) and a sensitivity of 48.89% (44/90). Patients with sensitive EGFR mutations in ctDNA had objective response and disease control rates (DCR) similar to patients who had sensitive EGFR mutations in tissues detected by ARMS when treated with icotinib (57.14% vs. 51.51% and 92.86% vs. 90.91%, respectively).
Conclusions
Patients with sensitive EGFR mutations in plasma specimens detected with ddPCR had a higher ORR and DCR compared with patients with sensitive EGFR mutations in tissue detected with the ARMS method.
Keywords: NSCLC, EGFR-tyrosine kinase inhibitors (EGFR-TKIs), T790M, ddPCR
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide and morbidity is expected to increase in the coming decades (1). Adenocarcinoma is the most common histologic type of lung cancer (2). Radical surgery is still the most effective method for treating early lung adenocarcinoma, but post-operative relapse occurs with or without tumor metastasis, and most patients with lung adenocarcinoma are diagnosed with advanced-stage disease. The response rate of platinum-based chemotherapy for advanced or recurrent patients is only 20–35%, and the median survival time is only 8–11 months (3,4). In recent years, it has been confirmed that EGFR-tyrosine kinase inhibitors (TKIs) prolong the median progression-free survival time to >8 months in lung adenocarcinoma patients with activating EGFR mutations, and the response rate has increased to 56–74% (5-9). Indeed, EGFR-TKIs are the first-line standard treatment for lung adenocarcinoma patients with activating EGFR mutations.
The relationship between EGFR mutation status and the therapeutic efficacy of EGFR-TKIs has been verified. Assessing the presence of EGFR mutations is a critical step in therapeutic decision-making for lung adenocarcinoma patients (10). Tumor tissue is still the recommended source for detecting EGFR, but because quality tissue samples are not always available it is not possible to conduct EGFR testing for every patient in need (11,12). Thus, alternative, easily acquired specimens from patients and feasible methods of detection are essential. Recently, circulating tumor DNA (ctDNA) has been shown to be a suitable alternative sample because ctDNA can be obtained non-invasively (13). Several technologies have been developed for EGFR detection using ctDNA (14-19). Of these technologies, the droplet digital polymerase chain reaction (ddPCR) is a method with the following characteristics: high sensitivity; quantitative analysis; and easy detection of gene mutations.
Using EGFR mutation-positive cell DNA, ddPCR has expanded to 0.02% sensitivity (20). EGFR detection by ddPCR in plasma ctDNA could achieve a sensitivity of 70–81%, specificity of 85–100%, and concordance of 86–94% compared to tumor EGFR status (21-23); however more detailed clinical validation is needed to evaluate plasma specimens using ddPCR for EGFR detection, especially with respect to the diagnostic and prognostic significance compared to tissue specimens. In this study we compared the EGFR detection results and analyzed the clinical outcomes following icotinib treatment, as determined by ARMS for tissue specimens and ddPCR for matched plasma specimens. The purpose of the current study was to provide clinical evidence to verify the value of predicting EGFR mutation status detected by ddPCR in plasma.
Methods
Patients and treatment
In this study, patients who diagnosed with stage IV lung adenocarcinoma and sensitive EGFR mutations based on tissue specimens or ctDNA were enrolled from January 2016 to December 2017 at Zhejiang Rongjun Hospital were enrolled in the current study. The study was reviewed and approved by the Institutional Ethics Committee of this hospital. Informed consent was obtained from each patient prior to testing. Demographic, epidemiologic, and pathologic information were obtained from patient medical records. None of the patients received targeted therapy, radical surgery, or radiation treatment within 28 days prior to sample collection. The enrolled patients were given 125 mg of icotinib three times per day on the basis of the EGFR mutation detection results and patient preference. Chest CT scans were performed 1 month after beginning EGFR-TKI treatment. The response to EGFR-TKIs was evaluated according to the Response Evaluation Criteria in the Solid Tumor guideline (version 1.1).
DNA extraction
Plasma was collected from every patient before therapy. Ten milliliters of whole venous blood were collected into tubes with an anti-coagulant which also contained a ctDNA protective agent (Righton, Shanghai, China), stored at 4–25 °C, and centrifuged (1,800 g for 10 min at 4 °C) as soon as possible. A Qiamp® Circulating Nucleic Acid Kit (Qiagen, Duesseldof, Germany) was used to extract ctDNA from plasma specimens. Tissue DNA was extracted using an AmoyDx® Tissue DNA Kit (Amoydx, Xiamen, China). DNA quality was verified using a nucleic acid quantitative instrument (Thermos City, MA, USA).
Detection of EGFR mutations in tissue DNA and ct DNA
EGFR mutation status in tissue DNA was detected using the ARMS method. The tests were carried out according to the manufacturer’s protocol using the AmoyDx EGFR 29 Mutation Kit (Amoydx) with the MX3000P real-time PCR system (Stratagene, La Jolla, CA, USA). The results were identified according to the criteria defined by the manufacturer’s instructions. EGFR mutation status in ctDNA was detected using ddPCR. Scientists affiliated with the Righton Testing Institute, who conducted the ddPCR, were blinded to the tissue results.
Statistical and database analyses
The objective response rate (ORR) and disease control rate (DCR) after TKI treatment were analyzed by chi-square or Fisher’s exact tests. The identified mutation status (M+ or M−) in both samples was the basis of comparison of ctDNA versus tumor EGFR mutations before treatment. The data were analyzed using GraphPad Prism (version 7.0; GraphPad, city, CA, USA), and statistical significance was indicated by a P value <0.05.
Results
Clinical and pathological characteristics of patients
Sensitive EGFR mutations were detected in 100 patients from four hospitals. All 100 patients provided sufficient plasma specimens for EGFR detection, but tissue specimens were not available or insufficient for EGFR detection in four patients prior to treatment. The patient age range was 26–83 years and the median age was 59 years. The clinical and pathologic profiles of the enrolled patients are listed in Table 1.
Table 1. Clinical and pathological profiles of the enrolled patients.
| Variable | N (n=100) | Percentage (%) |
|---|---|---|
| Age | ||
| ≥60 | 42 | 42 |
| <60 | 58 | 58 |
| Gender | ||
| Male | 39 | 39 |
| Female | 61 | 61 |
| Smoking history | ||
| Non-smoker | 84 | 84 |
| Smoker | 16 | 16 |
EGFR mutation status
Sensitive EGFR mutations were detected in tissue specimens from 92 patients prior to treatment. Of the 92 patients, common sensitive mutations were detected in 87 patients; 49 patients had 19del mutations, 41 patients had L858R mutations, and three patients had both 19del and L858R mutations. Five patients had rare sensitive mutations; three patients had G719X mutations and two patients had L861Q mutations (Table 2). T790M mutations existed in two patients who had L858R mutations. An EML4-ALK rearrangement was detected in one patient with a 19del mutation. EGFR mutations were detected in plasma ctDNA from 100 patients by ddPCR prior to treatment. Fifty-eight patients had common activating EGFR mutations. Of the 58 patients, 33 had 19del mutations, 28 had L858R mutations, both 19del and L858R mutations existed in three patients, one patient had both T790M and L858R mutations, and two patients were shown to have no other mutations. No patients had rare sensitive mutations (Table 2).
Table 2. EGFR mutation status in tissue and plasma specimens.
| Mutation type | Tissue (n=96) | Plasma (n=100) | Overall in this study |
|---|---|---|---|
| 19del | 49 | 33 | 60 |
| L858R | 41 | 28 | 47 |
| G719X | 3 | 0 | 3 |
| L861Q | 2 | 0 | 2 |
| Total | 92 | 58 | 100 |
Comparison of EGFR mutation status in plasma and tumor tissue
EGFR mutation status was determined in the plasma and tissue specimens from 96 patients. For four patients, the EGFR status was determined in their plasma specimens, but not in their tissue specimens from four patients. The EGFR status was not detected in tissue specimens, but not plasma specimens from any patients.
Thirty-three plasma specimens were positive for 19del mutations. The matched tissue results were lost in two of the 33 cases, nine had false-positive results (nine patients had positive plasma specimen results, but negative tissue specimen results); the positive predictive value for 19del mutations was 70.97% (22/31) and the sensitivity was 44.90% (22/49). Of 28 plasma specimens with L858R mutations, there were no matched tissue results in two cases and four cases had L858R mutations in plasma specimens, but not in tissue specimens. The positive predictive value for L858R mutations was 84.62% (22/26) and the sensitivity was 53.66% (22/41). For the common sensitive EGFR mutations, the positive predictive value was 77.19% (44/57) and the sensitivity was 48.89% (44/90; Table 3).
Table 3. Comparison of EGFR mutation status in plasma specimens and matched tissue specimens.
| EGFR mutation status, plasma, n | EGFR mutation status, tissue, n | |
|---|---|---|
| Positive | Negative | |
| 19del | ||
| Positive | 22 | 9 |
| Negative | 27 | NA |
| Total | 49 | NA |
| L858R | ||
| Positive | 22 | 4 |
| Negative | 19 | NA |
| Total | 41 | NA |
NA, not available.
Relationship between EGFR mutation status in plasma and tissue specimens with EGFR-TKI efficacy
Of the 100 patients in whom plasma or tissue specimens were EGFR sensitive mutation-positive, 66 received icotinib treatment. All 66 patients had sensitive EGFR mutations in tissue specimens. Thirty-four patients had partial responses (PR) to icotinib treatment; of these patients, 16 had 19del mutations, 15 patients had L858R mutations, and the other three had both mutations. The total ORR was 51.52% (34/66). The total DCR was 90.91% (60/66). With the exception of patients who had a PR, 26 had disease stabilization (SD); specifically, 14 patients had 19del mutations, nine had L858R mutations, two had G719X mutations, and one patient had a L861Q mutation. Forty-two of 66 patients had sensitive EGFR mutations in plasma specimens. The total ORR and DCR was 57.14% (24/42) and 92.86% (39/42), respectively. Twenty-four of the 42 patients had a PR, including 12 patients with 19del mutations, 11 patients with L858R mutations, and one patient with both mutations. Nine patients with 19del mutations and six patients with L858R mutations had SD (Figure 1).
Figure 1.
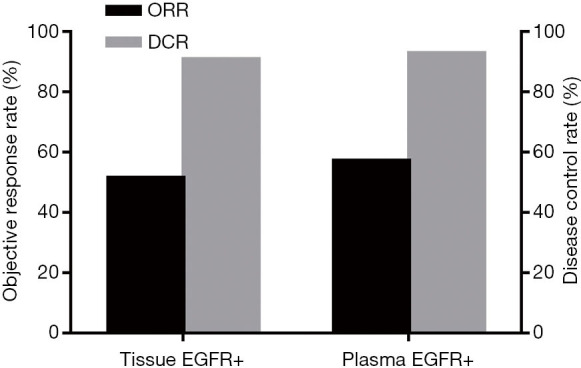
Comparison of ORR and DCR in the plasma mutation group and tissue mutation group. ORR, objective response rate; DCR, disease control rate.
Discussion
EGFR status is a significant prognostic factor for NSCLC patients who receive EGFR-TKI treatment. Plasma has been shown to be effective as an addition to tissue specimens for EGFR testing (24,25). In this study, EGFR status in plasma ctDNA detected by ddPCR was compared to matched tissues detected by the ARMS method, and the efficacy of icotinib on patients with sensitive EGFR mutations detected in plasma ctDNA was explored.
In this study, 100 plasma specimens and 98 matched tissue specimens were collected for EGFR detection by ddPCR and ARMS, respectively. The sensitivity and positive predictive value of 19del mutations detected in plasma by ddPCR were 44.90% (22/49) and 70.97% (22/31), respectively. We detected L858R mutations in plasma by ddPCR with a positive predictive value of 84.62% (22/26) and sensitivity of 53.66% (22/41). For the common sensitive EGFR mutations, ddPCR had a positive predictive value of 77.19% (44/57) and a sensitivity of 48.89% (44/90). The ddPCR method failed to detect rare sensitive mutations. Negative results existed in the five plasma specimens with rare sensitive EGFR mutations detected in matched tissue specimens.
In 13 patients, including nine with 19del mutations and four with L858R mutations, EGFR mutations were detected in plasma samples, but not in the paired tumor tissues. This finding may reflect tumor heterogeneity or biopsy bias (26,27). In another 46 patients (27 with 19del mutations and 19 with L858R mutations), EGFR mutations were present in tissue specimens, but not in plasma specimens. Tumors with EGFR mutations may not or rarely release mutant tumor DNA fragments into the blood; thus, no mutations cannot be detected in plasma (28).
The goal of EGFR mutation testing in the clinic is to select patients who might benefit from EGFR-TKIs. Therefore, we examined the efficacy of icotinib on patients who had sensitive EGFR mutations in plasma based on ddPCR. Compared with patients who had sensitive EGFR mutations in tissue based on the ARMS method, the patients who had sensitive EGFR mutations in plasma specimens had a higher ORR and DCR (57.14% vs. 51.51% and 92.86% vs. 90.91%, respectively), but the difference was not statistically significant.
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of China (31871402), Science and Technology Planning project of Zhejiang Province (LGF19H160002), Medical Scientific Research Foundation of Zhejiang Province of China (2019RC027), Scientific research foundation of Zhejiang Medical Association (2018ZYC-B5), Science and Technology Bureau Project of Jiaxing City of Zhejiang Province (2016AY23087, 2017BY1850, 2018AD32163, 2019AD32266, 2019AD32097), and Xisike-Hanson Cancer Research Foundation (Y-HS2019-20).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was reviewed and approved by the Institutional Ethics Committee of this hospital. Informed consent was obtained from each patient prior to testing.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.10.48). The authors have no conflicts of interest to declare.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014;14:535-46. 10.1038/nrc3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sweeney CJ, Zhu J, Sandler AB, et al. Outcome of patients with a performance status of 2 in Eastern Cooperative Oncology Group Study E1594: a Phase II trial in patients with metastatic nonsmall cell lung carcinoma. Cancer 2001;92:2639-47. [DOI] [PubMed] [Google Scholar]
- 4.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 2002;346:92-8. 10.1056/NEJMoa011954 [DOI] [PubMed] [Google Scholar]
- 5.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 6.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. 10.1016/S1470-2045(09)70364-X [DOI] [PubMed] [Google Scholar]
- 7.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. 10.1016/S1470-2045(11)70184-X [DOI] [PubMed] [Google Scholar]
- 8.Han JY, Park K, Kim SW, et al. First-SIGNAL: first-line single-agent iressa versus gemcitabine and cisplatin trial in never-smokers with adenocarcinoma of the lung. J Clin Oncol 2012;30:1122-8. 10.1200/JCO.2011.36.8456 [DOI] [PubMed] [Google Scholar]
- 9.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 10.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. 10.1126/science.1099314 [DOI] [PubMed] [Google Scholar]
- 11.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. 10.1158/1078-0432.CCR-12-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chouaid C, Dujon C, Do P, et al. Feasibility and clinical impact of re-biopsy in advanced non small-cell lung cancer: a prospective multicenter study in a real-world setting (GFPC study 12-01). Lung Cancer 2014;86:170-3. 10.1016/j.lungcan.2014.08.016 [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Han X, Hu X, et al. Clinical significance of pretreatment plasma biomarkers in advanced non-small cell lung cancer patients. Clin Chim Acta 2014;430:63-70. 10.1016/j.cca.2013.12.026 [DOI] [PubMed] [Google Scholar]
- 14.Kimura H, Kasahara K, Kawaishi M, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res 2006;12:3915-21. 10.1158/1078-0432.CCR-05-2324 [DOI] [PubMed] [Google Scholar]
- 15.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai H, Mao L, Wang HS, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol 2009;27:2653-9. 10.1200/JCO.2008.17.3930 [DOI] [PubMed] [Google Scholar]
- 17.Richardson AL, Iglehart JD. BEAMing up personalized medicine: mutation detection in blood. Clin Cancer Res 2012;18:3209-11. 10.1158/1078-0432.CCR-12-0871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brevet M, Johnson ML, Azzoli CG, et al. Detection of EGFR mutations in plasma DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors. Lung Cancer 2011;73:96-102. 10.1016/j.lungcan.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yung TK, Chan KC, Mok TS, et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 2009;15:2076-84. 10.1158/1078-0432.CCR-08-2622 [DOI] [PubMed] [Google Scholar]
- 20.Xu Q, Zhu Y, Bai Y, et al. Detection of epidermal growth factor receptor mutation in lung cancer by droplet digital polymerase chain reaction. Onco Targets Ther 2015;8:1533-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu G, Ye X, Dong Z, et al. Highly Sensitive Droplet Digital PCR Method for Detection of EGFR-Activating Mutations in Plasma Cell-Free DNA from Patients with Advanced Non-Small Cell Lung Cancer. J Mol Diagn 2015;17:265-72. 10.1016/j.jmoldx.2015.01.004 [DOI] [PubMed] [Google Scholar]
- 22.Zhu YJ, Zhang HB, Liu YH, et al. Association of mutant EGFR concentration in circulating cell-free DNA (cfDNA) by ddPCR with tumor burden and rognosis in NSCLC patients treated with EGFR-TKIs. J Clin Oncol 2016;34:abstr e23063.
- 23.Lee JY, Qing X, Xiumin W, et al. Longitudinal monitoring of EGFR mutations in plasma predicts outcomes of NSCLC patients treated with EGFR TKIs: Korean Lung Cancer Consortium (KLCC-12-02). Oncotarget 2016;7:6984-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Lu Y, Zhu G, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol 2013;66:1065-9. 10.1136/jclinpath-2013-201728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber B, Meldgaard P, Hager H, et al. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer 2014;14:294. 10.1186/1471-2407-14-294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai H, Wang Z, Wang Y, et al. Detection and clinical significance of intratumoral EGFR mutational heterogeneity in Chinese patients with advanced non-small cell lung cancer. PLoS One 2013;8:e54170. 10.1371/journal.pone.0054170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taniguchi K, Okami J, Kodama K, et al. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci 2008;99:929-35. 10.1111/j.1349-7006.2008.00782.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goto K, Ichinose Y, Ohe Y, et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol 2012;7:115-21. 10.1097/JTO.0b013e3182307f98 [DOI] [PubMed] [Google Scholar]


