Abstract
Background
Long non-coding RNA MIR17HG (miR-17-92 cluster host gene lncRNA) is synthesized from miR-17-92 cluster host gene due to pre-miRNA processing, and is referred as miRNA-host gene lncRNA (lnc-miRHG). Until now, the biologic function of most lnc-miRHGs in tumor progression is not well understood. This study aimed to investigate the expression and clinical significance of MIR17HG in colon cancer.
Methods
Quantitative real-time PCR analysis was used to evaluate MIR17HG expression in colon cancer tissues and paired adjacent normal mucosa. The levels of miR-17-92 cluster and important components of Wnt/β-catenin signaling pathway were also explored. Cell biology assays were used to investigate biologic consequences of MIR17HG in regulating cell proliferation, colony formation and invasion, as well as the roles in regulating epithelial-mesenchymal transition (EMT). Moreover, a colon tumor xenograft model was established to explore the in vivo effect of MIR17HG on cell proliferation.
Results
Expression of MIR17HG was found to be elevated in colon cancer (P<0.001). Upregulation of MIR17HG was significantly correlated with lymph node metastasis (P=0.005) and TNM stage (P<0.001), and also an independent prognostic indicator of overall survival and disease-free survival in colon cancer patients. Suppression of MIR17HG inhibited colon cancer cell viability, invasion, and EMT process, and significantly reduced the ability of the cells to form tumors in vivo. Moreover, miR-17 and miR-18a, two key components of miR-17-92 cluster, were suppressed after knockdown of MIR17HG. Furthermore, the Wnt/β-catenin signaling, activated by miR-17 and miR-18a, was required for the oncogenic role of MIR17HG in colon cancer.
Conclusions
MIR17HG mediated upregulation of miR-17 and miR-18a promotes the progression of colon cancer via activating Wnt/β-catenin signaling.
Keywords: MIR17HG, miR-17, miR-18a, colon cancer, Wnt/β-catenin signaling
Introduction
Colon cancer is one of the most common malignancies and the second leading cause of cancer-related deaths worldwide (1). Recently, many improvements have been made in screening, detection and therapy for colon cancer. However, the prognosis of patients with advanced stage colon cancer remains poor because of tumor recurrence and metastasis (2,3). Despite a large number of genes have been associated with colon cancer, the findings failed to translate to clinical practice effectively (4). Therefore, it is crucial to identify efficient novel biomarkers for better prediction, diagnosis and disease management.
Increasing evidence has revealed that long non-coding RNAs (lncRNAs), a newly identified transcripts longer than 200 nucleotides with limited protein coding potential, can participate in various physiologic or pathologic processes via diverse mechanisms (5-7). Dysregulated lncRNAs have also been indicated to modulate important cancer hallmarks, such as proliferation, metastasis, and drug-resistance (8-10). As novel heterogeneous class of non-coding RNAs, lncRNAs account for much of the transcribed genome and gain widespread concern. Some lncRNAs harbor miRNAs within their exonic or intronic sequences, and hence are categorized as miRNA-host gene lncRNAs (lnc-miRHGs) (11). Nowadays, the clinical relevance of lnc-miRHGs in colon cancer remains largely unexplored. A great deal of studies have demonstrated that miR-17-92 cluster (composed of 6 miRNAs including miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a) is significantly overexpressed in digestive tract malignancies, such as gastric cancer, colon cancer, carcinoma and pancreatic cancer, and contributes to tumorigenesis and progression (12-15). As a novel lnc-miRHG, MIR17HG (miR-17-92 cluster host gene lncRNA) is known to be synthesized from miR-17-92 cluster host gene due to pre-miRNA processing, but limited evidence has clarified its roles in malignancies (16). These clues intrigued us to ascertain the potential functions of MIR17HG in cancer biology. Thus, we were interested in determining the possible association between MIR17HG and miR-17-92 cluster in colon cancer.
In the present study, we investigated the expression and clinical significance of MIR17HG, and firstly demonstrated a novel MIR17HG-miR-17/18a-Wnt/β-catenin signaling axis that mediated the progression of colon cancer.
Methods
Specimens and cell lines
Primary tumor samples and matched normal mucosa were randomly collected according to the standard operation procedures from 60 colon cancer patients (33 males and 27 females) who underwent surgery without prior chemotherapy or radiotherapy. A single sample (taken from the tumor center) was used from each cancer, and the matched normal mucosa was taken at least 5 cm away from the edge of the tumor tissue. The diagnosis of all specimens was histopathologically confirmed by at least two pathologists. The research was designed and performed according to the principles of Helsinki Declaration and was approved by the Institutional Ethics Committee of Taian City Central Hospital (approval No. TACH-20170414).
Six colon cancer cell lines RKO, Caco2, SW480, SW620, HT29 and Hct116, and the human normal colon mucosa cell line FHC were purchased from Genechem Co., (Shanghai, China). Cells were cultured in DMEM medium (Hyclone, Logan, UT, USA) containing 10% FBS (Gibco, Carlsbad, USA) in humidified air at 37 °C with 5% CO2.
Plasmid construction
The short hairpin RNAs (shRNAs) used for MIR17HG knockdown were synthesized by OBiO (Shanghai, China). Stably modified cells were selected with puromycin after transfection. The mimics used for hsa-miR-17 and hsa-miR-18a overexpression, and negative control were purchased from RiboBio (Guangzhou, China). Cells were transfected with sh-RNA or miRNA mimics at a final concentration of 100 or 60 nM, respectively. Transfections were carried out using Lipofectamie 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA from primary tumors, adjacent normal mucosa or culture cells was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and then reversely transcribed to cDNA by using PrimeScript RT reagent Kit (Takara, Dalian, China) for mRNA and lncRNA, or by using miRNA cDNA First Strand Synthesis (Tiangen, Beijing, China) for miRNA, respectively. qRT-PCR was utilized to determine the levels of lncRNA, mRNA and miRNA by using a SYBR Green reagent (Takara, Dalian, China). The primer sequences can be found in Table S1. The relative RNA level was normalized to GAPDH or U6, and calculated by using 2−ΔΔCt method.
Table S1. shRNA and primer sequences.
| Name of primer | Primer sequences |
|---|---|
| MIR17HG qPCR forward primer | 5'-CTCTGGAGGCTGAGGCAGGAG-3' |
| MIR17HG qPCR reverse primer | 5'-TGGAGTGCAGTGGCAGGATCTC-3' |
| c-Myc qPCR forward primer | 5'-CGAGGAGAATGTCAAGAGGCGAAC-3' |
| c-Myc qPCR reverse primer | 5'-GCTTGGACGGACAGGATGTATGC-3' |
| Cyclin D1 qPCR forward primer | 5'-TACCGCCTCACACGCTTCCTC-3' |
| Cyclin D1 qPCR reverse primer | 5'-ACCTCCTCCTCCTCCTCTTCCTC-3' |
| Survivin qPCR forward primer | 5'-TCTGGAGGTCATCTCGGCTGTTC-3' |
| Survivin qPCR reverse primer | 5'-TCCTCACTGCGGCTGTCTGG-3' |
| sh-MIR17HG | 5'-GCCTCTCAACTGAGCAGTA-3' |
Cell proliferation, colony formation and invasion assays
Cell counting kit-8 (CCK-8) assay was performed to examine cell viability. SW480 and Hct116 cells were cultured in the 96-well plate and measured with a CCK-8 kit (Dojindo Laboratories, Japan) at appropriate time (24, 48, 72, 96 hours). The 450 nm OD value was detected by a Gen5 microplate reader (BioTek, Winooski, VT, USA).
For plate colony formation assay, 1,000 cells were cultured in 6 cm plates with DMEM medium containing 10% FBS. Cells were fixed by methyl alcohol for 25 min and dyed with Giemsa solution for 30 min after 2 weeks. Visible colonies were counted and imaged. Experiments were independently repeated in triplicate.
For cell invasion assay, transwell chamber (Corning Costar) with membrane coated with Matrigel was used; 2×105 cells were seeded in the upper transwell chamber containing serum-free medium. Medium containing 10% FBS was added to the lower chamber. After incubation for 48 hours, cells that invaded through the membrane were fixed with methanol and then stained with 0.1% crystal violet, and then imaged by a microscope (magnification: ×200). Experiments were independently repeated in triplicate.
Western blot
Antibodies against nuclear-β-catenin and Lamin B1 were purchased from Cell Signaling Technology (CST, MA, USA). Western blot procedures were performed as previously described (17). The protein signal was determined by using the ECL detection reagents (Millipore, USA).
Confocal immunofluorescent assay
Different transfected cells were incubated with antibodies specific for E-cadherin (1:1,000, CST, MA, USA) or Vimentin (1:1,000, CST, MA, USA), and then incubated with goat anti-rat IgG (1:200, Alexa Fluor 488, Invitrogen, Carlsbad, CA, USA) or goat anti-rat IgG (1:200, Alexa Fluor 647, Invitrogen, Carlsbad, CA, USA), cells were examined using a confocal microscope (Olympus) after adding DAPI (Southern Biotech, SBA, Birmingham, AL).
Immunohistochemistry
Immunohistochemistry analysis was carried out following the manufacturer’s instructions. Antibody specific to β-catenin was purchased from Cell Signaling Technology company (CST, MA, USA). The final stainings were scored as described previously. Nuclear β-catenin samples were defined by at least weak positive nuclear staining in ≥25% cells of tumor parenchyma (18).
Tumor xenografts
Six weeks old female BALB/C nude mice were housed under standard conditions. SW480 cells transduced with sh-MIR17HG or sh-NC (1.0×106 cells) were injected subcutaneously into the groin of nude mice (n=5 for each group). Tumor xenografts were harvested and weighed after 31 days. The procedures were performed following the Taian City Central Hospital Animal Care Guidelines.
Statistical analysis
All statistical assays were performed by using SPSS 19.0 statistical software package (SPSS, Chicago, IL, USA). The differences between two groups or multiple groups was used the Student’s t-test, one-way ANOVAs or χ2 tests. Overall survival (OS) curves and disease-free survival (DFS) curves were plotted using the Kaplan-Meier method.
The statistical differences between survival curves were estimated by log-rank test. Univariate and multivariate cox regression analyses were also performed. P<0.05 was considered statistically significant.
Results
MIR17HG is upregulated in colon cancer and predicts poor prognosis
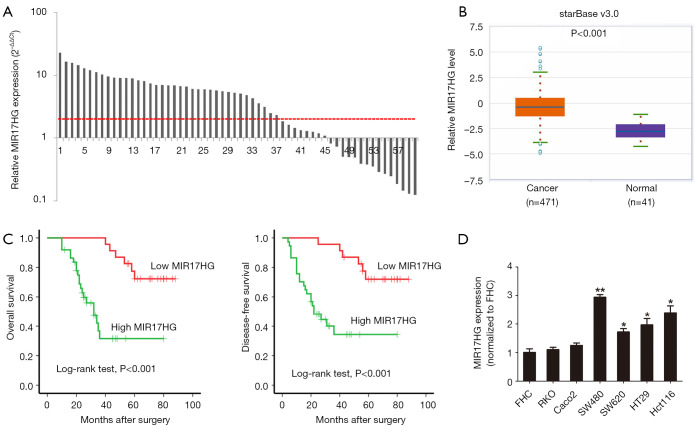
We detected the expression levels of MIR17HG in 60 paired colon cancer specimens. As shown in Figure 1A, 61.67% (37/60) colon cancer tissues showed a more than 2-fold increase in MIR117HG levels compared with adjacent normal mucosa (P<0.001). It is worth noting that MIR17HG was also found to be significantly upregulated in TCGA dataset of colon cancer samples (starBase v3.0, http://starbase.sysu.edu.cn/panCancer.php), which further supported our findings (Figure 1B). Further analysis of the 60 paired colon cancer samples cohort revealed that high MIR17HG expression in colon cancer tissues was associated with lymph node metastasis (P=0.005) and TNM stage (P<0.001), whereas no relationship was observed between MIR17HG expression and the age, gender, tumor location or differentiation (Table 1). Moreover, patients with higher MIR17HG levels exhibited worse overall survival (log-rank test, P<0.001) and disease-free survival (log-rank test, P<0.001) (Figure 1C). Multivariate analysis manifested that high MIR17HG level was an independent predictor for poor prognosis of colon cancer patients (Table 2). Next, we examined the expression of MIR17HG in a series of cell lines. Among the colon cancer cell lines, especially in SW480 and Hct116 cells, the MIR17HG levels were increased as compared with that in the normal colon mucosa cell line FHC (Figure 1D). These data suggested that MIR17HG was significantly overexpressed in colon cancer and associated with poor prognosis.
Figure 1.
MIR17HG expression levels in colon cancer and its clinical significance. (A) MIR17HG expression levels in 60 paired colon cancer tissues and adjacent noncancerous tissues. The results were normalized to adjacent noncancerous tissues (P<0.001). (B) MIR17HG expression levels in TCGA colon cancer cohort integrated by starBase v3.0 project (P<0.001). (C) Patients with higher MIR17HG levels exhibited worse overall survival and disease-free survival (P<0.001). (D) MIR17HG expression levels in normal colon mucosa cell line (FHC) and colon cancer cell lines. *P<0.05; **P<0.01.
Table 1. Association between MIR17HG expression and clinicopathological characteristics in colon cancer.
| Variable | Total (n=60) | Expression of MIR17HG | P | |
|---|---|---|---|---|
| High (n=37) | Low (n=23) | |||
| Age (years) | 0.082 | |||
| <65 | 23 | 11 | 12 | |
| ≥65 | 37 | 26 | 11 | |
| Gender | 0.471 | |||
| Male | 33 | 19 | 14 | |
| Female | 27 | 18 | 9 | |
| Location | 0.811 | |||
| Right | 22 | 14 | 8 | |
| Others | 38 | 23 | 15 | |
| pT stage | 0.062 | |||
| T1 | 6 | 1 | 5 | |
| T2 | 24 | 18 | 6 | |
| T3 | 21 | 12 | 9 | |
| T4 | 9 | 6 | 3 | |
| pN stage | 0.005* | |||
| N0 | 29 | 12 | 17 | |
| N1 | 12 | 9 | 3 | |
| N2 | 19 | 16 | 3 | |
| TNM stage | <0.001* | |||
| I/II | 25 | 8 | 17 | |
| III/IV | 35 | 29 | 6 | |
| Differentiation | 0.838 | |||
| Well | 32 | 19 | 13 | |
| Moderate | 21 | 13 | 8 | |
| Poor | 7 | 5 | 2 | |
| Vessel invasion | 0.327 | |||
| No | 41 | 27 | 14 | |
| Yes | 19 | 10 | 9 | |
*, P<0.05 represents the P values with significant differences.
Table 2. Univariate and multivariate analysis of overall survival and disease-free survival in colon cancer.
| Variable | Overall survival | Disease-free survival | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | ||||||||
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | ||||
| Age | 1.43 (0.63, 3.22) | 0.393 | – | – | 1.27 (0.58, 2.78) | 0.547 | – | – | |||
| Gender | 1.51 (0.70, 3.26) | 0.299 | – | – | 1.26 (0.60, 2.66) | 0.537 | – | – | |||
| Location | 0.97 (0.44, 2.14) | 0.937 | – | – | 1.07 (0.49, 2.33) | 0.859 | – | – | |||
| T stage | 1.01 (0.66, 1.54) | 0.960 | – | – | 0.96 (0.63, 1.44) | 0.829 | – | – | |||
| N stage | 2.76 (1.67, 4.55) | <0.001* | 2.23 (1.30, 3.83) | 0.004* | 2.18 (1.38, 3.45) | 0.001* | 1.84 (1.11, 3.05) | 0.019* | |||
| M stage | 16.60 (6.01, 45.89) | <0.001* | 9.43 (3.39, 26.27) | <0.001* | 10.67 (4.44, 25.66) | <0.001* | 6.65 (2.74, 16.10) | <0.001* | |||
| Differentiation | 1.30 (0.76, 2.23) | 0.345 | – | – | 1.12 (0.65, 1.92) | 0.681 | – | – | |||
| Vessel invasion | 0.60 (0.24, 1.49) | 0.268 | – | – | 0.70 (0.30, 1.66) | 0.419 | – | – | |||
| MIR17HG | 7.02 (2.55, 19.29) | <0.001* | 3.44 (1.07, 11.07) | 0.039* | 6.26 (2.30, 17.05) | <0.001* | 3.23 (1.02, 10.22) | 0.046* | |||
*, P<0.05 represents the P values with significant differences. HR, hazard ratio; 95% CI, 95% confidence interval.
Suppression of MIR17HG inhibits the progression of colon cancer in vitro and in vivo
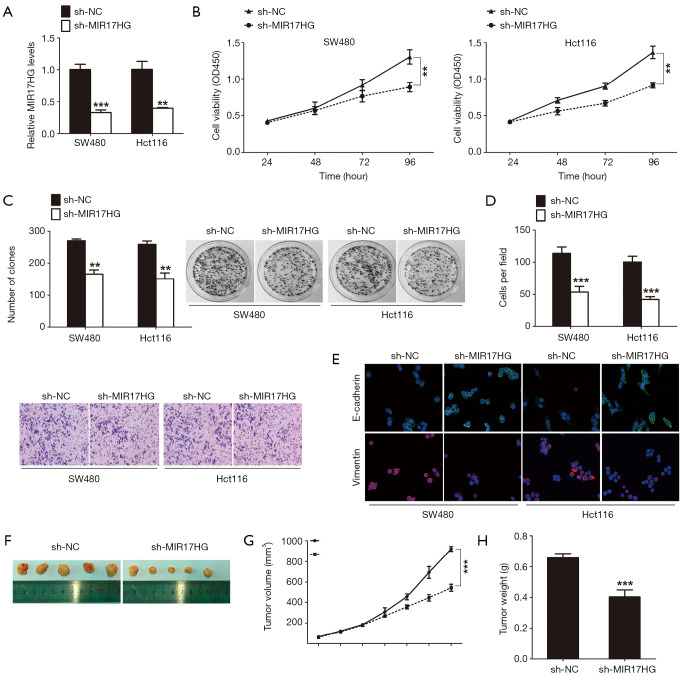
To assess the biological role of MIR17HG in colon cancer cells, we established stable MIR17HG knockdown models in SW480 and Hct116 cells. Successful knockdown of MIR17HG was confirmed via qRT-PCR analysis (Figure 2A). CCK-8 assays revealed that cell growth was significantly impaired in cells transfected with MIR17HG-shRNA compared with controls (Figure 2B). Similarly, knockdown of MIR17HG dramatically suppressed colon cancer cell colony formation abilities (Figure 2C). In addition, transwell assays showed that attenuation of MIR17HG inhibited cell invasion into Matrigel-coated transwell membranes (Figure 2D). Given that epithelial-mesenchymal transition (EMT) occurs during tumor progression, which provides cancer cells with metastatic properties, we next investigated the effect of MIR17HG knockdown on the expression levels of several EMT-related genes. As shown in Figure 2E, confocal immunofluorescent assays demonstrated that downregulation of MIR17HG increased the expression of epithelial marker E-cadherin with a simultaneous decrease in the expression of mesenchymal marker Vimentin. Furthermore, in vivo tumor xenografts experiment was also performed. We found that sh-MIR17HG transfected SW480 cells developed significantly smaller tumors than normal control group (Figure 2F,G,H). All these results indicated that suppression of MIR17HG inhibited the progression of colon cancer in vitro and in vivo.
Figure 2.
Suppression of MIR17HG inhibits the progression of colon cancer in vitro and in vivo. (A) Relative levels of MIR17HG in colon cancer cells after treatment with sh-MRI17HG or sh-NC. (B) CCK-8 assay was carried out to detect the viability of transfected colon cancer cells. (C) Plate colony formation assay was performed to examine the proliferation of transfected colon cancer cells. (D) Transwell assay was performed to determine the invasive ability of transfected colon cancer cells (×200). (E) Confocal immunofluorescent assay demonstrated that downregulation of MIR17HG increased the expression of epithelial marker E-cadherin with a simultaneous decrease in the expression of mesenchymal marker Vimentin (×400). (F,G,H) In vivo tumor xenografts experiment showed that sh-MIR17HG transfected SW480 cells developed significantly smaller tumors than normal control group. **P<0.01; ***P<0.001.
MIR17HG positively regulates the expression of miR-17 and miR-18a
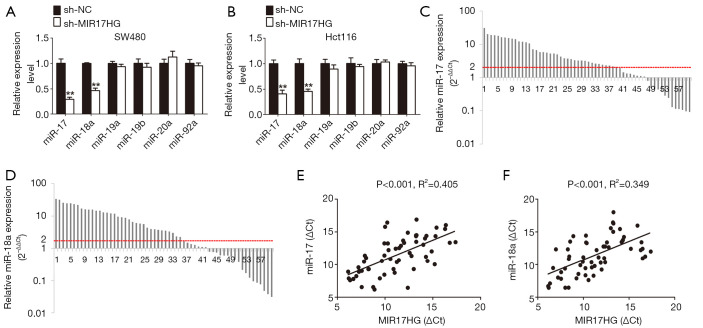
The lnc-miRHGs/miRNAs interaction may participate in the progression of human cancers (19). The miR-17-92 cluster, including miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a, has been associated with the development of a number of cancer diseases (20). Interestingly, MIR17HG was identified as a lnc-miRHG of miR-17-92 cluster (21). Therefore, to explore the possible effect of MIR17HG on miR-17-92 cluster members, we firstly profiled the expression levels of the six miR-17-92 family miRNAs in colon cancer cells in response to MIR17HG inhibition. As a result, miR-17 and miR-18a were significant downregulated in MIR17HG knockdown cells (Figure 3A,B). Furthermore, based on the 60 paired clinical samples, we found that miR-17 and miR-18a were markedly upregulated in colon cancer tissues compared with adjacent normal mucosa (Figure 3C,D). As expected, MIR17HG transcript level was positively correlated with miR-17 or miR-18a level, respectively (Figure 3E,F). These findings demonstrated a crucial role of MIR17HG in regulating miR-17 and miR-18a.
Figure 3.
MIR17HG positively regulates the expression of miR-17 and miR-18a in colon cancer. (A,B) Attenuation of miR-17 and miR-18a was observed in both SW480 and Hct116 cells after treatment with sh-MIR17HG. (C,D) miR-17 and miR-18a were markedly upregulated in colon cancer tissues compared with adjacent normal mucosa (n=60, P<0.001). (E,F) MIR17HG transcript level was positively correlated with miR-17 or miR-18a level in colon cancer tissues, respectively. **P<0.01.
MIR17HG promotes the proliferation, invasion and EMT process of colon cancer cells through upregulating miR-17 and miR-18a expression
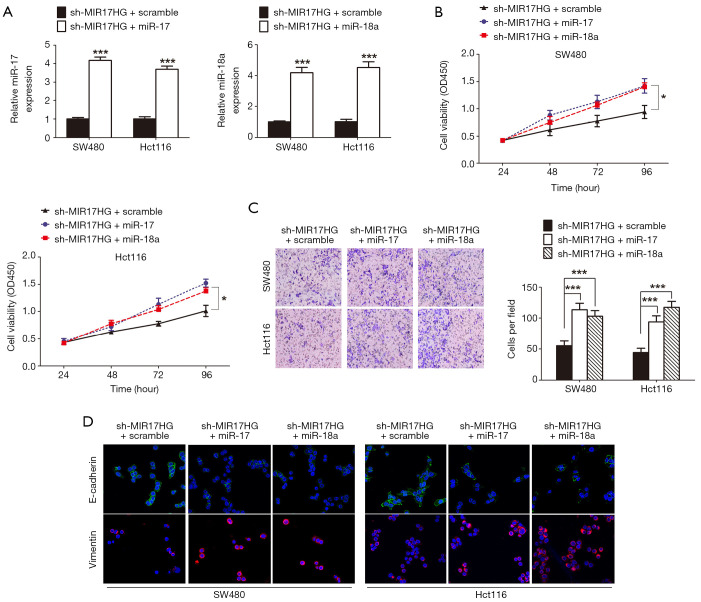
To ascertain whether the modulation of colon cancer progression by MIR17HG was dependent on miR-17 or miR-18a, we transduced miR-17 or miR-18a mimics to the MIR17HG-knockdown cells (Figure 4A). In CCK-8 assays, decreased cell viability was observed after inhibition of MIR17HG in both SW480 and Hct116 cells, which was reversed via overexpression of miR-17 or miR-18a (Figure 4B). Consistently, suppression of MIR17HG reduced cell invasive ability, but was abolished through ectopic expression of miR-17 or miR-18a (Figure 4C). Additionally, confocal immunofluorescent assays also revealed that depletion of MIR17HG increased E-cadherin expression, and induced loss of Vimentin, which was also abrogated by overexpression of miR-17 or miR-18a (Figure 4D). Taken together, these data confirmed the oncogenic role of MIR17HG in colon cancer via regulating miR-17 and miR-18a.
Figure 4.
MIR17HG promotes the proliferation, invasion and EMT process of colon cancer cells through upregulating miR-17 and miR-18a expression. (A) The expression of miR-17 or miR-18a was increased in MIR17HG-knockdown colon cancer cells after transduced with miR-17 or miR-18a mimic, respectively. (B) Elevated expression of miR-17 or miR-18a increased the viability of MIR17HG-knockdown colon cancer cells. (C) Ectopic expression of miR-17 or miR-18a abrogated the suppressive effects on cell invasion caused by MIR17HG-knockdown (×200). (D) Confocal immunofluorescent assay revealed that depletion of MIR17HG increased E-cadherin expression, and induced loss of Vimentin, which was also abrogated by overexpression of miR-17 or miR-18a (×400). *P<0.05; ***P<0.001.
Wnt/β-catenin signaling is responsible for the oncogenic role of MIR17HG-miR-17/miR-18a axis in colon cancer
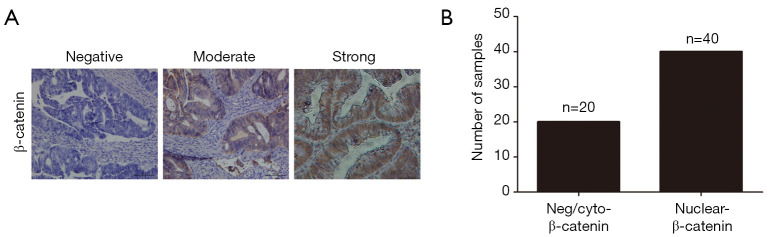
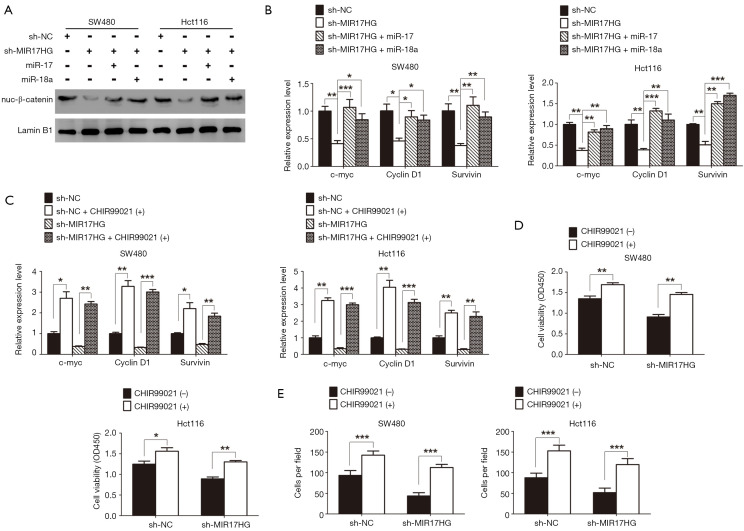
The redistribution of cytoplasmic β-catenin to nuclear localization which reflects the canonical signaling activation is known as one of the most vital mechanism in colon cancer (18). In the supplementary experiment, immunochemistry staining was performed to assess the β-catenin expression in our sample cohort. As expected, we found that nuclear accumulation of β-catenin was a frequent event in colon cancer tissues (Figure S1). As miR-17 and miR-18a could regulate Wnt/β-catenin signaling (22,23), we wondered whether this signaling confers the progression of colon cancer through involving in the MIR17HG-miR-17/miR-18a axis. Western blot analysis showed that depletion of MIR17HG in colon cancer cells resulted in decreased nuclear accumulation of β-catenin, whereas miR-17 or miR-18a overexpression abolished above effect (Figure 5A). Additionally, a series of downstream target genes of Wnt signaling, including c-Myc, Cyclin D1 and Survivin, were also downregulated in both SW480 and Hct116 cells after MIR17HG inhibition, whereas miR-17 or miR-18a overexpression abrogated the MIR17HG knockdown effect on these targets (Figure 5B). Moreover, CHIR99021, the activator of Wnt/β-catenin signaling, was utilized in further study. We found that induction of CHIR99021 re-activated the Wnt/β-catenin signaling in MIR17HG-knockdown cells (Figure 5C). Furthermore, in vitro assays revealed that depletion of MIR17HG decreased the proliferative and invasive abilities of SW480 and Hct116 cells, and these effects were rescued by activating Wnt/β-catenin signaling (Figure 5D,E). These findings suggested that Wnt/β-catenin signaling was required for the oncogenic role of MIR17HG-miR-17/miR-18a axis in colon cancer.
Figure S1.
Nuclear accumulation of β-catenin in colon cancer tissues. (A) Immunochemistry staining was performed to assess the β-catenin expression in colon cancer tissue samples (×200); (B) different expression levels of β-catenin in colon cancer tissues.
Figure 5.
Wnt/β-catenin signaling is responsible for the oncogenic role of MIR17HG-miR-17/miR-18a axis in colon cancer. (A,B) Depletion of MIR17HG in colon cancer cells caused the suppression of Wnt/β-catenin signaling, whereas miR-17 or miR-18a overexpression abrogated this effect. (C) The downstream target genes of Wnt/β-catenin signaling in sh-NC/sh-MIR17HG colon cancer cells were detected by qRT-PCR after treated with 10 µmol/L CHIR99021 for 36 hours. (D,E) Depletion of MIR17HG decreased the proliferative and invasive abilities of colon cancer cells, and these effects were rescued by activating Wnt/β-catenin signaling. *P<0.05; **P<0.01; ***P<0.001.
Discussion
In this study, we reported that lncRNA MIR17HG, which acts as the lnc-miRHG of miR-17-92 cluster, promotes colon cancer progression by upregulating miR-17 and miR-18a, two family members of miR-17-92 cluster, and then activating Wnt/β-catenin signaling. To our knowledge, this is the first report for MIR17HG-miR-17/18a-Wnt/β-catenin signaling axis.
LncRNAs are a group of non-coding RNAs with limited protein-coding capacity. Growing evidences have demonstrated a great deal of lncRNAs in regulation of various physiologic or pathologic processes (5,6). As a novel identified lncRNA, MIR17HG is categorized as miRNA-host gene lncRNA. Its gene locus is located on chromosome 13q31.3 and encodes the miR-17-92 gene cluster (21). The six mature miRNAs, including miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a, which derived from miR-17-92 cluster have been found to be amplified in plenty of malignancies and be linked to poor prognosis (24-26). Nevertheless, as the lnc-miRHG of miR-17-92 gene cluster, MIR17HG participating in the regulation of tumor progression are still seldom reported up to now. Here, we provide a new insight into the function of MIR17HG in colon cancer, and unveil that MIR17HG is clinically significant and might be used as a potential biomarker in colon cancer.
Few studies thus far have looked at the potential role of lnc-miRHGs/miRNAs interaction in tumor progression. For instance, as the lnc-miRHG of miR-31, LOC554202 promoted laryngeal squamous cell carcinoma by suppressing miR-31 expression (19). Moreover, miR-34 host gene lncRNA lncTAM34a positively regulated miR34a expression resulting in a decrease of several tumorigenic phenotypes (27). Additionally, LncRNA MIR100HG-derived miR-100 and miR-125b mediated cetuximab resistance, concomitant upregulation of MIR100HG and miR-100/125b occurs in cetuximab-resistant colorectal cancer and head and neck squamous cell cancer (28). Lastly, miR-22-3p was positively correlated with its host gene MIR22HG in hepatocellular carcinoma (29). All above studies intrigued us to wondered the potential correlation between MIR17HG and miR-17-92 gene cluster in colon cancer. Consistent with our speculation, miR-17-92 gene cluster family members (miR-17 and miR-18a) exhibited expression changes in sh-MIR17HG transfected colon cancer cells. MIR17HG influenced colon cancer progression via a miR-17/18a dependent manner.
Both miR-17 and miR-18a are involved in multiple progression of human cancers by activating Wnt/β-catenin signaling (22,23). The canonical Wnt/β-catenin signaling is known as one of the most vital pathways to accelerate the progression of colon cancer (18). Therefore, to determine whether MIR17HG contributes to colon cancer progression via miR-17/18a-Wnt/β-catenin signaling axis, we examined the activity of Wnt/β-catenin signaling in transfected cells, and the specific signaling activator CHIR99021 was also used in the research. Our findings revealed that MIR17HG mediated upregulation of miR-17 and miR-18a activates the Wnt/β-catenin signaling. Moreover, the suppressive effect of MIR17HG knockdown on colon cancer progression was significantly reversed by CHIR99021. These results indicated that MIR17HG induced colon cancer progression at least in part through modulating the miR-17/18a-Wnt/β-catenin signaling axis.
Since colon cancer is a heterogeneous tumor, it can be divided into different pathological types and molecular subtypes. A good understanding of these heterogeneities is important for the proper diagnosis and management of patients. On the other hand, some characteristics, such as MS status (mismatch repair) and KRAS/NRAS/PIK3CA mutation status, have been identified as the important features of colon cancer (30,31), and it would be interesting and significant to investigate their association with colon cancer. These are the focus of our future research.
Conclusions
In summary, we mainly focused on the downstream mechanism by which MIR17HG participates in colon cancer progression in this study. Our findings highlight the importance of lnc-miRHG/miRNA interaction in tumor progression. Taken together, the identification of MIR17HG and the involvement of lnc-miRHG/miRNA interaction in the aberrant MIR17HG-miR-17/18a-Wnt/β-catenin signaling axis may provide a promising option for investigation of tumors.
Acknowledgments
We thank Zhaofeng Zhu for his excellent technical assistance.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The present experiment was approved by the Institutional Ethics Committee of Taian City Central Hospital (approval No. TACH-20170414). All subjects were completely informed about the study goals and procedure and provided their signed informed consent.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.06.20). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Azvolinsky A. Colorectal cancer: to stack or sequence therapy? J Natl Cancer Inst 2015;107. doi: . 10.1093/jnci/djv138 [DOI] [PubMed] [Google Scholar]
- 3.Cunningham D, Atkin W, Lenz HJ, et al. Colorectal cancer. Lancet 2010;375:1030-47. 10.1016/S0140-6736(10)60353-4 [DOI] [PubMed] [Google Scholar]
- 4.Tejpar S, Bertagnolli M, Bosman F, et al. Prognostic and predictive biomarkers in resected colon cancer: current status and future perspectives for integrating genomics into biomarker discovery. Oncologist 2010;15:390-404. 10.1634/theoncologist.2009-0233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016;29:452-63. 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013;341:789-92. 10.1126/science.1240925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee S, Kopp F, Chang TC, et al. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 2016;164:69-80. 10.1016/j.cell.2015.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014;25:666-81. 10.1016/j.ccr.2014.03.010 [DOI] [PubMed] [Google Scholar]
- 9.Yue B, Cai D, Liu C, et al. Linc00152 Functions as a Competing Endogenous RNA to Confer Oxaliplatin Resistance and Holds Prognostic Values in Colon Cancer. Mol Ther 2016;24:2064-77. 10.1038/mt.2016.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun TT, He J, Liang Q, et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov 2016;6:784-801. 10.1158/2159-8290.CD-15-0921 [DOI] [PubMed] [Google Scholar]
- 11.Sun Q, Tripathi V, Yoon JH, et al. MIR100 host gene-encoded lncRNAs regulate cell cycle by modulating the interaction between HuR and its target mRNAs. Nucleic Acids Res 2018;46:10405-16. 10.1093/nar/gky696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kral J, Korenkova V, Novosadova V, et al. Expression profile of miR-17/92 cluster is predictive of treatment response in rectal cancer. Carcinogenesis 2018;39:1359-67. 10.1093/carcin/bgy100 [DOI] [PubMed] [Google Scholar]
- 13.Liu F, Cheng L, Xu J, et al. miR-17-92 functions as an oncogene and modulates NF-kappaB signaling by targeting TRAF3 in MGC-803 human gastric cancer cells. Int J Oncol 2018;53:2241-57. [DOI] [PubMed] [Google Scholar]
- 14.Quattrochi B, Gulvady A, Driscoll DR, et al. MicroRNAs of the mir-17~92 cluster regulate multiple aspects of pancreatic tumor development and progression. Oncotarget 2017;8:35902-18. 10.18632/oncotarget.16277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu H, Han C, Wu T. MiR-17-92 cluster promotes hepatocarcinogenesis. Carcinogenesis 2015;36:1213-22. 10.1093/carcin/bgv112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dews M, Fox JL, Hultine S, et al. The myc-miR-17~92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res 2010;70:8233-46. 10.1158/0008-5472.CAN-10-2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin Y, Chen W, Liu B, et al. MiR-200c Inhibits the Tumor Progression of Glioma via Targeting Moesin. Theranostics 2017;7:1663-73. 10.7150/thno.17886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yue B, Liu C, Sun H, et al. A Positive Feed-Forward Loop between LncRNA-CYTOR and Wnt/beta-Catenin Signaling Promotes Metastasis of Colon Cancer. Mol Ther 2018;26:1287-98. 10.1016/j.ymthe.2018.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang S, Wang J, Ge W, et al. Long non-coding RNA LOC554202 promotes laryngeal squamous cell carcinoma progression through regulating miR-31. J Cell Biochem 2018;119:6953-60. 10.1002/jcb.26902 [DOI] [PubMed] [Google Scholar]
- 20.Fang LL, Wang XH, Sun BF, et al. Expression, regulation and mechanism of action of the miR-17-92 cluster in tumor cells (Review). Int J Mol Med 2017;40:1624-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leng X, Ma J, Liu Y, et al. Mechanism of piR-DQ590027/MIR17HG regulating the permeability of glioma conditioned normal BBB. J Exp Clin Cancer Res 2018;37:246. 10.1186/s13046-018-0886-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu F, Lu Z, Huang K, et al. MicroRNA-17-5p-activated Wnt/beta-catenin pathway contributes to the progression of liver fibrosis. Oncotarget 2016;7:81-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong P, Xiong Y, Yu J, et al. Control of PD-L1 expression by miR-140/142/340/383 and oncogenic activation of the OCT4-miR-18a pathway in cervical cancer. Oncogene 2018;37:5257-68. 10.1038/s41388-018-0347-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Wang Y, Dou J, et al. Acetylation of AGO2 promotes cancer progression by increasing oncogenic miR-19b biogenesis. Oncogene 2019;38:1410-31. 10.1038/s41388-018-0530-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baumgartner U, Berger F, Hashemi Gheinani A, et al. miR-19b enhances proliferation and apoptosis resistance via the EGFR signaling pathway by targeting PP2A and BIM in non-small cell lung cancer. Mol Cancer 2018;17:44. 10.1186/s12943-018-0781-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K, Zhang L, Zhang M, et al. Prognostic value of high-expression of miR-17-92 cluster in various tumors: evidence from a meta-analysis. Sci Rep 2017;7:8375. 10.1038/s41598-017-08349-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serviss JT, Andrews N, Van den Eynden J, et al. An antisense RNA capable of modulating the expression of the tumor suppressor microRNA-34a. Cell Death Dis 2018;9:736. 10.1038/s41419-018-0777-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y, Zhao X, Liu Q, et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/beta-catenin signaling. Nat Med 2017;23:1331-41. 10.1038/nm.4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang DY, Zou XJ, Cao CH, et al. Identification and Functional Characterization of Long Non-coding RNA MIR22HG as a Tumor Suppressor for Hepatocellular Carcinoma. Theranostics 2018;8:3751-65. 10.7150/thno.22493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li SKH, Martin A. Mismatch Repair and Colon Cancer: Mechanisms and Therapies Explored. Trends Mol Med 2016;22:274-89. 10.1016/j.molmed.2016.02.003 [DOI] [PubMed] [Google Scholar]
- 31.Kovaleva V, Geissler AL, Lutz L, et al. Spatio-temporal mutation profiles of case-matched colorectal carcinomas and their metastases reveal unique de novo mutations in metachronous lung metastases by targeted next generation sequencing. Mol Cancer 2016;15:63. 10.1186/s12943-016-0549-8 [DOI] [PMC free article] [PubMed] [Google Scholar]