Abstract
Background
Studies have shown that S100A8 and S100A9 are highly expressed in a variety of tumors, including NPC, and are associated with tumor invasion and migration. MMPs are associated with the invasion and migration of tumor cells. To further investigate the mechanism by which S100A8 and S100A9 affect the invasion and migration of NPC cells, the present study examined the effects of S100A8 and S100A9 on MMPs in NPC CNE-2 cells.
Methods
Recombinant pEGFP-N1-S100A8 and recombinant pEGFP-N1-S100A9 overexpression vectors and S100A8 and S100A9 RNA interference (RNAi) vectors were constructed and transfected into NPC CNE-2 cells. The transfection efficiency in each group of cells was assessed, and the gene and protein expression of MMP7, MMP9 and MMP12 were determined.
Results
The transfection efficiency was approximately 60–70%. Compared with those in the control group, the expression levels of MMP7, MMP9 and MMP12 in the S100A8 and S100A9 overexpression groups was significantly higher (P<0.05), and the expression levels of MMP7, MMP9 and MMP12 in the S100A8-RNAi and S100A9-RNAi groups were significantly lower (P<0.05). The number of cells in S100A8 overexpression group and S100A9 overexpression group at 24, 48 and 72 h was higher than that in RNAi group, RNAi control group, overexpression control group and normal control group, with statistical significance; The cell doubling time in S100A8 and S100A9 overexpression group was significantly shorter than that in RNAi control group, overexpression control group and normal control group, with statistical significance.
Conclusions
High S100A8 and S100A9 expression may promote the expression of MMP7, MMP9 and MMP12, which are related to the invasion and metastasis of NPC cells.
Keywords: S100 calcium-binding protein A8 (S100A8), S100 calcium-binding protein A9 (S100A9), CNE-2 cells, matrix metalloproteinase (MMP), overexpression
Introduction
Nasopharyngeal carcinoma (NPC) is a highly malignant tumor derived from nasopharyngeal epithelial cells. Most NPC is characterized by high malignancy, rapid growth and easy metastasis. The proliferation and apoptosis of NPC cells play an important role in the occurrence, development, diagnosis, treatment and prognosis of NPC. S100 A8 and S100 A9 are active proteins with low molecular weight. They are members of S100 protein family, and form heterodimer complex S100A8/A9 through specific binding in cytoplasm. The combined complex can regulate the expression and activity of various proteins, and has effects on cell migration, cell maturation and apoptosis functional expression and other biological processes have obvious regulatory effect. Studies have shown that S100A8 and S100A9 are highly expressed in a variety of tumors (1,2) and have significant correlations with gastric mucosal infiltration, lymph node metastasis, and gastric cancer cell invasion (3). A preliminary study (4) found that S100A8 and S100A9 are highly expressed in nasopharyngeal carcinoma (NPC) tissue and are closely correlated to the differentiation degree, tumor, node and metastasis (TNM) staging and lymph node metastasis of NPC. Patients with positive S100A8 and S100A9 expression had the lowest 5-year survival rate, and the 5-year survival rate for patients who were negative for S100A8 and S100A9 expression was relatively high. The results showed that S100A8 and S100A9 were correlated with the invasion, metastasis and survival of NPC to a certain extent.
Matrix metalloproteinases (MMPs) can degrade a variety of proteins, such as extracellular matrix proteins, mucoprotein and proteoglycans. Therefore, MMPs are closely associated with tumor cell invasion and metastasis (5). In this study, the effects of S100A8 and S100A9 on the invasion and metastasis of NPC were investigated by upregulating or downregulating the expression of S100A8 and S100A9 and observing the effects on MMPs in NPC CNE-2 cells.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-21-441).
Methods
Reagents
The human NPC CNE-2 cell line was purchased from the Cell Bank Shanghai Institutes for biological research, Chinese Academy of Sciences, Chinese Academy of Sciences. Trypsin solution was purchased from Shanghai ZhiYou Biotechnology Co., Ltd. GE-HyClone fetal bovine serum was purchased from Shanghai Shenbang Biological Co., Ltd. S100A8 mouse anti-human IgG antibody, S100A9 mouse anti-human IgG antibody, MMP7 rabbit anti-human IgG antibody were purchased from Abcam Bioengineering Co., UK; Solarbio HRP-conjugated goat anti-mouse IgG, Solarbio HRP-conjugated mouse anti-rabbit IgG, and Solarbio HRP-conjugated goat anti-rabbit IgG were purchased from Shuanghai Jingkehuaxue Co., Ltd. DAB (Dako REALTM EnVisionTM Detection System) chromogenic reagent kits were purchased from Shanghai MSK Biological Co., Ltd. Recombinant pEGFP-N1-S100A8 and recombinant pEGFP-N1-S100A9 overexpression vectors and S100A8 and S100A9 RNA interference (RNAi) vectors were constructed by Shanghai GenePharma Co., Ltd . LipofectamineTM 2000 was purchased from Invitrogen. Primers were synthesized by GenePharma (Shanghai, China).
Primer design
According to the data of S100A8 gene (NM_002964) and S100A9 gene (NM_002965) in NCBI (national center for biotechnology information) database, open reading frame was used to design synthetic primers. Restriction sites were added to the 5 'end of the primers according to the expression vector. The primer sequence was shown in Table 1.
Table 1. S100A8 and S100A9 primer sequences.
| Genes | Primer | Primer sequence |
|---|---|---|
| S100A8 | Forward primer | 5'- CCGCTCGAGATAGTTGGCGGCAATGGGAC-3' XhoI |
| Reverse primer | 5'- GGAATTC TTAGGGATTCTTAGAAGTTCTACC -3' EcoRI | |
| S100A9 | Forward primer | 5'- CGGGATCCCCACATTGATCTGACTTTCAGTATAATC-3'BamHI |
| Reverse primer | 5'- GGAATTCCGATCGTAATAGCACATGAATGAGC C-3' EcoRI |
Cell culture and transfection
CNE-2 cells were cultured in RPMI (Roswell Park Memorial Institute)-1640 medium containing 10% fetal bovine serum in an incubator with 5% CO2. Cells in the logarithmic growth phase or late logarithmic growth phase were trypsinized, resuspended in complete culture medium, and seeded at a density of 6×105/well in a 6-well culture plate. Transfection was performed based on the LipofectamineTM 2000 product manual. Cells were divided into 7 groups: S100A8-RNAi, S100A8 overexpression, S100A9-RNAi, S100A9 overexpression, siRNA control, overexpression control and blank control.
Determination of the expression levels of related genes using reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted from the cells of each group using the TRIzol method. Total RNA was reverse transcribed into cDNA. The S100A8, S100A9, MMP7, MMP9 and MMP12 genes were detected using PCR. The primer sequences are shown in Table 2. The reaction conditions were as follows: 94 °C for 10 min, followed by 94 °C for 15 s and amplification, for 30 cycles. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal control. The data obtained from real-time fluorescence quantitative PCR were analyzed using the 2−ΔΔCT method. Before using 2−ΔΔCT method, we verified the amplification efficiency of target gene and reference gene. The amplification efficiency of target gene and reference gene is close to 100%, and the deviation between them is less than 5%. The expression levels of each gene were statistically analyzed.
Table 2. RT-PCR primer sequences.
| Genes | Primer | Primer sequence |
|---|---|---|
| MMP7 | Forward primer | 5'-GGAACAGGCTCAGGACTATCTC-3' |
| Reverse primer | 5'-CAACATCTGGCACTCCACA-3' | |
| MMP9 | Forward primer | 5'-CGAGGCTGTATCGAACACAGTC-3' |
| Reverse primer | 5'-CACAACATTCTGGCCCACAAT-3' | |
| MMP12 | Forward primer | 5'-ACAGGCTCGACTAGAGTAC-3' |
| Reverse primer | 5'-GCATCCATCCACTCGAGCTACA-3' | |
| GAPDH | Forward primer | 5'-GAGTCCACTGGCGTCTTCA-3' |
| Reverse primer | 5'-GGGGTGCTAAGCAGTTGGT-3' |
Western blot analysis of protein expression
Antibodies (primary antibodies) against S100A8, S100A9, MMP7, MMP9, MMP12 and other proteins were diluted in blocking solution and added to the appropriate plates. The plates were placed in an incubator at 4 °C overnight. After the reaction solution was discarded, the plates were rinsed with Tris-buffered saline containing 0.1% Tween® 20 (TBST) 3 times. The corresponding HRP-conjugated IgG antibody (secondary antibody) for each protein of interest was diluted 1,000-fold and added to the plates, which were then shaken at 50 rpm for 2 h. After the secondary antibody reaction solution was discarded, the plates were washed with TBST solution 3 times. Color development was performed according to the instructions provided with the DAB color development kit. Western blot bands were observed and recorded using a gel imaging system, and the protein expression levels of individual genes were analyzed using Bandscan 5.0 software.
Detect cell proliferation
The recombinant S100A8 and S100A9 overexpression vectors were constructed and transfected into human nasopharyngeal carcinoma cell line CNE-2. After confirming that the transfected cells in each group were in logarithmic growth period, prepare cell suspension. CNE-2 cells were added into each well of each well. After 24, 48 and 72 h of culture, MTT(3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) solution was added into the culture plate. The 492 nm absorbance of each well was measured by enzyme reader.
Detection of doubling time of cell
After confirming that the transfected cells in each group were in logarithmic growth period, cell suspension was prepared. The number of cells at 24, 36, 48 and 72 h was counted by blood cell counting board, and calculate the doubling time was.
Statistical analysis
SPSS 19.0 software was used for the statistical analyses. Measurement data are expressed as the mean ± standard deviations (). Intergroup comparisons were performed using analysis of variance (ANOVA). Pairwise comparisons between groups were performed using the t test. Count data are expressed as n (%), and intergroup comparisons were performed using the χ2 test. P<0.05 indicated that a difference was statistically significant.
Results
Results of cell transfection
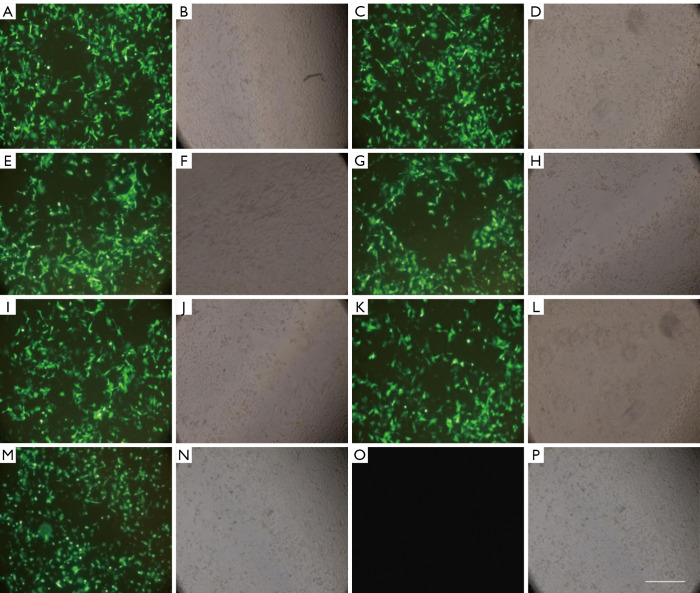
The transfection efficiency and success rate of cell transfection in each group were observed under an inverted fluorescence microscope. After transfection, the expression of fluorescent protein was relatively high in CNE-2 cells in the S100A8-RNAi group, S100A8 overexpression group, S100A9-RNAi group, S100A9 overexpression group, siRNA control group, and overexpression control group; the transfection efficiency was approximately 60–70%. No fluorescence was observed in the cells in the blank control group, indicating that cells in all other groups were successfully transfected. Detailed results are shown in Figure 1.
Figure 1.
CNE-2 cells in each group 24 h after transfection. (A,B) S100A8-siRNA; (C,D) S100A8-overexpression; (E,F) control siRNA; (G,H) S100A9-siRNA; (I,J) S100A9-overexpression; (K,L) control siRNA; (M,N) control overexpression; (O,P) normal control; bar indicates 100 µm. S100A8, S100 calcium-binding protein A8; S100A9, S100 calcium-binding protein A9.
Effects of S100A8 and S100A9 on the expression of MMPs
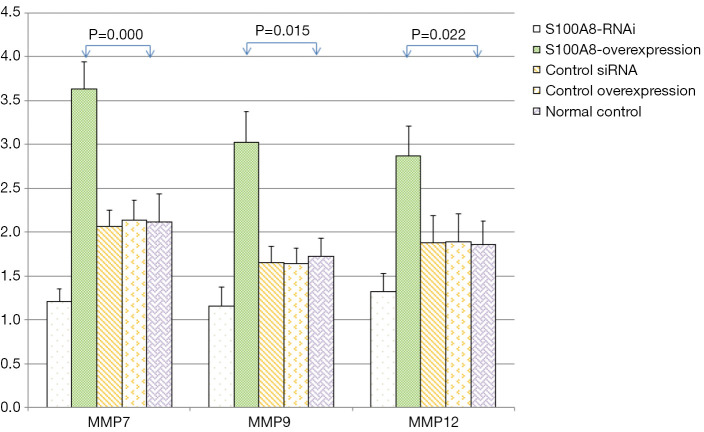
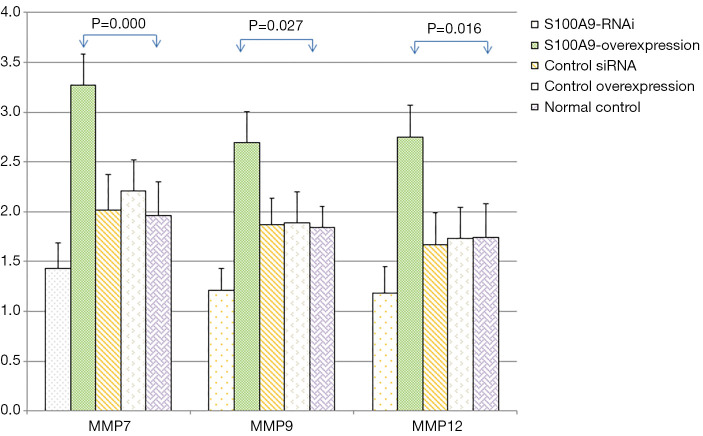
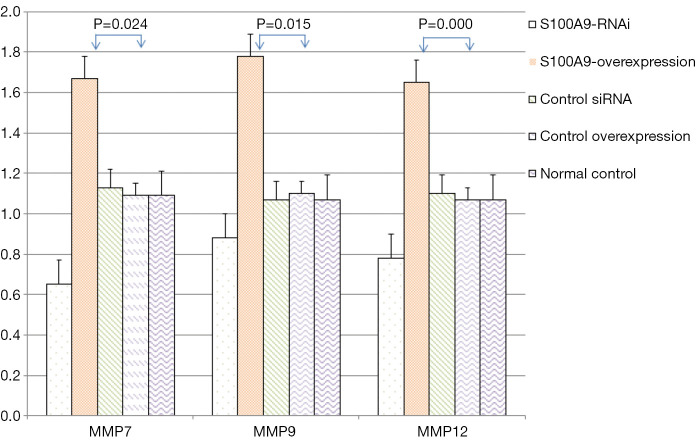
The expression levels of MMP7, MMP9, MMP12, and GAPDH in the cells in each S100A8 and S100A9 group were analyzed. The results showed that there was no significant difference in GAPDH expression among the groups (P>0.05). The expression levels of MMP7, MMP9, and MMP12 in the cells in the S100A8-RNAi group and S100A9-RNAi group were significantly lower than those in the cells in the control group (P<0.05). The expression levels of MMP7, MMP9 and MMP12 in the S100A8-overexpression group and S100A9-overexpression group were significantly higher than those in the control group (P<0.05). The expression levels of MMP7, MMP9 and MMP12 in the siRNA control group and overexpression control group were not significantly different from those in the blank control group (P>0.05), suggesting that the high expression levels of S100A8 and S100A9 might promote the expression of MMP7, MMP9, and MMP12. The detailed results are shown in Figure 2 and Figure 3.
Figure 2.
Effects of S100A8 on the expression levels of related genes (). Compared with the normal control group, the difference was significant (P<0.05). S100A8, S100 calcium-binding protein A8.
Figure 3.
Effects of S100A9 on the expression levels of related genes (). Compared with the normal control group, the difference was significant (P<0.05). S100A9, S100 calcium-binding protein A9.
Effects of S100A8 and S100A9 on the expression levels of related proteins
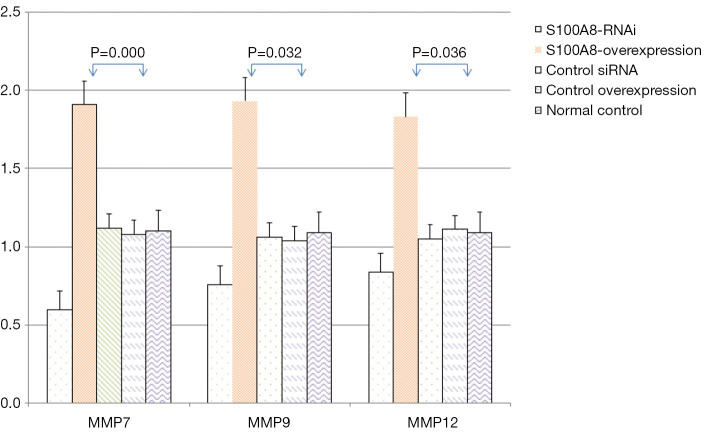
The protein expression levels of MMP7, MMP9 and MMP12 in CNE-2 cells in the S100A8 and S100A9 groups were assessed by Western blot. The results showed that there was no significant difference in GAPDH protein expression among the groups (P>0.05). The protein expression levels of MMP7, MMP9, and MMP12 in the S100A8-RNAi group and S100A9-RNAi group were lower than those in the siRNA control group, the overexpression control group, and the normal control group. The protein expression levels of MMP7, MMP9, and MMP12 in the S100A8 overexpression group and the S100A9 overexpression group were higher than those in the control groups. These findings indicate that the protein expression levels of MMP7, MMP9, and MMP12 were positively correlated with S100A8 and S100A9 expression. The detailed results are shown in Figure 4 and Figure 5.
Figure 4.
Effects of different levels of S100A8 on the expression of related proteins. Compared with the normal control group, the difference was significant (P<0.05). S100A8, S100 calcium-binding protein A8.
Figure 5.
Effects of different levels of S100A9 on the expression of related proteins. Compared with the normal control group, the difference was significant (P<0.05). S100A9, S100 calcium-binding protein A9.
By comparing the effects of S100A8 and S100A9 on the expression of related genes and proteins, it was found that the interference effect of S100A8 on MMP7, MMP9 and MMP12 expression might be stronger than that of S100A9.
Proliferation of nasopharyngeal carcinoma CNE-2 cells
At 0 h, the OD492 (absorbance) of CNE-2 cells in each group was 0.15±0.02, and there was no significant difference between each group (P>0.05). In the experiment, with the extension of culture time, the number of cells in each group with long culture time increased compared with that in the group with short time, and the number of cells in S100A8-RNAi group and S100A9-RNAi group increased at 24, 48 and 72 h. Compared with RNAi control group, overexpression group, overexpression control group and normal control group, the number of cells in S100A8 overexpression group and S100A9 overexpression group was significantly lower at 24, 48 and 72 h. The OD492 values in S100A8-RNAi group, S100A9-RNAi group, S100A8 overexpression group and S100A9 overexpression group were higher than those in RNAi group, RNAi control group, overexpression control group and normal control group at 24, 48 and 72 h, respectively. Compared with the normal control group at the same time, the difference was statistically significant (P<0.05). The RNAi control group and overexpression control group were cultured for 24, 48 and 72 h respectively After 24 hours, OD492 measured values were compared with the normal control group, and the differences were not statistically significant (P>0.05). It suggested that the proliferation rate of cells in S100A8-RNAi group and S100A9-RNAi group was significantly slower than that of normal CNE-2 cells. Compared with the control group, the proliferation rate of cells in S100A8 overexpression group and S100A9 overexpression group after intervention was significantly lower. The cell proliferation rate was significantly faster than that of the former. The detailed results are shown in Table 3.
Table 3. The effects of S100A8 and S100A9 on proliferation of CNE-2 cells ().
| Group | 0 h | 24 h | 48 h | 72 h |
|---|---|---|---|---|
| S100A8-RNAi | 0.15±0.02 | 0.23±0.03* | 0.31±0.02* | 0.37±0.03* |
| S100A8-Overexpression | 0.15±0.02 | 0.32±0.04* | 0.41±0.03* | 0.51±0.03* |
| S100A9-RNAi | 0.15±0.02 | 0.22±0.03* | 0.26±0.02* | 0.31±0.02* |
| S100A9-Overexpression | 0.15±0.02 | 0.29±0.03* | 0.37±0.03* | 0.52±0.03* |
| Control siRNA | 0.15±0.02 | 0.27±0.04 | 0.36±0.03 | 0.43±0.04 |
| Control Overexpression | 0.15±0.02 | 0.28±0.03 | 0.36±0.03 | 0.44±0.04 |
| Normal control | 0.15±0.02 | 0.28±0.03 | 0.37±0.03 | 0.44±0.03 |
*, compared with the normal control group, the difference was statistically significant (P<0.05).
Doubling time of nasopharyngeal carcinoma CNE-2 cells
Compared with RNAi control group, overexpression control group and normal control group, cell doubling time of S100A8-RNAi group and S100A9-RNAi group was relatively longer, and cell doubling time of S100A8 and S100A9 overexpression groups was significantly shorter than that of RNAi control group, overexpression control group and normal control group. The doubling time of S100A8-RNAi group, S100A9-RNAi group, S100A8 overexpression group and S100A9 overexpression group was significantly different from that of normal control group (P<0.05). There was no significant difference in doubling time among RNAi control group, overexpression control group and normal control group (P>0.05). These results suggest that with the increase of S100A8 and S100A9 expression, the proliferation rate of CNE-2 cells is increased. The detailed results are shown in Table 4.
Table 4. Effects of S100A8 and S100A9 on doubling time and cell cycle of CNE-2 cells.
| Group | Doubling time (h) |
|---|---|
| S100A8-RNAi | 42.24±3.47* |
| S100A8-Overexpression | 24.63±5.23* |
| S100A9-RNAi | 56.17±5.34* |
| S100A9-Overexpression | 29.23±5.51* |
| Control siRNA | 36.27±5.52 |
| Control overexpression | 33.28±5.06 |
| Normal control | 32.35±4.73 |
*, compared with the normal control group, the difference was statistically significant (P<0.05).
Discussion
The S100A8 and S100A9 genes are located in the long arm of chromosome 1 (1q21 band), and the 1q21-q23 band is a target region for the activation of malignant tumor gene transcription. The stability of this region is highly vulnerable to external stimuli and to changes in the internal environment and in some cytokine levels and is prone to mutations, such as deletions and translocations. Studies have shown that chromosome alterations in this region are related to tumorigenesis (6) and that the S100A8 and S100A9 proteins are closely correlated to the development, progression, infiltration, metastasis, and recurrence of a variety of tumors (7).
Researchers have examined the protein expression levels of S100A8 and S100A9 in NPC by immunohistochemistry, ELISA, and Western blot, and the levels of S100A8 and S100A9 proteins were significantly higher than those in normal tissues (8). A preliminary study (4) indicated that this abnormally high expression was positively correlated with higher TNM staging and a poor clinical prognosis, suggesting that S100A8 and S100A9 play roles in tumor invasion. Lim et al. (9) showed that monocytes/macrophages in the metastatic hepatoma microenvironment can enhance the invasion of metastatic hepatoma by inducing the high protein expression of S100A8 and S100A9 in tumor cells and promoting the proliferation and angiogenesis of tumor cells; additionally, blocking S100A8 or S100A9 protein expression reduced the migration and invasion potential of cells in tissue culture, as well as the formation of metastatic hepatoma and the metastasis and invasion of hepatocytes.
In this study, recombinant S100A8 and S100A9 RNAi expression vectors and recombinant S100A8 and S100A9 overexpression vectors were transfected into CNE-2 cells, and the gene and protein expression levels of MMP7, MMP9 and MMP12 significantly decreased when the expression levels of S100A8 and S100A9 were decreased. The expression levels of MMP7, MMP9 and MMP12 also significantly increased in CNE-2 cells after an increase in S100A8 and S100A9 expression. The interference effect of S100A8 on the expressions of MMP7, MMP9 and MMP12 might be stronger than that of S100A9, indicating that S100A8 has substantial biological activity in the S100A8/A9 complex, while S100A9 plays a role in maintaining structural stability (10). S100A8 and S100A9 form the S100A8/A9 heterodimeric complex (11) in a calcium-dependent manner. S100A8 is the main biologically active protein, while S100A9 ensures the functional stability of the S100A8/A9 heterodimeric complex by regulating S100A8 (12). Nishimura et al. (13) proposed that S100A8 and S100A9 proteins and S100A8/A9 dimers had significant regulatory effects on cell invasion and migration. We found that S100A8 and S100A9 both promoted the expression of MMP7, MMP9 and MMP12. MMP7, MMP9, and MMP12 are representative MMPs that can destroy the basal lamina of cells and promote the infiltration and/or metastasis of tumor cells by degrading a variety of proteins, such as mucoprotein and proteoglycans. Therefore, it is speculated that S100A8 and S100A9 can regulate the expression of MMP to play a role in the invasion and metastasis of NPC cells. This may be one of the reasons that S100A8 and S100A9 are closely correlated to the differentiation degree, TNM stage and lymph node metastasis of NPC.
MMPs are a family of enzymes that are dependent on Ca2+ and Zn2+ and can degrade all components of extracellular matrix proteins. MMP7 can not only specifically bind to a variety of substrate proteins but can also hydrolyze proteins in the basal lamina (14,15) and mutually promote the expression of vascular endothelial growth factor in the tumor microenvironment to promote the formation and growth of blood vessels therein, ultimately promoting tumor infiltration, metastasis, and recurrence through these pathways (16). MMP9 exerts a regulatory function on cell adhesion abilities and effects, which can not only loosen inter tissue adhesion but also condense the tissue structure, thereby regulating tumor metastasis and releasing a large amount of MMP9 when the tissue breaks down. These processes can form a positive feedback loop, which can further promote tumor metastasis and infiltration (17,18). MMP12 can specifically bind to elastin in the arterial wall as a substrate and degrade proteins such as type IV collagen, fibronectin, and laminin in the extracellular matrix. Studies (19) have suggested that MMP12 is closely related to cell metastasis, invasion and apoptosis of some malignant tumors.
Tumor invasion and metastasis are complex processes. Degradation of the extracellular matrix is a critical step for tumor cell invasion and migration. MMPs are key enzymes for the degradation of many proteins, including mucoprotein, extracellular matrix proteins and proteoglycans. S100A8 and S100A9 can regulate the expression of MMP2 and MMP9 by regulating the intracellular calcium concentration (20). A study of SNU484 human gastric cancer cells showed that when the expression of S100A8 or S100A9 was inhibited using siRNA interference, the invasion and migration phenotypes of SNU484 cells were significantly inhibited, and the expression of MMP2 was also significantly inhibited, indicating that S100A8 and S100A8 were the genes necessary for the transcription and activation of the MMP2 gene in SNU484 cells. Some studies (21) transfected CNE1 cells with siRNA-S100A8 or siRNA-S100A9 to downregulate the expression of S100A8 or S100A9 and found that compared with the control group, siRNA-S100A8 inhibited MMP7 expression by 63.57% and siRNA-S100A9 inhibited MMP7 expression by 41.36%, significantly inhibiting the migration-promoting effects of MMP7 on CNE1 cells. And researchers found that by regulating the level of MMPs, it can inhibit the growth of lung cancer cells (22).
Kwon et al. (23) studied gastric cancer cells and determined that S100A8 and S100A9 activated nuclear factor kappa B (NF-κB) through p38 mitogen-activated protein kinase (MAPK) phosphorylation. The activation of p38 MAPK and NF-κB induced the invasion and migration of gastric cancer cells, and p38 MAPK and NF-κB might be involved in the upregulation of MMP2 and MMP12 in gastric cancer cells transfected with S100A8 and S100A9. Other studies (24) have shown that S100A9 upregulated MMP9 expression through the Wnt/β-catenin signaling pathway and promoted the migration of rectal cancer cells and that S100A8 and S100A9 upregulated the Wnt/β-catenin signaling pathway by enhancing the expression of β-catenin and promoting the transcription of the signaling pathway target genes c-myc and MMP7.
In summary, we believe that the overexpression of S100A8 and S100A9 promotes the expression of MMPs through multiple signaling pathways, activating their functions. By degrading the extracellular matrix proteins, the invasive and metastatic capabilities of CNE-2 cells are enhanced, resulting in NPC cells that are prone to lymph node metastasis or hematogenous metastasis. S100A8 and S100A9 play roles in the regulation of the development, invasion, and metastasis of NPC cells by altering the conditions of the tumor microenvironment. This provides a theoretical basis for S100A8 and S100A9 as targets for NPC treatment.
Acknowledgments
Funding: Project supported by fund of the Health commission of Hubei Province (No: WJ2019F039).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-21-441
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-21-441
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-21-441). The authors have no conflicts of interest to declare.
References
- 1.Reeb AN, Li W, Sewell W, et al. S100A8 is a novel therapeutic target for anaplastic thyroid carcinoma. J Clin Endocrinol Metab 2015;100:E232-42. 10.1210/jc.2014-2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jo S H, Heo W H, Son H Y, et al. S100A8/A9 mediate the reprograming of normal mammary epithelial cells induced by dynamic cell–cell interactions with adjacent breast cancer cells. Scientific reports 2021;11:1337. 10.1038/s41598-020-80625-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwon CH, Moon HJ, Park HJ, et al. S100A8 and S100A9 promotes invasion and migration through p38 mitogen-activated protein kinase-dependent NF-κB activation in gastric cancer cells. Mol Cells 2013;35:226-34. 10.1007/s10059-013-2269-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu W, Wang G, Ye L, et al. Correlation of expressions of S100A8 and S100A9 and its prognostic potential in nasopharyngeal carcinoma. Trop J Pharm Res 2017;16:2577-83. 10.4314/tjpr.v16i11.2 [DOI] [Google Scholar]
- 5.Hu M, Sun X. Research progress of the relationship between extracellular matrix, matrix metalloproteinase and malignant tumor. J Cancer Pharmacy 2016;6:26-30. [Google Scholar]
- 6.Srikrishna G. S100 A8 and S100 A9: new insights into their roles in malignancy. J Innate Immun 2012;4:31-40. 10.1159/000330095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou Y, Guo Y, Tang Y, et al. Transcriptional regulation of human S100A8 / A9 gene. Chemistry of life 2016;36:293-7.
- 8.Han R, Huang Y, Chen L, et al. Determination and clinical significance of S100A8 and S100A9 Protein in plasma of patients with nasopharyngeal carcinoma. Journal of Clinical Laboratory 2014;32:252-4. [Google Scholar]
- 9.Lim SY, Yuzhalin AE, Gordon Weeks AN, et al. Tumor infiltrating monocytes/ macro-phages promote tumor invasion and migration by upregulating S100A8 and S100 A9 expression in cancer cells. Oncogene 2016;35:5735-45. 10.1038/onc.2016.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNeill E, Hogg N. S100A9 has a protective role in inflammation-induced skin carcinogenesis. Int J Cancer 2014;135:798-808. 10.1002/ijc.28725 [DOI] [PubMed] [Google Scholar]
- 11.Nordal HH, Brokstad K A, Solheim M, et al. Calprotectin (S100A8/A9) has the strongest association with ultrasound-detected synovitis and predicts response to biologic treatment: results from a longitudinal study of patients with established rheumatoid arthritis. Arthritis Res Ther 2017;19:3. 10.1186/s13075-016-1201-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khammanivong A, Sorenson BS, Ross K F, et al. Involvement of calprotectin (S100A8/A9) in molecular pathways associated with HNSCC. Oncotarget 2016;7:14029-47. 10.18632/oncotarget.7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishimura Y, Komatsu S, Ichikawa D, et al. Overexpression of YWHAZ relates to tumor cell proliferation and malignant outcome of gastric carcinoma. Br J Cancer 2013;108:1324-31. 10.1038/bjc.2013.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu L, Ma GQ, Liu XD, et al. Correlation between GDF15, MMP7 and gastric cancer and its prognosis. Eur Rev Med Pharmacol Sci 2017;21:535-41. [PubMed] [Google Scholar]
- 15.Qin C, Yang X, Wu Y, et al. Matrix metalloproteinases sensitive multifunctional micelles for inhibition of metastatic tumor growth and metastasis. Powder Technol 2019;358:3-12. 10.1016/j.powtec.2018.08.045 [DOI] [Google Scholar]
- 16.Banday MZ, Sameer AS, Mir AH, et al. Matrix metalloproteinase (MMP) -2, -7 and -9 promoter polymorphisms in colorectal cancer in ethnic Kashmiri population - A case-control study and a mini review. Gene 2016;589:81-9. 10.1016/j.gene.2016.05.028 [DOI] [PubMed] [Google Scholar]
- 17.Guo W, Gao X, Zhan R, et al. Tricolor imaging of MMPs to investigate the promoting roles of inflammation on invasion and migration of tumor cells. Talanta 2021;222:121525. 10.1016/j.talanta.2020.121525 [DOI] [PubMed] [Google Scholar]
- 18.Limoge M, Safina A, Beattie A, et al. Tumor-fibroblast interactions stimulate tumor vascularization by enhancing cytokine-driven production of MMP9 by tumor cells. Oncotarget 2017;8:35592-608. 10.18632/oncotarget.16022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klupp F, Neumann L, Kahlert C, et al. Serum MMP7, MMP10 and MMP12 level as negative prognostic markers in colon cancer patients. BMC Cancer 2016;16:494. 10.1186/s12885-016-2515-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Rai V , Agrawal DK. The role of damage- and pathogen-associated molecular patterns in inflammation-mediated vulnerability of atherosclerotic plaques. Can J Physiol Pharmacol 2017;95:1245-53. 10.1139/cjpp-2016-0664 [DOI] [PubMed] [Google Scholar]
- 21.Yan LL, Huang YJ, Yi X, et al. Effects of silencing S100A8 and S100A9 with small interfering RNA on the migration of CNE1 nasopharyngeal carcinoma cells. Oncol Lett 2015;9:2534-40. 10.3892/ol.2015.3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan Y, Johnson M, Zhou J, et al. Antrodia cinnamomea Inhibits Growth and Migration of Lung Cancer Cells through Regulating p53-Bcl2 and MMPs Pathways. Am J Chin Med 2020;48:1941-53. 10.1142/S0192415X20500974 [DOI] [PubMed] [Google Scholar]
- 23.Kwon CH, Moon HJ, Park HJ, et al. S100 A8 and S100 A9 promotes invasion and migration through p38 mitogen- activated protein kinase- dependent NF-κB activation in gastric cancer cells. Mol Cells 2013;35:226-34. 10.1007/s10059-013-2269-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan L, Wu R, Ye L, et al. S100A8 and S100A9 are associated with colorectal carcinoma progression and contribute to colorectal carcinoma cell survival and migration via Wnt/β-catenin pathway. PLoS One 2013;8:e62092. 10.1371/journal.pone.0062092 [DOI] [PMC free article] [PubMed] [Google Scholar]