Abstract
Background
Chemotherapy is the preferred treatment in many types of cancer including lung cancer. However, most of patients resist chemotherapy resulting in disease progressive and recurrence. Titin (TTN) mutation is proved as a beneficial role in lung squamous carcinoma (LUSC), but the predictive role on chemotherapy resistance of lung cancer is still limited and discussable.
Methods
Clinical information and related somatic mutation profiles were obtained from The Cancer Genome Atlas (TCGA) database and analyzed by R-Studio using R-package. Overall survival (OS) curve and the association between gene mutation and clinical features were determined by GraphPad 6.0 software.
Results
Available data including 563 lung adenocarcinoma (LUAD) and 505 LUSC subjects were included in this study. Among all patients, 205 out of 563 LUAD and 326 out of 505 LUSC patients displayed TTN gene mutation. When comparing the clinical features in TTN-mutated patients to patients without TTN mutation who received chemotherapy, the tumors were always located in the upper lung in LUAD patients with TTN mutation and most of TTN-mutated subjects were at low pathological stage, which was not observed in LUSC patients. However, patients with TTN-mutation, particularly missense mutation, had a higher chemosensitivity and longer OS period than that patients without TTN mutation in both LUAD and LUSC. Of note, LUAD and LUSC patients possessed favorable OS and better chemotherapy response benefiting from TTN/tumor protein 53 (TP53) double mutation compared to TTN and TP53 mutation alone, respectively. Additionally, TTN/TP53 double mutation-initiated high rate of chemotherapy response were largely concentrated within LUAD and LUSC patients whose anatomic neoplasm subdivision were located in the upper lung.
Conclusions
Collectively, TTN/TP53 co-mutation is possibly served as an effective predictor for OS and chemotherapy response in lung cancer.
Keywords: Lung adenocarcinoma (LUAD), lung squamous carcinoma (LUSC), titin (TTN), tumor protein 53 (TP53), gene mutation, chemotherapy response
Introduction
Lung cancer is the leading cause of cancer mortality both in men and women worldwide (1). Lung cancer is classified to two broad histologic classifications including small-cell lung carcinomas (SCLCs) and non-small cell lung carcinomas (NSCLCs) (2). Patients with NSCLCs account for more than 85% of all lung cancer cases and NSCLCs patients always have a high mortality rate (3). Clinically, platinum-based chemotherapy is also the standard therapy for advanced or recurrent NSCLCs (4). Nonsurgical patients in stage I or II and metastatic subjects always are considered for the combination of radiotherapy and adjuvant chemotherapy (ACT) (5). Unfortunately, most of NSCLCs patients display pre-existing resistant cells and/or acquisition of chemotherapy resistance during treatment procedure (6), which are the primary determinant in NSCLCs treatment failure and relapse (7).
Detection of gene mutation is mandatory to guide the choice of treatment for chemoresistant NSCLCs patients, that is because selection based only on clinicopathologic features is inadequate. For example, tyrosine kinase inhibitors (TKIs) are particularly effective in tumors that harbor activating tyrosine kinase domain mutations of epidermal growth factor receptor (EGFR) gene, and TKI instead of chemotherapy is the best choice of treatment for lung cancer patients with EGFR mutation (8). Additionally, tumor protein 53 (TP53) and KRAS genes always display significant mutations and have been extensively served as the predictive and prognostic gene targets in lung adenocarcinoma (LUAD) (9,10). Patients with tumors harboring concomitant KRAS and TP53 mutations present a poor clinical outcome after receiving cisplatin-based ACT (11). Therefore, sensitizing mutations have a great impact on available treatment options on lung cancer which differ for LUAD and lung squamous carcinoma (LUSC) (12). The longest-known gene TTN encoding for TITIN protein is a common mutated gene in diverse types of tumor including LUAD and LUSC (13). Mutated TTN is frequently observed in solid tumors and closely correlated with the raising tumor mutational burden (TMB) and objective response to immune checkpoint blockade (ICB) (14). Actually, TTN truncating mutations are firstly proved strongly correlating to the muscular diseases, but not in cancer (15). Truncating TTN variants are also associated with chemotherapy-induced cardiomyopathy (CCMP), showing a poor prognosis with a 60% mortality rate at 2 years in patients with truncating TTN variants (16). Although TP53 or TTN mutations alone were not associated with chemotherapy response or relapse in triple-negative breast cancers (TNBC), patients with TTN/TP53 co-mutation is completely mutually exclusive in those subjects with AKT1, NCOA3, ARID1A or MAP3K1 mutations who enjoy a favorable prognosis after the chemotherapy (17). Thus, TTN and/or TP53 mutations possibly affect the chemotherapy response. A recent study has proved that NSCLCs patients with TTN/TP53 double mutation harbor a favorable overall survival (OS) and disease-free survival (DFS) (18). However, whether TTN/TP53 mutations have an impact on chemotherapeutic efficacy in NSCLCs remains vague and discussable.
In the present study, we focused on the correlation between TTN and/or TP53 mutations and clinical outcome after receiving chemotherapy in LUAD and LUSC patients. Further analyses were an attempt to explore the association between chemotherapy sensitivity and anatomic neoplasm subdivision in TTN/TP53 double or alone-mutated NSCLCs patients. Our research possibly provided the predicting role of concomitant TTN/TP53 mutations for predicting chemotherapy response in NSCLCs.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2568).
Methods
Data collection and correlation analysis
Clinical data and mutation data of LUAD and LUSC patients were downloaded from The Cancer Genome Atlas (TCGA) database (https://www.genome.gov/Funded-Programs-Projects/Cancer-Genome-Atlas). Patients with incomplete information including sample type, follow-up information, mutation type, therapy method and treatment response were excluded from the downloaded data. Ultimately, 563 LUAD subjects and 505 LUSC subjects were enrolled in this study, respectively. Then the overall mutation information and Top mutation genes were analyzed by R-Studio using “maftools” package. Subsequently, the clinical information of LUAD and LUSC patients who receiving chemotherapy was collected carefully, the correlation between TTN mutation alone or TTN/TP53 double mutation and clinical features including gender, race, anatomic neoplasm subdivision, tumor stages and treatment response was determined by GraphPad 6.0 using chi-square and Fisher’s exact tests. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All experiments were approved by the Ethics Committee of Fujian Medical university union Hospital (FJMU-2017-077). Informed consent is not needed. The downloaded data and non-randomized analysis were analyzed and evaluated by three independent scientists.
Survival analysis
For the association between TTN mutation and OS of chemotherapy-treated LUAD and LUSC patients, the follow-up information obtained from clinical data was downloaded from TCGA database using R-package. Then OS curve was analyzed and created by GraphPad 6.0 software on the basis of TTN mutation alone, TTN/TP53 double mutation and TP53 mutation alone.
Statistical analyses
All analyses were performed at least thrice by three independent scientists. The association between clinical features and gene mutation was analyzed by GraphPad 6.0 using chi-square and Fisher’s exact tests. The comparison of OS in different group was compared by Graph Pad Prism using log-rank (Mantel-Cox) test. P<0.05 was considered statistically significant.
Results
Patients on chemotherapy with TTN mutation display a favorable clinical outcome in chemotherapy-challenged NSCLCs
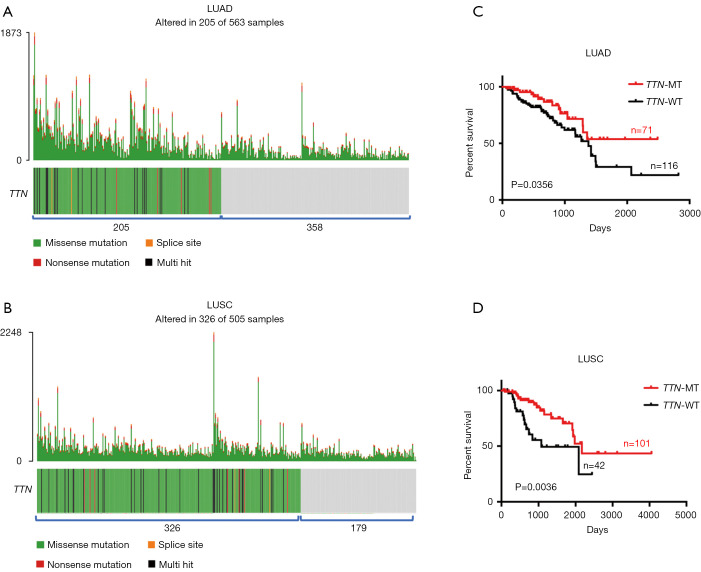
One thousand and sixty-eight cases NSCLCs subjects including LUAD (n=563) and LUSC (n=505) were available for correlation analysis in this study. Both somatic mutation profiles and clinical data were obtained from TCGA database. Using “maftools” R-package, we observed that 205 out of 563 (36%) LUAD samples and 326 out of 505 (65%) LUSC samples displayed multiple types of mutants in TTN including missense mutation, splice site, nonsense mutation and multi hit (Figure 1A,B and Table S1). As shown in Figure 1A,B, most mutants centered on missense mutation of TTN in LUAD and LUSC patients. To explore the association between TTN mutation and chemotherapy sensitivity, 187 LUAD and 143 LUSC patients who received chemotherapy were picked up from the 1,068 cases. Among these subjects, 71 out of 187 LUAD and 101 out of 143 LUSC patients harbored mutation of TTN (Table S1). Combined with the follow-up information, we found that patients on chemotherapy with TTN mutation (n=71) had a longer period of OS than that in patients without TTN mutation (n=116) (P=0.0356) (Figure 1C). Meanwhile, TTN mutation was often more strongly correlated with good prognosis in LUSC patients who received chemotherapy, showing a favorable OS in TTN-mutated group (n=101) compared to that group without TTN mutation (n=42) (P=0.0036) (Figure 1D). In addition to that, the association between different types of TTN mutation and drug response in chemotherapy-challenged NSCLCs patients also were further investigated. Actually, among 71 chemotherapy-challenged LUAD patients, 62 patients harbored with missense mutation, 7 with multi hit mutation, 1 with nonsense mutation and 1 with splice site. In addition to that, 88 out of 101 chemotherapy-challenged LUSC patients harbored with missense mutation, 10 with multi hit mutation, 3 with nonsense mutation and none with splice site (Table S2). We did not perform association analysis in patients who harbored with nonsense mutation and splice site due to the limited sample size. In LUAD patients on chemotherapy with TTN missense mutation (n=62), a longer period of OS were observed compared with that patients without TTN mutation (n=116) (P=0.0471). Meanwhile, TTN missense mutation was often more strongly correlated with good prognosis in LUSC patients on chemotherapy, showing a favorable OS in the group with missense mutation (n=88) compared to that group without TTN mutation (n=42) (P=0.0017) (Figure S1A,B). By contrast, no positive association between multi hit mutation with drug response was found in LUAD and LUSC patients who received with chemotherapy (Figure S1C,D).
Figure 1.
The association between TTN mutation and OS in LUAD and LUSC patients on chemotherapy. (A) Number of patients with TTN mutation in LUAD. (B) Number of patients with TTN mutation in LUSC. (C) The difference in OS between patients with TTN mutation and TTN-WT in LUAD treated with chemotherapy. (D) The difference in OS between patients with TTN mutation and TTN-WT in LUSC treated with chemotherapy. TTN, titin; OS, overall survival; LUAD, lung adenocarcinoma; LUSC, lung squamous carcinoma; MT, mutant-type; WT, wild-type.
Next, the association between different chemotherapy drugs and outcome were further analyzed. In TTN-mutated LUAD patients, 52 out of 71 patients received with platinum treatment, 2 with Vinca alkaloids, 3 with Taxanes, 2 with Gemcitabine-related drugs, and 12 with other types of drug. Due to the limitation of sample size, patients on platinum were only used to assess whether different chemotherapeutic drugs affected the above results. As shown in Figure S2A, patients on platinum with TTN mutation (n=52) had a longer period of OS than that platinum-challenged patients without TTN mutation (n=59) (P=0.0233) (Figure S2A). Meanwhile, in TTN-mutated LUSC patients, patients on platinum were only used to perform the further analysis (62 out of 101 with platinum, 10 with Vinca alkaloids, 10 with Taxanes, 10 with Gemcitabine-related drugs, and 9 with other types of drug). The results showed that patients on platinum with TTN mutation (n=62) had a better OS curve than that platinum-challenged patients without TTN mutation (n=23) (P=0.0233) (Figure S2B). Possibly, the mutation of TTN might be an independent significant favorable prognostic indicator in LUAD and LUSC patients on chemotherapy.
Mutated TTN forebodes a good response to chemotherapy in LUAD and LUSC
Next, we analyzed the relationship between TTN mutation and chemotherapy response in LUAD and LUSC. In the patients on chemotherapy, there were no any association between patient’s gender or race and TTN mutation in both LUAD and LUSC (Table 1). However, by contrast to patients without TTN mutation, tumors that harboring TTN mutation mainly appeared in the upper lung (46/71) in chemotherapy-treated LUAD patients (P=0.038). Additionally, pathological grade of LUAD patients with TTN mutation mainly gathered together in stage I to II (n=51), while patients with TTN wild-type (TTN-WT) gathered in late stage (n=96) (P=0.0327) (Table 1). Although there were no any association between TTN mutation and anatomic neoplasm subdivision and tumor stage in LUSC, we observed that 51 out of 71 LUAD and 76 out of 101 LUSC patients who harbored the mutation of TTN were active responders after chemotherapy, which had a low response rate in patients without TTN mutation (P=0.0001 and P=0.0156) (Table 1). Possibly, detection of TTN mutation contributes to the assessment of chemosensitivity of NSCLCs.
Table 1. Chi-squared analysis of contingency table between TTN mutation and clinicopathological characteristics of patients with LUAD and LUSC.
| Clinicopathological characteristics | LUAD | LUSC | |||||
|---|---|---|---|---|---|---|---|
| TTN-MT | TTN-WT | P value | TTN-MT | TTN-WT | P value | ||
| Gender | |||||||
| Female | 39 | 62 | 0.8436 | 27 | 8 | 0.3303 | |
| Male | 32 | 54 | 74 | 34 | |||
| Race | |||||||
| Black | 14 | 25 | 0.7646 | 22 | 10 | 0.7911 | |
| White | 57 | 91 | 79 | 32 | |||
| Anatomic neoplasm subdivision | |||||||
| Upper | 46 | 57 | 0.0368* | 61 | 24 | 0.8080 | |
| Lower | 25 | 59 | 50 | 18 | |||
| Stage | |||||||
| I | 24 | 20 | 0.0327* | 21 | 7 | 0.8495 | |
| II | 27 | 52 | 52 | 23 | |||
| III–IV | 20 | 44 | 28 | 12 | |||
| Treatment response | |||||||
| Response | 51 | 44 | <0.0001** | 76 | 23 | 0.0156* | |
| No response | 20 | 72 | 25 | 19 | |||
*, P<0.05; **, P<0.01. TTN, titin; LUAD, lung adenocarcinoma; LUSC, lung squamous carcinoma; MT, mutant-type; WT, wild-type.
TTN/TP53 double mutation is positively associated with OS in NSCLC
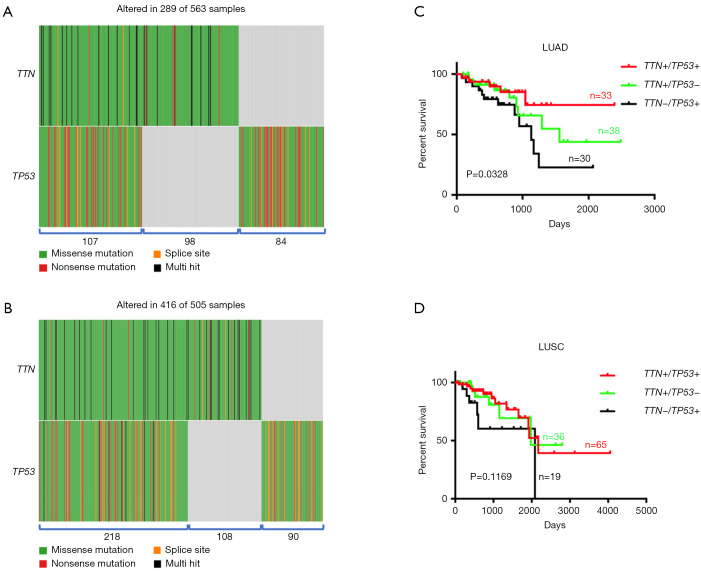
Since TTN/TP53 co-mutation was related with chemotherapy response, we next investigated whether double mutation in TTN/TP53 also affected the clinical outcomes in NSCLCs patients on chemotherapy. Using TCGA database and R-package, we discovered that patients with TTN and/or TP53 mutations accounted for approximately 51% (289/563) in LUAD patients. Of these patients, 107 patients harbored with TTN/TP53 co-mutation, 98 with TTN mutation alone and 84 with TP53 mutation alone (Figure 2A and Table S3). In LUSC subjects, there were 218 cases of TTN/TP53 co-mutated patients, 108 cases of TTN mutation alone and 90 cases of TP53 mutation alone (Figure 2B and Table S3). We also picked up the chemotherapy-treated patients to assess the role of double mutation of TTN/TP53 on chemosensitivity of NSCLCs (Table S3). OS analysis showed that patients with TTN/TP53 double mutation (n=33) displayed the longest median survival period than those patients with TTN (n=48) or TP53 (n=30) mutation alone in LUAD (P=0.0328) (Figure 2C). However, there were no difference in the OS period between patients with TTN mutation alone and patients with TP53 mutation alone, as well as between patients with TTN/TP53 double mutation and patients with TTN mutation alone in LUAD (Figure S3A,B). In LUSC patients on chemotherapy, we did not observe the overall difference in OS analysis among patients with TTN/TP53 double mutation (n=65), TTN mutation alone (n=36) and TP53 mutation alone (n=19) (P=0.1169) (Figure 2D). Meanwhile, there showed no difference in the median period of OS between TTN and TP53 mutation alone, or TTN/TP53 double mutation and TTN mutation alone in LUSC (Figure S3C,D). Of note, in comparison to patients with TP53 mutation alone, co-mutated TTN and TP53 showed a significant favorable OS in chemotherapy-treated LUAD (P=0.0348) and LUSC patients (P=0.0337) (Figure S4A,B). Therefore, patients with TTN/TP53 double mutation might have a longer median survival period after chemotherapy.
Figure 2.
Effects of TTN/TP53 double mutation on OS of LUSC patients on chemotherapy. (A) Patients with TTN/TP53 double mutation, TTN and TP53 single mutation in LUAD. (B) Number of patients with TTN/TP53 double mutation, TTN and TP53 single mutation in LUSC. (C) The difference in OS period between TTN/TP53 double mutation and single mutation of TTN or TP53 in LUAD. (D) The difference in OS period between TTN/TP53 double mutation and single mutation of TTN or TP53 in LUSC. “+” indicates MT; “–” indicates WT. TTN, titin; TP53, tumor protein 53; OS, overall survival; LUAD, lung adenocarcinoma; LUSC, lung squamous carcinoma; MT, mutant-type; WT, wild-type.
TTN/TP53 double mutation harbors a good chemosensitivity in LUAD and LUSC
Then, we also assessed the difference in chemotherapy response between patients with double mutation and single mutation. As shown in Table 2, we observed that most of patients with TTN/TP53 double mutation were the active responders both in LUAD (24 out of 33) and LUSC (40 out of 65), while most of samples with single mutation were non-responders (P=0.0241 and P=0.0212). When compared the clinicopathological characteristics (gender, race, anatomic neoplasm subdivision and stage) of LUAD and LUSC patients on chemotherapy, we did not find any difference among patients with TTN/TP53 double mutation or single mutation, which implied that the significant variation in treatment response to chemotherapy might be caused by genetic factors (Table 2). Interestingly, we discovered that tumors harboring double mutation or single mutation mainly appeared in the upper lung, no matter LUAD or LUSC (Table 2). Given tumor heterogeneity in different anatomic site, we next explored whether the higher chemosensitivity in patients with TTN/TP53 double mutation was related with the anatomic neoplasm subdivision in LUAD and LUSC. The results showed that tumors of the responders were basically located in the upper lung in TTN/TP53 co-mutated patients, showing 19 out of 24 in LUAD and 29 out of 40 in LUSC (P=0.0025 and P=0.0216) (Table 3). In LUSC subjects, we also observed the significant difference in chemotherapy response between the upper and lower lung of patients with TTN mutation alone, showing higher chemosensitivity in upper lung than that tumors located in lower (P=0.0368), while that discrepancy was not found in LUAD patients (Table 3). And there was no distinction in chemosensitivity between different anatomic neoplasm subdivision in patients with TP53 mutation alone. The data suggested that high rate of chemotherapy response in patients with TTN/TP53 double mutation mainly gathered in LUAD and LUSC subjects whose anatomic neoplasm subdivision was located in the upper lung.
Table 2. Chi-squared analysis of contingency table between TTN and/or TP53 mutation and clinicopathological characteristics of LUAD and LUSC patients on chemotherapy.
| Clinicopathological characteristics | LUAD | LUSC | |||||||
|---|---|---|---|---|---|---|---|---|---|
| TTN+/TP53+ | TTN+/TP53– | TTN–/TP53+ | P value | TTN+/TP53+ | TTN+/TP53– | TTN–/TP53+ | P value | ||
| Gender | |||||||||
| Female | 17 | 22 | 19 | 0.6366 | 21 | 6 | 3 | 0.1324 | |
| Male | 16 | 16 | 11 | 44 | 30 | 16 | |||
| Race | |||||||||
| Black | 8 | 6 | 6 | 0.6718 | 12 | 10 | 3 | 0.4567 | |
| White | 25 | 32 | 24 | 53 | 26 | 16 | |||
| Anatomic neoplasm subdivision | |||||||||
| Upper | 21 | 25 | 21 | 0.8637 | 40 | 21 | 11 | 0.9320 | |
| Lower | 12 | 13 | 9 | 25 | 15 | 8 | |||
| Stage | |||||||||
| I–II | 22 | 24 | 16 | 0.5328 | 46 | 27 | 15 | 0.7498 | |
| III–IV | 11 | 14 | 14 | 19 | 9 | 4 | |||
| Treatment response | |||||||||
| Response | 24 | 16 | 14 | 0.0241* | 40 | 17 | 5 | 0.0212* | |
| No response | 9 | 22 | 16 | 25 | 19 | 14 | |||
“+” indicates MT; “–” indicates WT. *, P<0.05. TTN, titin; TP53, tumor protein 53; LUAD, lung adenocarcinoma; LUSC, lung squamous carcinoma; MT, mutant-type; WT, wild-type.
Table 3. Chi-squared analysis of contingency table between TTN/TP53 mutation and anatomic neoplasm subdivision of LUAD and LUSC patients receiving chemotherapy.
| Therapeutic response | Anatomic neoplasm subdivision | Anatomic neoplasm subdivision | |||||
|---|---|---|---|---|---|---|---|
| LUAD | P value | LUSC | P value | ||||
| Upper | Lower | Upper | Lower | ||||
| TTN+/TP53+ | |||||||
| Response | 19 | 5 | 0.0025** | 29 | 11 | 0.0216* | |
| No response | 2 | 7 | 11 | 14 | |||
| TTN+/TP53– | |||||||
| Response | 11 | 5 | 0.7429 | 13 | 4 | 0.0368* | |
| No response | 14 | 8 | 8 | 11 | |||
| TTN–/TP53+ | |||||||
| Response | 9 | 5 | 0.5229 | 2 | 3 | 0.3451 | |
| No response | 12 | 4 | 9 | 5 | |||
“+” indicates MT; “–” indicates WT. *, P<0.05; **, P<0.01. TTN, titin; TP53, tumor protein 53; LUAD, lung adenocarcinoma; LUSC, lung squamous carcinoma; MT, mutant-type; WT, wild-type.
Discussion
Although therapies targeting driver oncogenic mutations are more effective than chemotherapy for patients with EGFR- or TP53-mutation-positive tumors, chemotherapy remains dominant for treating early stage or locally advanced tumor disease (19). Patients on chemotherapy with KRAS and/or TP53 mutation has a poor OS compared to NSCLCs patients with the WT TP53 and KRAS (20). It is to say that detection of gene mutation contributes to the prediction of chemotherapeutic efficacy. In the present study, we demonstrated that patients with TTN mutation had a favorable OS and a good chemotherapy response compared to those patients with TTN-WT. Furthermore, TTN/TP53 double-mutant tumors had an active response to chemotherapy compared to NSCLCs patients with single mutation of TTN or TP53. The finding demonstrates that coexisting TTN and TP53 mutations may be a good indicator for chemotherapy in NSCLCs patients.
Somatic mutation of TTN is proved to be frequent in most cancer types including lung cancer (13). TTN mutation is an early genetic alteration event and the key driver risk in smoking-related lung cancer (21,22). TTN is one of the most commonly individualized HLA Class II presented mutant peptides, possibly as prognostic immunological biomarker for LUAD (23). Our data further proved the frequent mutation of TTN in LUAD (36%) and higher mutation rate in LUSC (65%). Using the deep learning model to predict drug response (DeepDR), Chiu et al. has indicated that vinorelbine possesses obviously anti-tumor effects on TTN-mutated tumors, which implies that detection of TTN mutation may facilitate the prediction of chemotherapeutic drug response (24). In this study, we observed that patients with TTN mutation appeared a better drug response than that patients with TTN-WT. Of note, this active chemotherapy efficiency maintains for long term in TTN-mutated tumors which was reflected in the comparison of OS between the two tumor types, showing a better OS in patients with TTN mutation than NSCLCs patients with TTN-WT. These findings indicated the significant predictive effect of TTN mutation on NSCLCs treated with chemotherapy. Recent evidence confirms that missense mutation of TTN may serve as a beneficial determinant in LUSC but not LUAD (18). Although patients with TTN missense variation and multi hit mutation (green/black color in Figures 1,2) were closely related with patients on chemotherapy, the association between other types of TTN mutation (nonsense mutation/splice site) and clinical outcome or chemotherapy response in NSCLCs should be further investigated using a large scale of subjects. Additionally, Cisplatin-treated patients present a good prognosis in patients with TTN mutation compared to that patients with TTN-WT. However, the enrolled subjects receive the different drug such as Paclitaxel, Tarceva and so on. Possibly, TTN mutation should present different chemosensitivity in various chemotherapeutics. These should be further investigated in the future using a large scale of subjects.
The presence of TP53 mutation in EGFR mutation is associated with inferior response to targeted therapy and poor OS in NSCLCs patients (25). KRAS-mutated lung cancers always enrich with genomic alterations in TP53 which is closely related with treatment failure and poor prognosis (26). In ALK-rearranged NSCLC, co-occurring TP53 mutation predicts a poor outcome of systemic therapy (27). By contrast, we found that most responders to chemotherapy always accompany by the TP53 mutation in TTN-mutated tumors. However, there was no difference in drug response between patients with TTN mutation and TP53 mutation alone. Additionally, TTN/TP53 double-mutant tumors had a significant better OS than LUAD patients with single mutation of TTN/TP53, and patients with single mutation of TP53 in both LUAD and LUSC. Thus, TP53 mutation might be a beneficial factor for antitumor chemotherapeutics in TTN-mutated tumors. Mutational heterogeneity in cancer contributes to enable the identification of genes truly associated with cancer (28). In colon cancer, amounts of differences in molecular expression have been reported between right-sided and left-sided colon cancer (29). The tumor located in the upper right lobe is positive for a TP53 c.659A > G mutation, while the tumor from the upper left lobe is positive for TP53 c.725G > T mutation, which improves diagnostic accuracy in patients with multiple lung tumors (30). The data indicate that mutational heterogeneity may be frequent and an important indicator for lung cancer diagnosis and treatment. Here, we found that mutation of TTN was predispositional to localized the upper lung. Further analysis proved that TTN/TP53 double mutated tumors located in upper lung had a good response to chemotherapy compared to that tumors located in lower lung. We speculated that the common mutation of TTN/TP53 double mutation in the upper lung may associate to the tumor initiating cells which are closely related with tumor location. Therefore, a better effectiveness of chemotherapy is also related with the anatomic neoplasm subdivision of LUAD and LUSC patients with TTN/TP53 double mutations. However, in cohort with TTN mutation, TP53 status (mutation or non-mutation) did not affect the association between anatomic neoplasm subdivision and drug response in LUSC. In addition to that, whether TTN/TP53 double mutations also have predictive value on other types of lung cancer should also be further investigated. Potentially, TTN mutation alone is a better diagnostic factor for chemotherapy effectiveness in LUSC, whereas TTN/TP53 double mutation is a preferable predictive factor for drug response in LUAD.
Conclusions
In summary, TTN mutation alone predicted a beneficial OS and good drug response in NSCLC. Moreover, TTN/TP53 double mutation had a better clinical outcome and chemosensitivity than TTN and/or TP53 mutation alone. And the improvement of chemosensitivity in tumors with TTN/TP53 double mutation always located in the upper lung. The data provide the clinical diagnosis value of TTN mutation for chemotherapy treatment. Possibly, detection of TTN/TP53 co-mutation may be used to predict the response to chemotherapy in NSCLCs patients, particularly in patients whose anatomic neoplasm subdivision was in the upper lung. In addition, mutational heterogeneity of TTN and/or TP53 also facilitates the choice of treatment and provides the novel method for searching new cancer-associated genes.
Acknowledgments
We thanked M. R. Elphick for checking and editing the manuscript.
Funding: The present study was supported by the Joint Funds for the Innovation of Science and Technology, Fujian Province, China (grant no. 2016Y9028, 2017Y9031 and 2018Y9038).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All experiments were approved by the Ethics Committee of Fujian Medical university union Hospital (FJMU-2017-077). Informed consent is not needed.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2568
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2568). The authors have no conflicts of interest to declare.
References
- 1.Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. 10.1016/S0140-6736(16)30958-8 [DOI] [PubMed] [Google Scholar]
- 2.Lemjabbar-Alaoui H, Hassan OU, Yang YW, et al. Lung cancer: biology and treatment options. Biochim Biophys Acta 2015;1856:189-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veluswamy RR, Levy B, Wisnivesky JP. Chemotherapy in elderly patients with nonsmall cell lung cancer. Curr Opin Pulm Med 2016;22:336-43. 10.1097/MCP.0000000000000286 [DOI] [PubMed] [Google Scholar]
- 4.da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol 2011;6:49-69. 10.1146/annurev-pathol-011110-130206 [DOI] [PubMed] [Google Scholar]
- 5.Duma N, Santana-Davila R, Molina JR. Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc 2019;94:1623-40. 10.1016/j.mayocp.2019.01.013 [DOI] [PubMed] [Google Scholar]
- 6.Zalcman G, Bergot E, Lechapt E. Update on nonsmall cell lung cancer. Eur Respir Rev 2010;19:173-85. 10.1183/09059180.00006610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galluzzi L, Senovilla L, Vitale I, et al. Molecular mechanisms of cisplatin resistance. Oncogene 2012;31:1869-83. 10.1038/onc.2011.384 [DOI] [PubMed] [Google Scholar]
- 8.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. 10.1056/NEJMoa040938 [DOI] [PubMed] [Google Scholar]
- 9.Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Brit J Cancer 2005;92:131-9. 10.1038/sj.bjc.6602258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olivier M, Taniere P. Somatic mutations in cancer prognosis and prediction: lessons from TP53 and EGFR genes. Curr Opin Oncol 2011;23:88-92. 10.1097/CCO.0b013e3283412dfa [DOI] [PubMed] [Google Scholar]
- 11.Shepherd FA, Lacas B, Le Teuff G, et al. Pooled analysis of the prognostic and predictive effects of TP53 comutation status combined with KRAS or EGFR mutation in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol 2017;35:2018-27. 10.1200/JCO.2016.71.2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan BA, Hughes BGM. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res 2015;4:36-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim N, Hong Y, Kwon D, et al. Somatic mutaome profile in human cancer tissues. Genomics Inform 2013;11:239-44. 10.5808/GI.2013.11.4.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia Q, Wang J, He N, et al. Titin mutation associated with responsiveness to checkpoint blockades in solid tumors. JCI Insight 2019;4:e127901. 10.1172/jci.insight.127901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schafer S, de Marvao A, Adami E, et al. Titin-truncating variants affect heart function in disease cohorts and the general population. Nat Genet 2017;49:46-53. 10.1038/ng.3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felker GM, Thompson RE, Hare JM, et al. Underlying causes and long-term survival in patients with initially unexplained cardiomyopathy. N Engl J Med 2000;342:1077-84. 10.1056/NEJM200004133421502 [DOI] [PubMed] [Google Scholar]
- 17.Kim YA, Madan S, Przytycka TM. WeSME: uncovering mutual exclusivity of cancer drivers and beyond. Bioinformatics 2017;33:814-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng X, Yin H, Fu J, et al. Aggregate analysis based on TCGA: TTN missense mutation correlates with favorable prognosis in lung squamous cell carcinoma. J Cancer Res Clin Oncol 2019;145:1027-35. 10.1007/s00432-019-02861-y [DOI] [PubMed] [Google Scholar]
- 19.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 20.Tomasini P, Mascaux C, Jao K, et al. Effect of coexisting KRAS and TP53 mutations in patients treated with chemotherapy for non-small-cell lung cancer. Clin Lung Cancer 2019;20:e338-45. 10.1016/j.cllc.2018.12.009 [DOI] [PubMed] [Google Scholar]
- 21.Yu XJ, Chen G, Yang J, et al. Smoking alters the evolutionary trajectory of non-small cell lung cancer. Exp Ther Med 2019;18:3315-24. 10.3892/etm.2019.7958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donner I, Katainen R, Sipila LJ, et al. Germline mutations in young non-smoking women with lung adenocarcinoma. Lung Cancer 2018;122:76-82. 10.1016/j.lungcan.2018.05.027 [DOI] [PubMed] [Google Scholar]
- 23.Cai W, Zhou D, Wu W, et al. MHC class II restricted neoantigen peptides predicted by clonal mutation analysis in lung adenocarcinoma patients: implications on prognostic immunological biomarker and vaccine design. BMC Genomics 2018;19:582. 10.1186/s12864-018-4958-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiu YC, Chen HIH, Zhang TH, et al. Predicting drug response of tumors from integrated genomic profiles by deep neural networks (vol 12, pg 18, 2019). Bmc Med Genomics 2019;12:143-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aggarwal C, Davis CW, Mick R, et al. Influence of TP53 mutation on survival in patients with advanced EGFR-mutant non-small-cell lung cancer. JCO Precis Oncol 2018;2018:PO.18.00107. [DOI] [PMC free article] [PubMed]
- 26.Wang M, Han J, Marcar L, et al. Radiation resistance in KRAS-mutated lung cancer is enabled by stem-like properties mediated by an osteopontin-EGFR pathway. Cancer Res 2017;77:2018-28. 10.1158/0008-5472.CAN-16-0808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kron A, Alidousty C, Scheffler M, et al. Impact of TP53 mutation status on systemic treatment outcome in ALK-rearranged non-small-cell lung cancer. Ann Oncol 2018;29:2068-75. 10.1093/annonc/mdy333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214-8. 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee GH, Malietzis G, Askari A, et al. Is right-sided colon cancer different to left-sided colorectal cancer? - a systematic review. Eur J Surg Oncol 2015;41:300-8. 10.1016/j.ejso.2014.11.001 [DOI] [PubMed] [Google Scholar]
- 30.Eguren-Santamaria I, Sanchez-Bayona R, Patino-Garcia A, et al. Targeted DNA sequencing for assessing clonality in multiple lung tumors: a new approach to an old dilemma. Lung Cancer 2018;122:120-3. 10.1016/j.lungcan.2018.05.029 [DOI] [PubMed] [Google Scholar]