Abstract
Background
DICER is a key RNase III enzyme that cleaves processes miRNAs into their mature form. It is remarkably down-regulated in most epithelial ovarian cancer (EOC) and the down-regulation is associated with high grade malignancy as well as poor clinical outcomes. In this study, we aimed to discover a lncRNA interacting with DICER to participate in the process of microRNAs maturation and as a result promoting development of EOC.
Methods
We conducted a RIP-seq aimed at DICER and we obtained its chromosomal location by aligning the sequence in PubMed. And the lncRNA was named DATOC-1 (DICER Associated Transcript 1 in Ovarian Cancer). We tested relative expression of DATOC-1 in 53 EOC and 7 adjacent ovarian samples by qRT-PCR. Then the expression in EOC patients derived from The Cancer Genome Atlas (TCGA) were analysed to identify DATOC-1-based signatures for EOC prognosis with survival analysis. Lastly, shRNA Screening and lentiviral transduction was used to determine the function of DATOC-1 in vitro and in vivo.
Results
We identified the lncRNA RP5-1120P11.1 as DATOC-1, which highly expressed in EOC tissues than in adjacent. And Kaplan-Meier analysis indicated that the patients with EOC that expressed high levels of DATOC-1 had a worse prognosis and shorter disease OS compared with DATOC-1 low-expressed patients. In addition, DATOC-1 were further identified participating in EOC cell proliferation, cell cycle regulation and cell invasion. And knockdown of DATOC-1 inhibits tumor progression in vivo. Furthermore, knockdown of DATOC-1 increased the expression of miR7. The evidence showed that miR7 functioning as a tumor suppressor gene in EOC.
Conclusions
Our research shows that DATOC-1 can inhibit the development of EOC and is a promising therapeutic target.
Keywords: LncRNA DATOC-1, epithelial ovarian cancer (EOC), DICER, target, miR7
Introduction
Ovarian cancer (OC) is the third most commonly diagnosed malignant cancer among females and the leading cause of cancer death in women (1). And approximately 90% ovarian tumors are of epithelial origin. The poor prognosis is not only because of the aggressive nature of epithelial ovarian cancer (EOC) but also for that metastases are often present when diagnosed (2). The standard treatment for advanced EOC is cytoreduction surgery and chemotherapy; however, the overall survival rate remains unsatisfactory (3). Therefore, better understanding of the pathogenesis and molecular changes of EOC will help improve the treatment of OC.
DICER is a key RNase III enzyme that cleaves processes microRNA (miRNA) into their mature form (4). As an upstream modulator of miRNAs, the down-regulation of Dicer can promote cellular transformation and tumorigenesis by a global decrease in miRNA expression (5). And recent work has shown that dysregulation of miRNAs is related to the pathogenesis, tumor phenotype and chemosensitivity of OC (6). Merritt et al. demonstrate that Dicer is remarkably down-regulated in 60% of detected patients with invasive EOC and the down-regulation is associated with high grade malignancy as well as poor clinical outcomes (7). Besides this, low-expression of DICER in EOC is involved in chemotherapy resistance (8). Similarity, it has been reported that compared to normal tissues, Dicer mRNA expression is down-regulated in the majority of ovarian tumors and it may be the reason of dysregulation of miRNAs (9). However, the underlying mechanism of Dicer down-regulation in EOC remains largely unclear.
Mammalian genomes encode a large number of noncoding RNAs including small regulatory RNAs such as miRNAs and long noncoding RNAs, RNA species greater than 200 bp in length (10,11). LncRNAs mediate several cancer-associated processes, including epigenetic regulation, signal transduction pathways, miRNA silencing, DNA damage and cell cycle control, and hormone-driven disease (12). To date, some research suggest that they may be potential drivers of cancer biology and serve as clinical biomarkers (13-16), and also some were demonstrated in OC (17-19). It is demonstrated that lncRNAs may regulate gene functions by binding with miRNAs (20-22). Then, may lncRNA regulates microRNA expression in microRNA production such as transcription and maturation stage? To date, there is litter report about lncRNA regulate cancer progress by act in miRNA maturation, and if there any lncRNA act in DICER Function process has not yet be reported.
In this study, we aimed to discover a lncRNA interacting with DICER to participate in the process of microRNAs maturation and as a result promoting development of EOC. Finally, we found the clinical significance of the expression of lncRNA RP5-1120P11.1 in EOC, which interacted with DICER, correlated with prognoses and recurrence rate.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-189).
Methods
Cell lines and culture conditions
Human ovarian epithelial carcinoma cell line HO8910, SKOV3 and A2780 (obtained from ATCC) were grown in 5% CO2 at 37 °C in RPMI 1640-glutamax (Hyclone, America) supplemented with 10% FBS (Gibco, America) and 1% Penicillin-Streptomycin Solution (Hyclone, America).
RIP-seq
RIP was carried out using the Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore). The antibodies against DICER (D5F2, rabbit monoclonal), and GAPDH (D16H11, rabbit monoclonal) IgG (7074s, rabbit monoclonal) were from Cell Signaling Technology. Coimmunoprecipitated RNA was extracted using Trizol LS (Invitrogen, Life Technologies) and Qiagen RNeasy columns, treated with TURBO DNAfree, and then analyzed by clone sequencing.
Patients specimens
Fifty three patients suffering from EOC received surgical resection at Changzheng Hospital in Shanghai, China. Patients participating in this study did not receive any treatment. All patients had read and signed informed consent before taking the samples. According to the World Health Organization classification, the 53 EOC cases were pathologically confirmed and histologically graded. Tissue samples were fresh-frozen in liquid nitrogen after surgery.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of the Second Military Medical University Changzheng Hospital. Informed consent was obtained before surgery and biopsy.
RNA Isolation and qRT-PCR
Real-time quantitative PCR was performed with SYBR Green (Toyobo; Osaka, Japan) following the manufacturer’s protocol. Quantification was normalized to the GAPDH, and the results were expressed as log10(2-∆∆Ct). The primer sequences for LncRNA DATOC-1 detection were as follows: forward: 5’-TAACACTCACTACCCCTCACG-3’ and reverse: 5’-CTATTTCCAACTACGGTCACATC-3. The sequences of the GAPDH primers were as follows: forward: 5’-TGACTTCAACAGCGACACCCA-3’ and reverse: 5’-CACCCTGTTGCTGTAGCCAAA-3’.
TCGA ovarian cancer and DATOC-1 datasets download and processing
The TCGA epithelial ovarian cancer data (level 3 RNA sequencing) of 426 individuals with clinical information were extracted from the TCGA database, and 88 normal ovarian tissues from the GTEX project. The exclusion criteria are as follows: (I) EOC was ruled out by histological diagnosis; (II) the existence of another malignancy with EOC; and (III) patient death due to unrelated causes. Only patients who were followed up for longer than one month were included in the analysis. Since the data is downloaded from a public database, ethical approval does not apply in this case. The data processing procedures met the policies of TCGA data access and human subject protection http://cancergenome.nih.gov/publications/publication guidelines).
ShRNA screening and lentiviral transduction
Lentiviral vector (pLKO.1) and packaging vectors were transfected into 293T cells. The medium was changed every 8 hours after transfection, and collected 48 hours later. Cancer cells were infected with lentivirus in the presence of 8 mg/mLpolybrene. The target sequences for NC was as follows: TTCTCCGAACGTGTCACGT. The target sequences for KD1 was as follows: AGGTACATAAACTGTGGCATA. The target sequences for KD2 was as follows: TAGGATAGTAGCTAGGACTA. The target sequences for KD3 was as follows: GCAGCTATGAGCCGAGAAATA. The target sequences for KD4 was as follows: GTGGAAACGAAGCACATATA.
Cell proliferation assay
To evaluate the effect of DATOC-1 expression on the proliferation of HO-8910 cells, Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan) was used to assess cell viability according to the manufacturer’s instructions. Cells were transfected with 20 nM shRNA targeted to DATOC-1 (sh-DATOC-1) or scrambled negative control (shRNA-NC). Cell survival rate (%) = experimental group OD value-blank group OD value/control group OD value-blank group OD value.
Cell cycle analysis and apoptosis assay
Cells were transfected with 20 nM sh-DATOC-1 or sh-NC for 48 h. Then two group of cells were stained for 10 min with annexin V and PI using the FITC Annexin V Apoptosis Detection Kit II. Cell cycle analysis was performed using an LSR flow cytometer (Becton Dickinson; San Jose, CA, USA) with ModFit LT software (Verity Software House; Topsham, ME, USA). Cell apoptosis was measured by flow cytometry with a BD FACS Caliber instrument (BD Biosciences, San Jose, CA, USA).
Matrigel invasion assays
A cell invasion assay was carried out using modified Boyden chambers (BD Biosciences, San Jose, CA, USA). The transfected cells were seeded in the top chamber of transwell assay inserts, with the lower chamber with RPMI 1640 with 20% FBS. 48 h later, cells in the upper chamber were scrubbed while invading cells were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet. Last, cells in two chambers were examined, counted, and imaged using digital microscopy.
In vivo xenograft experiments
The experimental animals that BALB/c nude mice, male, approximately 6 weeks old and 20 g were purchased from the Shanghai Experimental Animal Center of the Chinese Academic of Sciences (Shanghai, China). The mice were maintained in a pathogen-free environment (four per cage) and allowed to acclimate for at least 1 week before tumor implantation. The tumor-bearing mice model was set up as follows: the BALB/c nude mice were inoculated subcutaneously on the right of their armpit with 5×106 EOC cells (in 100 µL culture medium) to develop xenograft tumors. After about 2 weeks, the volume of the tumors reached about 50 mm3. The tumor-bearing mice was grouped randomly as follows: negative control group (n=4) and the DATOC-1 knockdown group (n=4). All mice were executed after 35 days, and tumor tissues were collected with capsule.
All experiments were performed under a project license granted by the Institutional Review Board of the Second Military Medical University Changzheng Hospital, in compliance with the national or institutional guidelines for the care and use of animals.
Statistical analysis
All graph values represent means ± SD from three independent experiments with each measured in triplicate. The differences between two groups were analyzed with the unpaired two-sided Student’s t test. The log-rank test was used to test for differences between curves using SPSS 17.0 Statistical Software (SPSS Inc., Chicago, IL). All statistical tests were two-sided. A P value of less than 0.05 was considered statistically significant and indicated with asterisks, as described in figure legends.
Results
Identify lncRNA DATOC-1 which interacts with DICER and is dysregulated in EOC
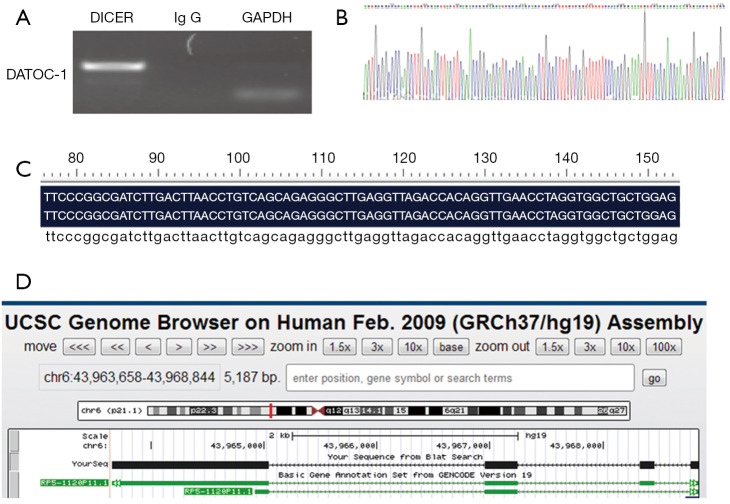
To find out the most interesting candidate that combine directly with DICER, we conducted a RIP-seq aimed at DICER. We named the resulting fragment DATOC-1 (DICER Associated Transcript 1 in Ovarian Cancer) (Figure 1A,B). And we obtained its chromosomal location as RP5-1120P11.1 by aligning the sequence in PubMed (Figure 1C,D).
Figure 1.
Determination the DATOC-1 sequence and location by performing RIP-seq on DICER. (A) DICER IP fragment cloning; (B) sequencing of the fragment; (C) sequence alignment; (D) RP5-1120P11.1 (DATOC-1) position on the chromosome. DATOC-1, DICER Associated Transcript 1 in Ovarian Cancer.
Expression of DATOC-1 is upregulated in EOC
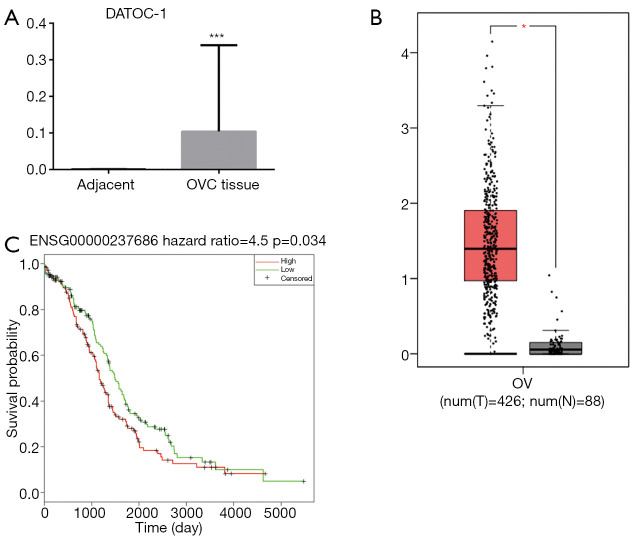
To determine the role of DATOC-1 in EOC, we detected the expression levels of DATOC-1 in 53 EOC and 7 adjacent ovarian samples using qPCR. We found that DATOC-1 is highly expressed in EOC than in adjacent tissue (Figure 2A, P<0.001). To further validate these findings, we extracted the expression data of RP5-1120P11.1 in TCGA and GTEX project and analyzed the lncRNA SCNAs in EOC. RNASeq includes sequencing data of 426 EOC patients and sequencing data of 88 normal tissues. The expression data of gene ENSG00000237686 in which RP5-1120P11.1 is showed that the expression level of this gene was also significantly up-regulated in EOC tissues in the database (Figure 2B, P<0.05).
Figure 2.
Upregulation of DATOC-1 in epithelial ovarian cancer tissues. (A) Relative expression of DATOC-1 in 53 EOC and 7 adjacent ovarian samples by qRT-PCR; (B) relative expression of DATOC-1 in 426 epithelial ovarian cancer patients and 88 normal tissues on TCGA and GTEX project; (C) Kaplan-Meier survival analysis for patients with different levels of DATOC-1 in EOC. DATOC-1 expression was normalized against GAPDH expression. *, P<0.05; ***, P<0.001. EOC, epithelial ovarian cancer; TCGA, The Cancer Genome Atlas; GTEx, Genotype-Tissue Expression.
According to the median value of DATOC-1 expression in EOC, the 423 EOC patients were divided into two groups: high expression (n=211) and low expression (n=212) groups. Then we found that patients with higher DATOC-1 expression had significantly reduced overall survival compared with patients with lower DATOC-1 expression, which suggested by the Kaplan-Meier survival analysis (Figure 2C, P=0.034). These results demonstrated DATOC-1 was upregulated in EOC tumors, supporting a potential role in EOC progression.
Effect of DATOC-1 on cell function in EOC cells
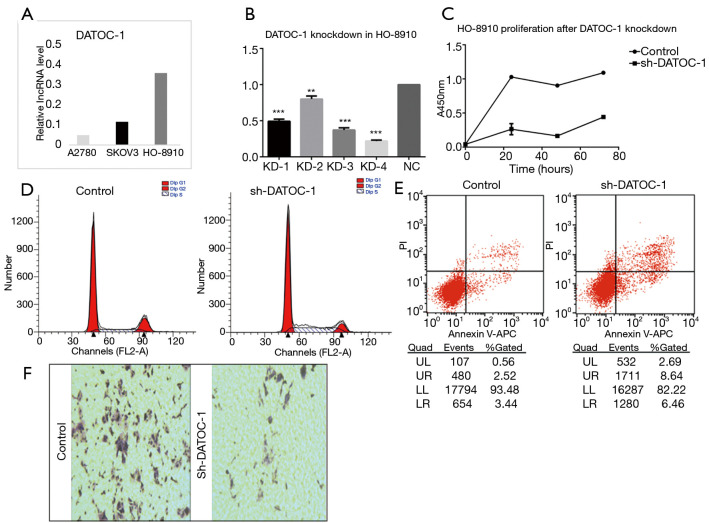
As the upstream modulator of miRNAs, the down-regulation of Dicer can promote tumorigenesis by decreasing the expression of miRNA (5). To determine whether DATOC-1, which combined directly with DICER, are involved in the DICER function, the effect of DATOC-1 in EOC cells was examined using shRNA Screening and lentiviral transduction. The expression of DATOC-1 in diverse EOC cell lines was analyzed by qRT-PCR. The three lines we tested had different expression levels of DATOC-1, with A2780 showing the lowest and HO-8910 showing the highest (Figure 3A). Next, quantification analysis showed the DATOC-1 expression level in HO-8910 after the lentiviral transduction (Figure 3B).And we choose KD4 as the most efficient interfering lentiviral following the infection result. HO-8910 cells were classified into two groups by DATOC-1 expression level (DATOC-1 knockdown and NC as control).
Figure 3.
Silencing of DATOC-1 OC cells suppressing cell proliferation. (A) Relative expression of DATOC-1 in A2780, skov3 and HO-8910 cells; (B) knockdown efficiency ascertained by qPCR in HO-8910 transfected with sh-NC or sh-DATOC-1; (C) knockdown DATOC-1 inhibited cell proliferation in HO-8910 by CCK-8 assay; (D) cell-cycle profiles of HO-8910 cells expressing control and sh-DATOC-1; (E) knockdown DATOC-1 promoted tumor cells apoptosis in HO-8910 by flow cytometry; (F) transwell assay testified silencing of DATOC-1 restrained cell invasion and metastasis. DATOC-1 expression was normalized against GAPDH expression. **, P<0.01; ***, P<0.001. sh-NC, sh-negative control; sh-DATOC-1, sh-DATOC-1 group.
The effect of DATOC-1 expression in cell proliferation was evaluated by CCK-8 assay and inhibited when DATOC-1 low expression compared with control cells (Figure 3C). In addition, the effect of DATOC-1 on cell cycle and apoptosis was analyzed by flow cytometry. Silencing of DATOC-1 blocked the cell cycle in the G1/S and S/G2 phase and promoted cell apoptosis in HO-8910 (Figure 3D,E). To further identify the functional role of DATOC-1 in cell migration and invasion, transwell assay were also performed in HO-8910. Knockdown of DATOC-1 decreased cell migration compared with the controls (Figure 3F). Taken together, these findings demonstrated that DATOC-1 can function as an oncogene.
DATOC-1 promote tumor progression in vivo
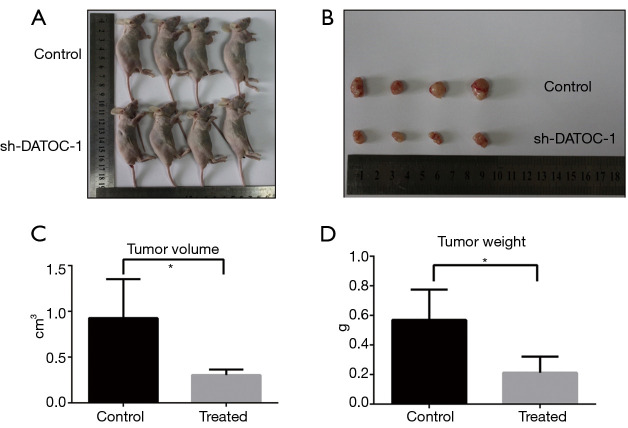
After analyzing the function of DATOC-1 in vitro, we then proceed to perform experiments in vivo. To cover the role of DATOC-1 in tumorigenicity in vivo, we constructed a xenograft tumor model. We stably transfected sh-NC and sh-DATOC-1 cells with lentiviral and injected them into the peritoneum of nude mice to establish abdominal epithelial ovarian tumors. All mice were executed after 35 days, and the growth of tumors in the DATOC-1 shRNA group was significantly slower than those in the control group (Figure 4A,B). Then we measured the volume and weight of tumors of sh-DATOC-1 group and sh-NC group. Tumor volume were significantly reduced in the treatment group which DATOC-1 was low expressed than in control (Figure 4C), so as the tumor weight (Figure 4D).
Figure 4.
Silencing of DATOC-1 suppressing tumor progression in vivo. (A,B) Tumors induced by HO-8910-shDATOC-1 cells and HO-8910-NC cells were excised from nude mice after 35 days; (C,D) the tumors were smaller and lighter in HO-8910-shDATOC-1 mice. *, P<0.05.
DATOC-1 associates with DICER protein and regulates expression of miRNA7
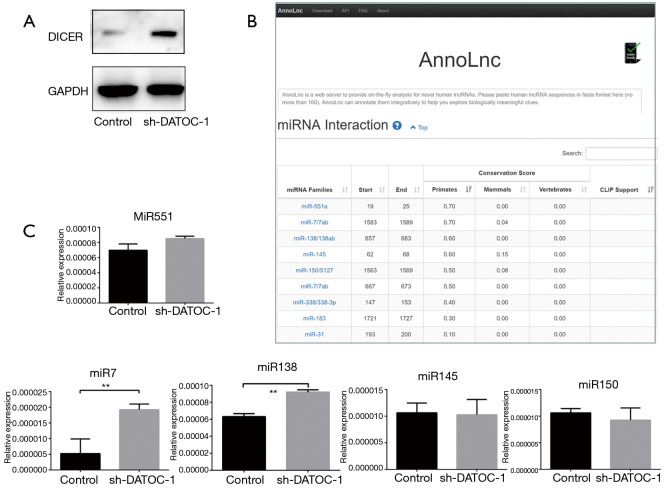
To identify the lncRNA that combined with DICER, we have employed a RIP-seq aimed at DICER and discovered lncRNA RP5-1120P11.1 (DATOC-1) by sequencing comparison. Then to further verify the relationships between DATOC-1 and DICER, we examined DICER expression levels in control and sh-DATOC-1 EOC cells. By comparing the expression levels of DICER in two groups, we found that the expression of DICER increased with the decrease of DATOC-1 expression in HO-8910 (Figure 5A).
Figure 5.
DATOC-1 associates with DICER protein and regulates expression of miRNA7. (A) Expression of DICER increased in sh-DATOC-1 group; (B) the possible target miRNAs we predicted by “AnnoLnc”; (C) the expression of miRNA7 and miR138 were increased in sh-DATOC-1 group. DATOC-1 expression was normalized against GAPDH expression. **, P<0.01.
It is known that DATOC-1 can regulate the expression of DICER. Then, we predict that DATOC-1 can further regulate the expression level of miRNA downstream of DICER, and thus play a role in tumorigenesis and development. We used the online tool “AnnoLnc” published by Peking University in BMC Genomics (23) to predict the possible target miRNAs of DATOC-1. Then we got the following miRNAs such as miR-551, miR-7, miR-150, miR-138, miR-145 (Figure 5B). We validated the expression of predicted miRNAs with a primate over than 50%. And we discovered that the expression of miRNA7 was increased significantly with the knockdown of DATOC-1 (Figure 5C). These data suggest that DATOC-1 may regulate EOC progress by affecting miR7 by acting on DICER.
Discussion
In this work, we identified the lncRNA RP5-1120P11.1 as DATOC-1 (DICER Associated Transcript 1 in Ovarian Cancer), which differentially regulated in EOC cells and tissues. And Kaplan-Meier analysis indicated that the patients with EOC that expressed high levels of DATOC-1 had a worse prognosis and shorter disease EOS compared with DATOC-1 low-expressed patients. In addition, DATOC-1 were further identified participating in EOC cell proliferation, cell cycle regulation and cell invasion. Lastly, we show that knockdown of DATOC-1 inhibits tumor progression in vivo. As a result, we hypothesize that DATOC-1 may be a significant therapeutic target for EOC.
DICER has been reported to be down-regulated in EOC, and its low expression may be responsible for changes in microRNA expression profiles (9,24). MicroRNAs intra-cellular are mainly processed to maturation by DICER. It has been classically reported that DICER expression is suppressed by hypoxia and subsequently, reduced miRNA processing leads to derepression of the miR-200 target ZEB1, stimulates the epithelial to mesenchymal transition in human mammary epithelial cells (25). While, we put forward lncRNA as other mechanism in DICER regulation for the first time. We showed the interaction between DATOC-1 and DICER and confirmed the segment that they combined. Further data supported that the interactions between DATOC-1 and DICER might be required for the regulation of DATOC-1 targets miRNAs. And we demonstrated that DATOC-1 expression promotes EOC development by modulation the expression of miRNA7.
MicroRNA7, as a member of the miRNA family, is mainly located on human chromosome 9q21. miR7 is highly conserved in function and delivers stability in different regulatory mesh systems. Studies had shown that miR7 is significantly low-expressed in various tumor tissues such as human lung cancer, gastric cancer, and malignant schwannomas (26-28). It participates in the occurrence and development of various tumors by inhibiting the expression of EGFR, ACK1 and other genes, thereby functioning as a tumor suppressor gene (26,28,29). Xiaodan Meng found that serum levels of miR-7 (P=0.001) were upregulated in EOC patients compared with healthy women (30). Sun showed that the proliferation and invasion ability of SKOV3 cells were increased in the miR-7 inhibitor group. And the inhibition of miR-7 on the proliferation and invasion of OC might be related to downregulating the expression of EGFR (31). Thus, the expression level of miR7 is closely related to the development, invasion and metastasis of OC. Consistently, we found that miR7 expression increased in HO-8910 cells with DATOC-1 knockdown, which may be the mechanism that DATOC-1 inhibits the progression of EOC. These results strongly indicate that DATOC-1 can become an effective target for the diagnosis and treatment of EOC, which opens a new path for the treatment and prognosis of EOC.
Conclusions
Our research shows that DATOC-1 can inhibit the development of EOC and is a promising therapeutic target.
Acknowledgments
Funding: The study was funded by Top young talents of ten thousand talents plan in Yunnan Province (2019), the medical reserve personnel training program of Yunnan province (H-201609), the General Research Projects in Yunnan Province (2019FB113) and Shanghai Municipal Health Commission (202040242).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of the Second Military Medical University Changzheng Hospital. Informed consent was obtained before surgery and biopsy. All experiments were performed under a project license granted by the Institutional Review Board of the Second Military Medical University Changzheng Hospital, in compliance with the national or institutional guidelines for the care and use of animals.
Footnotes
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-189
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-189
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-189). The authors have no conflicts of interest to declare.
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Biki B, Mascha E, Moriarty DC, et al. Anesthetic technique for radical prostatectomy surgery affects cancer recurrence: a retrospective analysis. Anesthesiology 2008;109:180-7. 10.1097/ALN.0b013e31817f5b73 [DOI] [PubMed] [Google Scholar]
- 3.Qiu JJ, Lin YY, Ye LC, et al. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol 2014;134:121-8. 10.1016/j.ygyno.2014.03.556 [DOI] [PubMed] [Google Scholar]
- 4.Bernstein E, Caudy AA, Hammond SM, et al. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001;409:363-6. 10.1038/35053110 [DOI] [PubMed] [Google Scholar]
- 5.Kumar MS, Lu J, Mercer KL, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 2007;39:673-7. 10.1038/ng2003 [DOI] [PubMed] [Google Scholar]
- 6.Mezzanzanica D, Bagnoli M, De Cecco L, et al. Role of microRNAs in ovarian cancer pathogenesis and potential clinical implications. Int J Biochem Cell Biol 2010;42:1262-72. 10.1016/j.biocel.2009.12.017 [DOI] [PubMed] [Google Scholar]
- 7.Merritt WM, Lin YG, Han LY, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med 2008;359:2641-50. 10.1056/NEJMoa0803785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuang Y, Cai J, Li D, et al. Repression of Dicer is associated with invasive phenotype and chemoresistance in ovarian cancer. Oncol Lett 2013;5:1149-54. 10.3892/ol.2013.1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pampalakis G, Diamandis EP, Katsaros D, et al. Down-regulation of dicer expression in ovarian cancer tissues. Clin Biochem 2010;43:324-7. 10.1016/j.clinbiochem.2009.09.014 [DOI] [PubMed] [Google Scholar]
- 10.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet 2005;14 Spec No 1:R121-32. 10.1093/hmg/ddi101 [DOI] [PubMed] [Google Scholar]
- 11.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012;482:339-46. 10.1038/nature10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904-14. 10.1016/j.molcel.2011.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320-6. 10.1158/0008-5472.CAN-11-1021 [DOI] [PubMed] [Google Scholar]
- 14.Pandey GK, Mitra S, Subhash S, et al. The Risk-Associated Long Noncoding RNA NBAT-1 Controls Neuroblastoma Progression by Regulating Cell Proliferation and Neuronal Differentiation. Cancer Cell 2014;26:722-37. 10.1016/j.ccell.2014.09.014 [DOI] [PubMed] [Google Scholar]
- 15.Yap KL, Li S, Muñoz-Cabello AM, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 2010;38:662-74. 10.1016/j.molcel.2010.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang F, Zhang L, Huo XS, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011;54:1679-89. 10.1002/hep.24563 [DOI] [PubMed] [Google Scholar]
- 17.Hu X, Feng Y, Zhang D, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 2014;26:344-57. 10.1016/j.ccr.2014.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards EJ, Permuth-Wey J, Li Y, et al. A functional variant in HOXA11-AS, a novel long non-coding RNA, inhibits the oncogenic phenotype of epithelial ovarian cancer. Oncotarget 2015;6:34745-57. 10.18632/oncotarget.5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Z, Guo J, Chen L, et al. A long noncoding RNA AB073614 promotes tumorigenesis and predicts poor prognosis in ovarian cancer. Oncotarget 2015;6:25381-9. 10.18632/oncotarget.4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Y, Meng H, Liu S, et al. LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum Mol Genet 2015;24:841-52. 10.1093/hmg/ddu502 [DOI] [PubMed] [Google Scholar]
- 21.Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010;465:1033-8. 10.1038/nature09144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia T, Liao Q, Jiang X, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep 2014;4:6088. 10.1038/srep06088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou M, Tang X, Tian F, et al. AnnoLnc: a web server for systematically annotating novel human lncRNAs. Bmc Genomics 2016;17:931. 10.1186/s12864-016-3287-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Köbel M, Gilks CB, Huntsman DG. Dicer and Drosha in ovarian cancer. N Engl J Med 2009;360:1150-1; author reply 1151. 10.1056/NEJMc090085 [DOI] [PubMed] [Google Scholar]
- 25.van den Beucken T, Koch E, Chu K, et al. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun 2014;5:5203. 10.1038/ncomms6203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao X, Dou W, He L, et al. MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene 2013;32:1363-72. 10.1038/onc.2012.156 [DOI] [PubMed] [Google Scholar]
- 27.Xiong S, Zheng Y, Jiang P, et al. MicroRNA-7 inhibits the growth of human non-small cell lung cancer A549 cells through targeting BCL-2. Int J Biol Sci 2011;7:805-14. 10.7150/ijbs.7.805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saydam O, Senol O, Wurdinger T, et al. miRNA-7 Attenuation in Schwannoma Tumors Stimulates Growth by Upregulating Three Oncogenic Signaling Pathways. Cancer Res 2011;71:852-61. 10.1158/0008-5472.CAN-10-1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peeters M, Price T, Van Laethem JL. Anti-Epidermal Growth Factor Receptor Monotherapy in the Treatment of Metastatic Colorectal Cancer: Where Are We Today? Oncologist 2009;14:29-39. 10.1634/theoncologist.2008-0167 [DOI] [PubMed] [Google Scholar]
- 30.Meng X, Joosse SA, Müller V, et al. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br J Cancer 2015;113:1358-66. 10.1038/bjc.2015.340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun LB, Li GH, Cao DH. Effect of miR-7 targeting EGFR gene on the proliferation and invasion of ovarian cancer SKOV3 cells. Progress of Anatomical Sciences 2017;23:360-2, 366.