Abstract
Streptococcus uberis and Streptococcus parauberis reference strains and isolates obtained from routine diagnostics were investigated by PCR with oligonucleotide primers designed according to species-specific parts of the 16S rRNA gene, the 23S rRNA gene, and the 16S-23S rRNA intergenic spacer region of both species. All three primer pairs allowed an identification of 67 isolates as S. uberis and 4 isolates as S. parauberis.
The use of PCR as a diagnostic tool for the detection of bacterial pathogens has become more frequent during the past few years. The PCR technique allows the amplification of preselected, species-specific DNA regions which can be used for genotypic identification of bacteria. A molecule most suited for these purposes appears to be the gene encoding the 16S rRNA. According to Bentley et al. (4) and Bentley and Leigh (6), the sequence variability of the V2 region of the 16S rRNA allows a differentiation of 31 species of the genus Streptococcus, including the species S. uberis and S. parauberis. Both species, formerly classified as S. uberis types I and II (19), are well known as causative agents of bovine mastitis. According to biochemical and serological characteristics, the two species are almost indistinguishable (19). A molecular identification of both species can be performed by analysis of restriction fragment length polymorphisms (RFLPs) of the 16S rRNA gene (13, 14, 18) or by the use of species-specific oligonucleotide probes (5, 6). A major disadvantage of these procedures is the additional manipulation of the samples subsequent to the PCR and the time-consuming preparation of the gene probes. In the present study, species-specific regions of the 16S and 23S rRNA genes and the 16S-23S rRNA intergenic spacer region of S. uberis and S. parauberis were used to construct species-specific oligonucleotide primers. These oligonucleotide primers were used to identify and differentiate both species.
A total of 17 S. uberis and 2 S. parauberis strains were used in this study, all of which were obtained from the Institute's strain collection. The cultures were identified and differentiated into both species as described previously (15, 18). In addition, 51 streptococci obtained from routine mastitis diagnostics and also S. parauberis strain 94/16 were included. The latter, kindly obtained from J. F. Fernández-Garayzábal (Facultad de Veterinaria, Universidad Complutense de Madrid, Madrid, Spain), was originally isolated from a diseased turbot (9). The cultures isolated from routine diagnostics were preliminarily identified as S. uberis by using conventional methods (15, 16). These methods included carbohydrate fermentation tests for arabinose, fructose, inulin, lactose maltose, mannitol, raffinose, ribose, saccharose, salicin, sorbitol, and trehalose; esculin and sodium hippurate hydrolysis; determination of arginine hydrolysis; and analysis of the enzymes β-d-glucuronidase, pyrrolydonylaminopeptidase, and hyaluronidase. The β-d-glucuronidase and pyrrolydonylaminopeptidase enzyme activities were investigated with diagnostic tablets (Rosco, Hiss Diagnostics, Freiburg, Germany) as substrates, and the enzyme hyaluronidase activity was determined with a plate decapsulation test with cultivation of the isolates in close proximity to a growing mucoid Streptococcus equi subsp. zooepidemicus strain. The 51 isolates were additionally investigated by RFLP analysis of the 16S rRNA gene, using the restriction enzymes RsaI and AvaII (18). For control purposes, streptococci of the species S. pyogenes (n = 4), S. agalactiae (n = 12), S. equi subsp. equi (n = 2), S. equi subsp. zooepidemicus (n = 3), S. dysgalactiae serogroups C (n = 4), G (n = 4), and L (n = 4), S. canis (n = 12), and S. porcinus (n = 4) were included.
Based on the sequence analysis of the V2 region of the 16S rRNA gene reported by Bentley and Leigh (6) and the sequence data of the variable region of the 23S rRNA gene given by Harland et al. (12), species-specific primers were designed using the primer design program OLIGO 4.0.
The primer design of the species-specific part of the 16S-23S rRNA intergenic spacer region was performed after sequencing of the genes of S. uberis and S. parauberis reference strains. For the intergenic spacer region of S. uberis, the primer sequences recommended by Forsman et al. (10) were used.
For sequencing, the intergenic spacer regions of the S. parauberis reference strain NCDO 2020 and the S. uberis reference strain NCDO 2038 were amplified with the oligonucleotide primers c (5′-3′, TTGTACACACCGCCCGTCA) and b (5′-3′, GGTACCTTAGTTTCAGTTC) described by Chanter et al. (7). The primers were derived from conserved regions within the 16S rRNA (primer c) and the 23S rRNA (primer b) genes. The oligonucleotide primers were synthesized by ARK-Biosystem (Darmstadt, Germany). The sequencing was performed with the MegaBACE 1000 DNA sequencing system (Amersham Pharmacia Biotech, Europe, Freiburg, Germany) according to the protocols described by the manufacturer. The sequence data were further studied and analyzed with the computer program MegAlign 1993-97 (DNASTAR Inc., Constance, Germany).
The target genes, the oligonucleotide primers used, and the sizes of the amplicons are summarized in Table 1. For PCR, the reaction mixture (30 μl) contained 1 μl (16S rRNA gene, 16S-23S rRNA spacer region) or 0.8 μl (23S rRNA gene) of primer 1 (10 pmol/liter), 1 μl or 0.8 μl of primer 2 (10 pmol/liter), 0.6 μl of deoxynucleotide triphosphate (10 mmol; MBI Fermentas, St. Leon-Rot, Germany), 3 μl of 10× thermophilic buffer (Promega, Boehringer, Ingelheim, Germany), 1.8 μl (16S rRNA gene, 16S-23S rRNA spacer region) or 1.5 μl (23S rRNA gene) of MgCl2 (25 mmol; Promega), 0.1 μl of Taq DNA polymerase (5 U/μl; Promega), and 20.0 μl or 20.7 μl of distilled water. Finally, 2.5 μl of DNA preparation was added to each reaction tube. For DNA preparations, 5 to 10 colonies of the bacteria were first suspended in 100 μl of TE buffer (10 mmol/liter Tris-HCl, 1 mmol/liter EDTA [pH 8.0]) containing 5 μl of mutanolysin (50 U; Sigma, Deisenhofen, Germany) for 60 min at 37°C, and then the bacteria were suspended in 10 μl of proteinase K (14.8 mg/ml; Boehringer) for 120 min at 56°C. After boiling for 10 min at 100°C, the suspension was centrifuged (10,000 × g, 5 s) and subsequently cooled before use. The tubes were then placed in a thermal cycler (Techne-Progene; Thermodux, Wertheim, Germany) with the programs summarized in Table 1.
TABLE 1.
Oligonucleotide primers and PCR programs for amplification of species-specific parts of the genes encoding the 16S rRNA, 23S rRNA and 16S-23S rRNA intergenic spacer region of S. uberis and S. parauberis
| Species | Target rRNA gene | Oligonucleotide primer | Sequence (5′–3′) | Length (nt) | Size of PCR product (bp) | PCR programa |
|---|---|---|---|---|---|---|
| S. uberis | 16S | ub-I | CGC ATG ACA ATA GGG TAC A | 19 | 445 | 1 |
| ub-II | GCC TTT AAC TTC AGA CTT ATC A | 22 | ||||
| 23S | ub-23S-I | CGT ATT TAA AAT TGA CTT TAG CC | 23 | 451 | 2 | |
| ub-23S-II | AAT TTC TCC GCT ACC CAC | 18 | ||||
| 16S-23S intergenic spacer | STRU-UbIb | TAA GGA ACA CGT TGG TTA AG | 20 | 330 | 3 | |
| STRU-UbIIb | TCC AGT CCT TAG ACC TTC T | 19 | ||||
| S. parauberis | 16S | paraub-I | CAT GAC AAT TAA GTA CTC ATG TAC TA | 26 | 884 | 1 |
| paraub-II | CAC CAC CTG TCA CCT CTG TC | 20 | ||||
| 23S | paraub-23S-I | AAA ATA GTA AAT GAC TCT AGC AGT | 24 | 478 | 2 | |
| paraub-23S-II | CGG AGA GAA CCA GCT ATC | 18 | ||||
| 16S-23S intergenic spacer | paraub-16S-23S-I | AAA TGG AAG CAC GTT AGG AAA | 21 | 201 | 4 | |
| paraub-16S-23S-II | GCA AGC CGA ACA TCT CTT TG | 20 |
1 = 30× (94°C 60 s, 58°C 90 s, 72°C 90 s); 2 = 30× (94°C 45 s, 64°C 45 s, 72°C 45 s); 3 = 30× (94°C 30 s, 55°C 30 s, 72°C 30 s); 4 = 30× (94°C 10 s, 58°C 10 s, 72°C 10 s).
Forsman et al. (10).
The presence of PCR products was determined by electrophoresis of 12 μl of the reaction product in a 2% agarose gel with Tris acetate-electrophoresis buffer (TAE; 40 mmol of Tris-HCl per liter, 1.14 mol of glacial acetic acid per liter, 1 mmol of EDTA per liter [pH 8.0]) with a 100-bp DNA ladder (Gibco BRL, Eggenstein, Germany) as molecular size markers.
The oligonucleotide primers designed from the species-specific part of the 16S rRNA and 23S rRNA genes allowed a molecular identification of 67 isolates as S. uberis and 4 isolates as S. parauberis. The latter also included the S. parauberis strain 94/16, which was originally isolated from a diseased turbot (9). Both PCR systems produced identical results (Fig. 1). The identity of the S. uberis and S. parauberis isolates could additionally be confirmed by RFLP analysis (data not shown) and by the use of primer pairs amplifying a species-specific region of the 16S-23S rRNA intergenic spacer region. The consensus sequences of the intergenic spacer regions of both species are given in Fig. 2. All control strains of various species and serogroups were negative throughout. As reported earlier (13, 18), the occurrence of S. parauberis as a mastitis-causing pathogen appears to be rare. This species seems to be more frequent among British dairy herds (11). Isolation of S. parauberis from diseased fish was described by Doménech et al. (9). According to the results of the present study, the species-specific regions of the described target genes of S. uberis and S. parauberis did not show any sequence variations. Corresponding to these results, no intraspecies variations were reported in the 16S rRNA genes of S. agalactiae (1, 17) and S. porcinus (3). However, a variation in the sequence of the 16S rRNA gene was observed for S. suis (8) and for S. equi subsp. zooepidemicus (2).
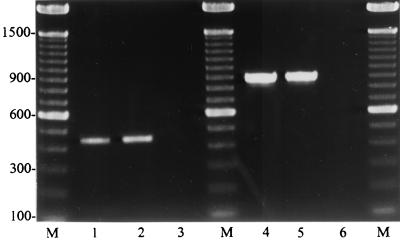
FIG. 1.
Typical amplicons of S. uberis (lanes 1 and 2) with a size of 445 bp and with S. parauberis (lane 3) as negative control, using the S. uberis 16S rRNA-specific primers ub-I and ub-II, and amplicons of S. parauberis (lanes 4 and 5) with a size of 884 bp and S. uberis (lane 6) as negative control, using the S. parauberis 16S rRNA-specific primers paraub-I and paraub-II. M, 100-bp ladder as size marker.
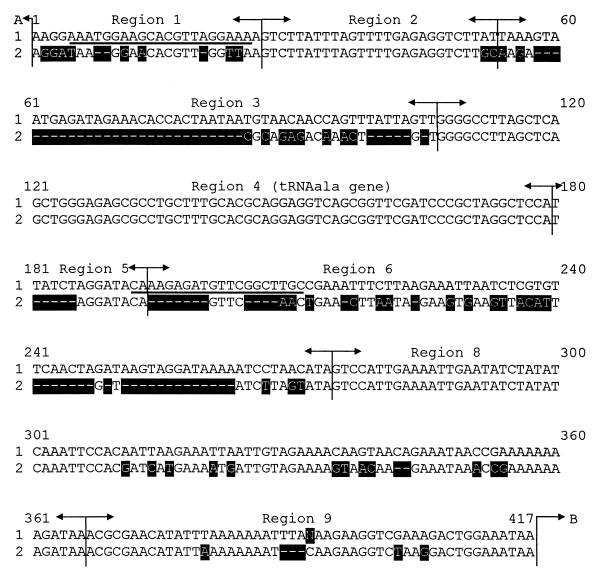
FIG. 2.
Alignment of the consensus gene sequences of the 16S-23S rRNA intergenic spacer region of S. parauberis NCDO 2020 (417 bp) (accession no. AF255656) and S. uberis NCDO 2038 (338 bp) (accession no. AF255657). The selected S. parauberis-specific oligonucleotide primers are underlined; the marked areas indicate the differences in nucleotide sequences. The region arrangement was performed according to Chanter et. al. (7); stuffer regions inserted to achieve alignment are indicated by –––––. Sequence 1, S. parauberis NCDO 2020; sequence 2, S. uberis NCDO 2038. A (top), end of 16S rRNA gene; B (bottom right), start of 23S rRNA gene.
The PCR amplification of species-specific sequences of S. uberis and S. parauberis in the present study offers a rapid and sensitive method by which to identify both biochemically and serologically almost indistinguishable species. This might help in determining the prevalence of both species in dairy herds.
REFERENCES
- 1.Abdulmawjood A, Lämmler C. Amplification of 16S ribosomal RNA gene sequences for the identification of streptococci of Lancefield group B. Res Vet Sci. 1999;67:159–162. doi: 10.1053/rvsc.1998.0298. [DOI] [PubMed] [Google Scholar]
- 2.Abdulmawjood A, Lämmler C. Determination of intraspecies variations of the V2 region of the 16S rRNA gene of Streptococcus equi subsp. zooepidemicus. Res Vet Sci. 2000;68:33–39. doi: 10.1053/rvsc.1999.0332. [DOI] [PubMed] [Google Scholar]
- 3.Abdulmawjood A, Weiß R, Lämmler C. Species identification of Streptococcus porcinus by restriction fragment length polymorphism analysis of 16S ribosomal DNA. Res Vet Sci. 1998;65:85–86. doi: 10.1016/s0034-5288(98)90033-9. [DOI] [PubMed] [Google Scholar]
- 4.Bentley R W, Leigh J A, Collins M D. Intrageneric structure of Streptococcus based on comparative analysis of small-subunit rRNA sequences. Int J Syst Bacteriol. 1991;41:487–494. doi: 10.1099/00207713-41-4-487. [DOI] [PubMed] [Google Scholar]
- 5.Bentley R W, Leigh J A, Collins M D. Development and use of species-specific oligonucleotide probes for differentiation of Streptococcus uberis and Streptococcus parauberis. J Clin Microbiol. 1993;31:57–60. doi: 10.1128/jcm.31.1.57-60.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley R W, Leigh J A. Development of the PCR-based hybridization protocol for identification of streptococcal species. J Clin Microbiol. 1995;33:1296–1301. doi: 10.1128/jcm.33.5.1296-1301.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chanter N, Collin N, Holmes N, Binns M, Mumford J. Characterization of the Lancefield group C Streptococcus 16S–23S rRNA gene intergenic spacer and its potential for identification and sub-specific typing. Epidemiol Infect. 1997;118:125–135. doi: 10.1017/s0950268896007285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatellier S, Harel J, Zhang Y, Gottschalk M, Higgins R, Devriese L A, Brousseau R. Phylogenetic diversity of Streptococcus suis strains of various serotypes as revealed by 16S rRNA gene sequence comparison. Int J Syst Bacteriol. 1998;48:581–589. doi: 10.1099/00207713-48-2-581. [DOI] [PubMed] [Google Scholar]
- 9.Doménech A, Fernández-Garayzábal J F, Pascual C, Garcia J A, Cutuli M T, Moreno M A, Collins M D, Dominguez L. Streptococcosis in cultured turbot, Scophthalmus maximus (L.), associated with Streptococcus parauberis. J Fish Dis. 1996;19:33–38. [Google Scholar]
- 10.Forsman P, Tilsala-Timisjärvi A, Alatossava T. Identification of streptococcal causes of bovine mastitis using 16S–23S rRNA spacer regions. Microbiology. 1997;143:3491–3500. doi: 10.1099/00221287-143-11-3491. [DOI] [PubMed] [Google Scholar]
- 11.Garvie E I, Bramley A. Streptococcus uberis: an approach to its classification. J Appl Bacteriol. 1979;46:295–304. doi: 10.1111/j.1365-2672.1979.tb00855.x. [DOI] [PubMed] [Google Scholar]
- 12.Harland N M, Leigh J A, Collins M D. Development of gene probes for the specific identification of Streptococcus uberis and Streptococcus parauberis based upon large subunit rRNA gene sequences. J Appl Bacteriol. 1993;74:526–531. [PubMed] [Google Scholar]
- 13.Jayarao B M, Doré J J F, Jr, Baumbach G A, Matthews K R, Oliver S P. Differentiation of Streptococcus uberis from Streptococcus parauberis by polymerase chain reaction and restriction fragment length polymorphism analysis of the 16S ribosomal DNA. J Clin Microbiol. 1991;29:2774–2778. doi: 10.1128/jcm.29.12.2774-2778.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayarao B M, Doré J J E, Jr, Oliver S P. Restriction fragment length polymorphism analysis of 16S ribosomal DNA of Streptococcus and Enterococcus species of bovine origin. J Clin Microbiol. 1992;30:2235–2240. doi: 10.1128/jcm.30.9.2235-2240.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lämmler C. Biochemical and serological properties of Streptococcus uberis. J Vet Med. 1991;38:737–742. doi: 10.1111/j.1439-0450.1991.tb00937.x. [DOI] [PubMed] [Google Scholar]
- 16.Lämmler C, Hahn G. Streptokokken. In: Blobel H, Schließer T, editors. Handbuch der bakteriellen Infektionen bei Tieren. Band II/2: Streptokokken-Infektionen und Rotlauf. 2. Auflage. Jena, Germany: Gustav Fischer Verlag; 1994. [Google Scholar]
- 17.Lämmler C, Abdulmawjood A, Weiß R. Properties of serological group B streptococci of dog, cat and monkey origin. J Vet Med. 1998;B45:561–566. doi: 10.1111/j.1439-0450.1998.tb00827.x. [DOI] [PubMed] [Google Scholar]
- 18.Lämmler C, Abdulmawjood A, Danic G, Vaillant S, Weiß R. Differentiation of Streptococcus uberis and Streptococcus parauberis by restriction fragment length polymorphism analysis of the 16S ribosomal RNA gene and further studies on serological properties. Med Sci Res. 1998;26:177–179. [Google Scholar]
- 19.Williams A M, Collins M D. Molecular taxonomic studies on Streptococcus uberis types I and II. Description of Streptococcus parauberis sp. nov. J Appl Bacteriol. 1990;68:485–490. doi: 10.1111/j.1365-2672.1990.tb02900.x. [DOI] [PubMed] [Google Scholar]