Abstract
Background
We sought to determine the differences with respect to the proteome of nasopharyngeal tissues between patients with nasopharyngeal carcinoma (NPC) and healthy controls by using sequential windowed acquisition of all theoretical fragment ion mass spectra (SWATHTM-MS) and ingenuity pathway analysis (IPA). Our primary purpose was to identify specific protein markers that can be applied for diagnosis or treatment of NPC.
Methods
The CNE-1, CNE-2 and H1299 cell lines were cultured in stable isotope labeling of amino acids in cell culture (SILAC) medium for 10 generations to obtain labeled proteins. Thirty samples of NPC and 30 healthy control nasopharyngeal tissues were collected from the Department of Otolaryngology of the First Affiliated Hospital of Jinan University. Proteome of the nasopharyngeal tissues were analyzed and compared by SWATH-MS to identify differently expressed proteins. Further, extraction of target proteins and biological pathways was performed by IPA. Super-SILAC technique and liquid chromatography-tandem mass spectrometry were used to verify the reliability of the data obtained using SWATH-MS.
Results
We identified 1,415 differentially expressed proteins between NPC patients and healthy controls. On IPA analysis, EIF2AK2 and MAPK1 proteins were found to be enriched in multiple biological pathways and functional networks.
Conclusions
The differentially expressed proteins EIF2AK2 and MAPK seem to play an important role in the biological network of NPC or may help discover the specific functional proteins of NPC. Further studies are required to identify the pathways and molecular mechanisms that underlie NPC.
Keywords: Nasopharyngeal carcinoma (NPC), proteomics, biomarkers, sequential windowed acquisition of all theoretical fragment ions mass spectrometry (SWATH-MS), ingenuity pathway analysis (IPA)
Introduction
Nasopharyngeal carcinoma (NPC) is a distinct type of head and neck cancer which originates from nasopharyngeal epithelial cells, and is highly prevalent in Southeast Asia and Southern China (1,2). Typical characteristics of NPC include concealed primary site, poor differentiation, and a tendency for early local invasion and metastatic spread to the head and neck lymph nodes. Histopathological diagnosis of NPC requires an invasive procedure which makes it unsuitable for large-scale screening and routine examination; moreover, nasopharyngeal biopsy of small or deep lesions frequently yields false-negative results (3).
The rapid advances in molecular biology technology have enabled wider use of proteomic approaches. Over the last decades, liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) has become the technology of choice for high-throughput characterization of proteins and proteomes (4). Sequential windowed acquisition of all theoretical fragment ion mass spectra (SWATH-MS) is an emerging strategy that typically involves digestion of non-labeled protein samples with trypsin and analysis of the resulting peptides by liquid chromatography coupled to a tandem mass spectrometer operating in the so-called data-independent acquisition (DIA) mode. Advances in SWATH-MS technology have facilitated its application in the fields of personalized medicine, biomarker research, drug screens, genetic association studies, and systems biology (5). Comparative proteomics has helped identify a large number of potential biomarkers associated with different cancers, such as lung cancer (6), renal cancer (7), and NPC (8). Analysis of proteomic changes can accurately reveal the changes in body proteins and help identify the protein markers of diagnostic and therapeutic relevance.
In the present study, we compared the differentially expressed proteins in the nasopharyngeal tissues of NPC patients and healthy controls using SWATH-MS and performed ingenuity pathway analysis (IPA) to extract target proteins and biological pathways. Our objective was to identify potential biomarkers which can be used for early diagnosis, prognostic assessment, and for designing new targeted therapy for NPC.
Methods
Cell culture and tissue samples
The CNE-1, CNE-2 and H1299 cell lines were obtained from the China Center of Type Culture Collection (CCTCC). Cells were cultured in stable isotope labeling of amino acids in cell culture (SILAC) medium (Thermo Fisher, USA) supplemented with 73 mg/L Lys8, 42 mg/L Arg10, and 10% dialyzed fetal bovine serum (FBS, Life Technologies, USA).
Nasopharyngeal tissue specimens of 30 patients with NPC and 30 aged-matched healthy controls were collected from the Department of Otolaryngology. All tumor tissue specimens were histologically confirmed and retrieved from the Department of Pathology at the First Affiliated Hospital of Jinan University (Guangzhou, China). The present study was approved by the hospital Ethics Committee. The enrolled patients provided informed consent to participate in this study. All patients had primary tumor and were newly-diagnosed and untreated. Further, laboratory tests (routine blood tests, biochemical, chest X-ray, electrocardiogram, and B-ultrasound) were performed to rule out interference of other concomitant diseases.
Fresh nasopharyngeal mucosal tissue specimens were rinsed twice with cold phosphate buffer saline (PBS) and placed in an Eppendorf tube. Tissue samples were snap frozen in liquid nitrogen directly after excision, and stored at −80 °C.
Super-SILAC standard and protein sample preparation
CNE-1, CNE-2, and H1299 cells were cultured in SILAC medium. The medium was changed after every two days. Marking test was performed after 10 generations of cultivation. Cells were lysed with SDS buffer [4% SDS and 100 mM Tris/HCl (pH 7.6), 100 µL/1×106 cells] for 10 min in boiling water at 95 . These were sonicated for 60 s (amplitude 20%, interval 5 s), and cell pellets obtained after centrifugation at 16,500 ×g for 10 min. The protein concentration in the supernatant was measured using the BCA kit (Thermo Fisher, USA).
Processing of the tissues for protein estimation
The samples were from cold storage (−80 °C) and allowed to thaw at room temperature. A surgical blade was used to separate the target cells from the surrounding tissue. PBS washes (2×) were performed to remove excess of blood. The sample was shredded with scalpel and placed into a mortar previously cooled with liquid nitrogen, and then ground into powder in the liquid nitrogen environment. The grounded tissue was subsequently subjected to lysis.
Sample lysis
Each 20 mg wet weight tissue was lysed with 200 µL of SDS buffer for 30 min on ice (vortex shake for 1 min, interval 10 min) and centrifuged at 16,500 ×g for 10 min. The protein concentration in the supernatant was measured using the BCA kit.
Ultrafiltration tube enzymatic hydrolysis, isolation and purification with SAX stage tip
Peptide separation and purification were performed according to the procedure described elsewhere (9) and the technology roadmap (Figure 1) will give a clearer explanation.
Figure 1.
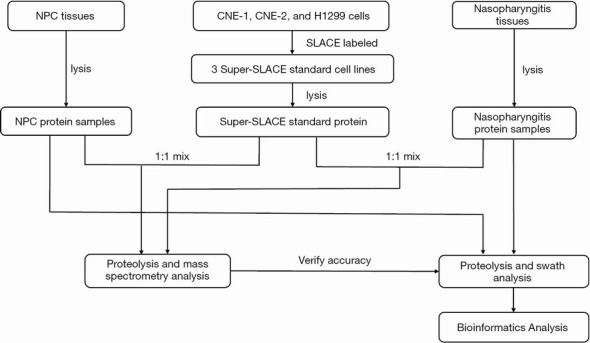
Processing of the tissues.
Mass spectrometry analysis
The sample was analyzed by LC-MS/MS for a duration of 4 h (120 min + 90 min + 30 min), as the mode is shown in Table 1.
Table 1. Analyzing mode.
| Mode | Analyzer |
|---|---|
| ProteinID-Bul-120 min | LC-MS/MS |
| SWATH-120 min | SWATH-MS |
| ProteinID-Bul-90 min | Elution |
| Autocal-2 uL-30 min | Correction |
LC-MS/MS, liquid chromatography coupled to tandem mass spectrometry; SWATH-MS, sequential windowed acquisition of all theoretical fragment ions mass spectrometry.
Each sample was eluted and corrected once. The protein database was selected from the UniProt Human and Epstein-Barr virus databases (downloaded on November 6, 2014).
Results
LC-MS/MS
A total of 3,974 proteins were detected from the 30 NPC tissues, which conformed to the confidence conditions (at least 1 peptide with 95% confidence score, unused score >1.3, and FDR <1%, verified by critical false discovery rates). A total of 4,703 proteins were detected from 30 normal nasopharyngeal tissue samples. A total of 3,550 identical proteins (Tables 2,3) were observed of which 1,814 proteins were up-regulated and 1,746 proteins were down-regulated in NPC tissues.
Table 2. Comparison of co-confidence protein content in NPC tissue and normal nasopharyngeal tissue (up-regulated most).
| Normal tissue | H:L | NPC tissue | H:L | NPC/normal | ↑1, ↓−1 |
|---|---|---|---|---|---|
| Q9UHJ6 | 78.66655731 | Q9UHJ6 | 0.01010315 | 7,786.339638 | 1 |
| Q9Y3D3 | 78.66655731 | Q9Y3D3 | 0.109218903 | 720.2650379 | 1 |
| Q13422 | 6.447723866 | Q13422 | 0.01010315 | 638.1894622 | 1 |
| Q15067 | 2.477478027 | Q15067 | 0.01010315 | 245.2183752 | 1 |
| P51159 | 78.66655731 | P51159 | 0.324296415 | 242.576093 | 1 |
| … |
NPC, nasopharyngeal carcinoma.
Table 3. Comparison of co-confidence protein content in NPC tissue and normal nasopharyngeal tissue (down-regulated most).
| Normal tissue | H:L | NPC tissue | H:L | NPC/normal | ↑1, ↓−1 |
|---|---|---|---|---|---|
| P31323 | 0.01 | P31323 | 0.88800931 | 0.011261143 | −1 |
| O95197 | 0.01 | O95197 | 0.968087196 | 0.010329648 | −1 |
| Q5T5Y3 | 0.968716025 | Q5T5Y3 | 100 | 0.00968716 | −1 |
| Q9P015 | 0.954339981 | Q9P015 | 100 | 0.0095434 | −1 |
| Q86YV0 | 0.094613113 | Q86YV0 | 10.05469036 | 0.009409848 | −1 |
| … |
NPC, nasopharyngeal carcinoma.
SWATH-MS
Thirty NPC and 30 normal nasopharyngeal protein samples were subjected to SWATH-MS (AB SCIEX, USA). The results were analyzed using the MaxQuant (AB SCIEX, USA) software. A total of 1,415 proteins were found to meet the confidence conditions. The content of confidence proteins in each sample is shown in Tables 4 and 5.
Table 4. Content of confidence proteins detected by SWATH-MS in normal nasopharyngeal tissue samples (partial data).
| Protein | C1 | C2 | C3 | C4 | C5 | … |
|---|---|---|---|---|---|---|
| Q09666 | 3,432,328 | 2,455,664 | 2,039,570 | 10,809.2 | 1,840,391 | |
| P35579 | 7,354,728 | 4,588,434 | 1.6E+07 | 49,017.7 | 3,181,416 | |
| Q15149 | 1,276,620 | 249,562 | 1,212,375 | 88,058.4 | 87,662.4 | |
| P21333 | 1,997,568 | 740,188 | 2,233,986 | 37,837.1 | 92,363.5 | |
| P13647 | 2.5E+07 | 3.6E+07 | 1.8E+07 | 976,931 | 3.3E+07 | |
| … |
SWATH-MS, sequential windowed acquisition of all theoretical fragment ions mass spectrometry.
Table 5. Content of confidence proteins detected by SWATH-MS in NPC tissue samples (partial data).
| Protein | N1 | N2 | N3 | N4 | N5 | … |
|---|---|---|---|---|---|---|
| Q09666 | 1,370,274 | 2,178,366 | 2,631,980 | 3,332,224 | 5,665,845 | |
| P35579 | 1,815,488 | 2,437,102 | 6,019,342 | 8,255,914 | 7,352,259 | |
| Q15149 | 222,778 | 225,434 | 292,301 | 3,092,402 | 1,086,874 | |
| P21333 | 102,282 | 595,178 | 753,666 | 1.1E+07 | 3,068,637 | |
| P13647 | 1.6E+07 | 2.3E+07 | 2.1E+07 | 8,076,338 | 1.1E+07 | |
| … |
SWATH-MS, sequential windowed acquisition of all theoretical fragment ions mass spectrometry; NPC, nasopharyngeal carcinoma.
IPA pathway analysis
The comparative data tool and interactive Venn diagram analysis showed that 1,286 of the 1,415 proteins identified by SWATH-MS were in agreement with those identified by LC-MS/MS (Figure 2). This indicated the reliability of results obtained by SWATH-MS.
Figure 2.
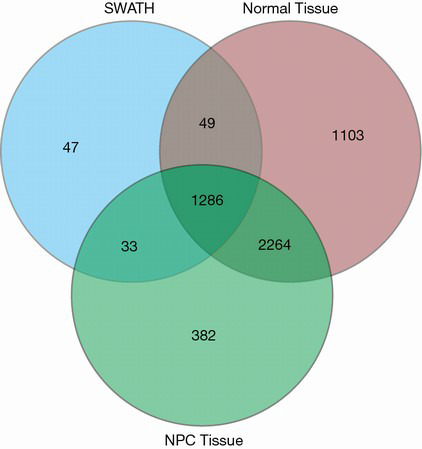
Comparison of the results of LC-MS/MS and SWATH-MS. Blue color represents the proteins detected by SWATH-MS; the red and green represent proteins detected by LC-MS/MS in normal nasopharyngeal tissue and NPC, respectively. LC-MS/MS, liquid chromatography coupled to tandem mass spectrometry; SWATH-MS, sequential windowed acquisition of all theoretical fragment ions mass spectrometry; NPC, nasopharyngeal carcinoma.
The protein data obtained by SWATH-MS was imported into the IPA software of Ingenuity® Systems for analysis (10). Most of these differential proteins were localized in the cells. Their main functions were post-transcriptional modification, cell growth and proliferation, protein synthesis, cell death, and regulation of gene expression (Figure 3); these have been implicated in infectious diseases, skin-related diseases, nervous system-related diseases, musculoskeletal diseases, and inflammatory diseases (Figure 4).
Figure 3.
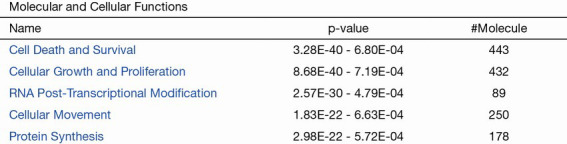
443 proteins are involved in cell death and survival, 432 proteins are involved in cell growth and proliferation, 89 proteins are involved in RNA post-transcriptional modification, 250 proteins are involved in cell movement, and 178 are involved in protein synthesis.
Figure 4.
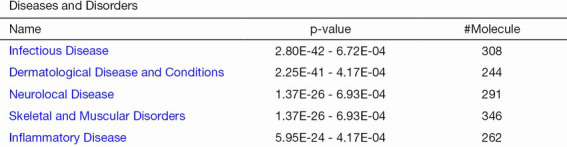
308 proteins are associated with infectious diseases, 244 proteins are associated with dermatological diseases and conditions, 291 proteins are associated with neurological diseases, 346 are associated with skeletal and muscular diseases, and 262 proteins are associated with inflammatory diseases.
Top disease and bio function analysis
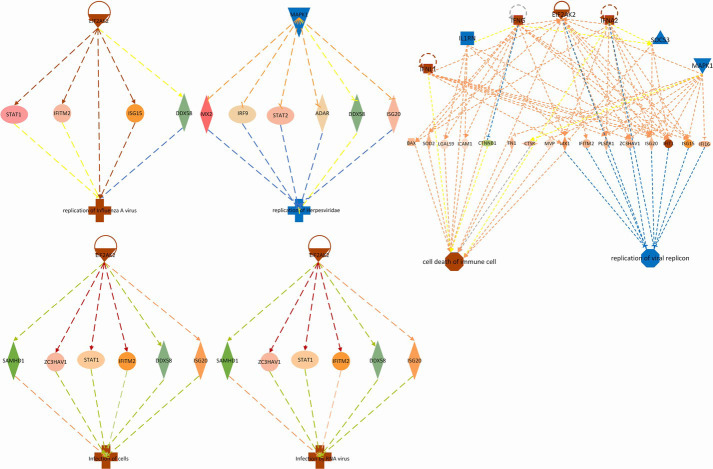
Different proteins form multiple biological function loops according to their functions and interactions (Figure 5). It is worth noting that EIF2AK2 and mitogen-activated protein kinase 1 (MAPK1) show their prominence in this analysis and participated in multiple biological networks. These function as promoters, mediators of death of immune cells, and are involved in replication of important DNA replicons, various viral infections, and replication of herpes virus (which is implicated in tumorigenesis). Therefore, further studies of EIF2AK2 and MAPK1 proteins may help identify specific biomarkers for NPC.
Figure 5.
MAPK1 and EIF2AK2 regulated proteins in SWATH-MS analyses. Up-regulated and down-regulated proteins are highlighted in red and green, respectively, and the color depth is correlated to the fold-change. Orange and blue dashed lines with arrows indicate indirect activation and inhibition, respectively. Yellow and gray dashed lines with arrows depict inconsistent effects and no prediction, respectively. SWATH-MS, sequential windowed acquisition of all theoretical fragment ions mass spectrometry.
Discussion
Carcinogenesis involves a series of genetic alterations which cause progressive derangement of the normal mechanisms that control cell growth (11). Investigation of protein profiles and their interactions and functions at the cellular level has evoked increasing attention in recent years. In particular, their biological behavior and regulatory mechanisms of gene expression in different physiological and pathological environments is an emerging area of research. Proteomics has been widely studied in various diseases and especially in the context of tumors. This has enabled a better understanding of the etiology, and the mechanisms of carcinogenesis and cancer progression (12). Proteomic approaches are useful for the study of human proteome of NPC (13). Many diagnostic and predictive biomarkers for NPC have been identified using the proteomic approach. However, further studies are required to unravel the mechanisms and perform clinical validation (8).
MS/MSALL combined with SWATH-MSTM acquisition is a powerful technology developed using fast scanning and high sensitivity features of the TripleTOFTM 5600+ system. It can be used for both qualitative and quantitative analysis (14).
In the present study, we applied the SILAC technique, LC-MS/MS and SWATH-MS to identify differentially expressed proteins between NPC and normal nasopharyngeal tissues. Further, we used LC-MS/MS to confirm the results of SWATH-MS; 92% proteins identified by SWATH-MS were also identified by LC-MS/MS. Thus, our results demonstrate the feasibility of use of SWATH-MS for conducting further experiments. The subjective analysis and Top Disease and Bio Function analysis of IPA bioanalytical software helps reveal the functional network map of the differentially expressed proteins, and provides more comprehensive and organic understanding for further research on NPC. The EIF2AK2 and MAPK1 proteins are known to be involved in multiple biological network functions and act as promoters. Our findings may help identify specific molecular biomarkers of NPC and specific functional proteins or facilitate discovery of specific proteins for NPC in future research.
EIF2AK2, also known as PRKR, is a eukaryotic translation initiation factor 2-alpha kinase 2 (eIF2α). There are four kinds of protein kinases of eIF2α in mammalian cells. These are composed of three subunits (α, β and γ), where phosphorylation of serine at the 51-regulated subunit inhibits the activity of eIF2B (15,16). It is an important signal node for initiation of translation, and for changing the signaling pathways of cell cycle, cell checkpoints, energy, and nutrients though regulation of translation. EIF2 can combine with ATP, bind double-stranded RNA, and activate eIF2α kinase to participate in cell apoptosis, translation, phosphorylation, growth, and proliferation; all of these processes require eIF2 for initiation of translation. Thus, defects in eIF2 are potentially fatal (17,18). These are currently known to be associated with a wide range of disorders, such as delayed sensitivity, contact dermatitis, impaired glucose tolerance, insulin resistance, Alzheimer's disease, myelodysplastic syndromes, and different types of cancers (breast cancer, leukemia, melanoma, hepatocellular carcinoma, and colon cancer) (19,20).
MAPK1 is also known as extracellular signal-regulated kinase 2 (ERK2) and has a molecular weight of 41,390 Da. It belongs to the MAPKs family, and acts as a major transmitter for transduction of extracellular signals into the nucleus; it is involved in many cellular responses to exogenous and endogenous stimuli and regulates many intracellular pathways. MAPK1 plays an important regulatory role in physiological processes such as cell proliferation and survival by combining with ATP. Further, it is involved in DNA binding, kinase binding and its activation, binding to transcription factors, and transferase activity in response to inflammation, growth, and differentiation of cells. The most widely studied MAPK pathways include ERK1/2 (for extracellular growth factors), p38MAPK, and JNK (for stress) (21). Intracellular aggregation of double-stranded RNA activates p38 MAPK and JNK, which is found mainly in lung cancer (22), cardiac hypertrophy, heart failure, hypertrophy, dedifferentiation, Q61 mutant melanoma, melanocarcinoma, amyotrophic lateral sclerosis, leiomyomas, renal cancer, renal clear cell adenocarcinoma, and epithelial cancer (23-26).
Conclusions
Both EIF2AK2 and MAPK1 proteins play an important role in the pathogenesis and progression of NPC.
The differently expressed proteins EIF2AK2 and MAPK play a special role in the biological network. These appear to be either specific proteins of NPC or may facilitate the discovery of other specific functional proteins of NPC. Therefore further research may be necessary to identify the pathways and molecular mechanisms underlying NPC.
Acknowledgments
Funding: This work was supported by The Macao Science and Technology Development Fund (025/2013/A).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the hospital Ethics Committee. The enrolled patients provided informed consent to participate in this study.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.11.14). The authors have no conflicts of interest to declare.
References
- 1.Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev 2006;15:1765-77. 10.1158/1055-9965.EPI-06-0353 [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 3.Brennan B. Nasopharyngeal carcinoma. Orphanet J Rare Dis 2006;1:23. 10.1186/1750-1172-1-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature 2016;537:347-55. 10.1038/nature19949 [DOI] [PubMed] [Google Scholar]
- 5.Iwamoto N, Shimada T. Recent advances in mass spectrometry-based approaches for proteomics and biologics: Great contribution for developing therapeutic antibodies. Pharmacol Ther 2018;185:147-54. 10.1016/j.pharmthera.2017.12.007 [DOI] [PubMed] [Google Scholar]
- 6.Cheung CHY, Juan HF. Quantitative proteomics in lung cancer. J Biomed Sci 2017;24:37. 10.1186/s12929-017-0343-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atrih A, Mudaliar MA, Zakikhani P, et al. Quantitative proteomics in resected renal cancer tissue for biomarker discovery and profiling. Br J Cancer 2014;110:1622-33. 10.1038/bjc.2014.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao L, Xiao T, Wang ZM, et al. Biomarker discovery of nasopharyngeal carcinoma by proteomics. Expert Rev Proteomics 2014;11:215-25. 10.1586/14789450.2014.897613 [DOI] [PubMed] [Google Scholar]
- 9.Hoedt E, Zhang G, Neubert TA. Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) for Quantitative Proteomics. Adv Exp Med Biol 2019;1140:531-9. 10.1007/978-3-030-15950-4_31 [DOI] [PubMed] [Google Scholar]
- 10.Puzar Dominkus P, Ferdin J, Plemenitas A, et al. Nef is secreted in exosomes from Nef.GFP-expressing and HIV-1-infected human astrocytes. J Neurovirol 2017;23:713-24. 10.1007/s13365-017-0552-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med 1988;319:525-32. 10.1056/NEJM198809013190901 [DOI] [PubMed] [Google Scholar]
- 12.Karimi P, Shahrokni A, Ranjbar MR. Implementation of proteomics for cancer research: past, present, and future. Asian Pac J Cancer Prev 2014;15:2433-8. 10.7314/APJCP.2014.15.6.2433 [DOI] [PubMed] [Google Scholar]
- 13.Chen ZT, Liang ZG, Zhu XD. A. Review: Proteomics in Nasopharyngeal Carcinoma. Int J Mol Sci 2015;16:15497-530. 10.3390/ijms160715497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen QY, Tang QN, Tang LQ, et al. Pretreatment Serum Amyloid A and C-reactive Protein Comparing with Epstein-Barr Virus DNA as Prognostic Indicators in Patients with Nasopharyngeal Carcinoma: A Prospective Study. Cancer Res Treat 2018;50:701-11. 10.4143/crt.2017.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue F, Chen S, Chunxiang B, et al. eIF2 alpha phosphorylation alleviates UVA-induced HO-1 expression in mouse epidermal cells. Free Radic Res 2018;52:1359-70. 10.1080/10715762.2018.1489127 [DOI] [PubMed] [Google Scholar]
- 16.Kashiwagi K, Ito T, Yokoyama S. Crystal structure of eIF2B and insights into eIF2-eIF2B interactions. FEBS J 2017;284:868-74. 10.1111/febs.13896 [DOI] [PubMed] [Google Scholar]
- 17.Kim E, Kim JH, Seo K, et al. eIF2A, an initiator tRNA carrier refractory to eIF2alpha kinases, functions synergistically with eIF5B. Cell Mol Life Sci 2018;75:4287-300. 10.1007/s00018-018-2870-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng J, Erdjument-Bromage H, Neubert TA. Quantitative Comparison of Proteomes Using SILAC. Curr Protoc Protein Sci 2019;95:e74. 10.1002/cpps.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang L, Cho HJ, Chan C, et al. Binding site multiplicity with fatty acid ligands: implications for the regulation of PKR kinase autophosphorylation with palmitate. Proteins 2014;82:2429-42. 10.1002/prot.24607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Cruz-Herrera CF, Campagna M, Garcia MA, et al. Activation of the double-stranded RNA-dependent protein kinase PKR by small ubiquitin-like modifier (SUMO). J Biol Chem 2014;289:26357-67. 10.1074/jbc.M114.560961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R, Wang J, Acharya D, et al. Antiviral responses in mouse embryonic stem cells: differential development of cellular mechanisms in type I interferon production and response. J Biol Chem 2014;289:25186-98. 10.1074/jbc.M113.537746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi Y, Zhang M, Li H, et al. MicroRNA-29b regulates ethanol-induced neuronal apoptosis in the developing cerebellum through SP1/RAX/PKR cascade. J Biol Chem 2014;289:10201-10. 10.1074/jbc.M113.535195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Zhang M, Qian YY, et al. Imbalanced expression of mitogen-activated protein kinase phosphatase-1 and phosphorylated extracellular signal-regulated kinases in lung squamous cell carcinoma. J Zhejiang Univ Sci B 2011;12:828-34. 10.1631/jzus.B1100165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang PT, Chen CH, Hsu IU, et al. Huntingtin-associated protein 1 interacts with breakpoint cluster region protein to regulate neuronal differentiation. PLoS One 2015;10:e0116372. 10.1371/journal.pone.0116372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishii A, Furusho M, Dupree JL. Role of ERK1/2 MAPK signaling in the maintenance of myelin and axonal integrity in the adult CNS. 2014;34:16031-45. [DOI] [PMC free article] [PubMed]
- 26.Gaestel M. MAPK-Activated Protein Kinases (MKs): Novel Insights and Challenges. Front Cell Dev Biol 2016;3:88. 10.3389/fcell.2015.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]