Abstract
Background
Recent evidence demonstrates that the long non-coding RNA (lncRNA) BLACAT1 is associated with the progression and development of various cancers; however, its effect on tumorigenesis of colorectal cancer (CRC) is still poorly understood. The aim of the present study was to investigate the expression and function of BLACAT1 in CRC.
Methods
Expression data from the GEO and GEPIA databases and results obtained from clinical samples/patients were used to determine the correlation between BLACAT1 expression, and CRC metastasis and overall survival (OS). Furthermore, we knocked down BLACAT1 using short interfering RNA (siRNA) and observed its biological functions using quantitative reverse transcription-polymerase chain reaction (qRT-PCR), cell counting kit-8 (CCK-8) assay, tumor cell clone formation, and Matrigel invasion assays in the HCT116 cell line.
Results
BLACAT1 level was higher in CRC tissues and cell lines than in normal colon mucosal tissues and cell lines. Correlation of data from the GEO and GEPIA databases with several clinical parameters revealed that CRC patients with high BLACAT1 expression showed poor OS. Multivariate analysis indicated that high BLACAT1 expression is an independent risk factor in patients with CRC. Furthermore, siRNA-mediated knockdown of BLACAT1 suppressed proliferation and invasion of CRC cells in vitro. This in turn was associated with reduced expression of cyclin D1, CDK6, and vimentin, and enhanced expression of E-cadherin.
Conclusions
BLACAT1 may play an important role in the progression and development of CRC, and may serve as a potential therapeutic target for patients with CRC.
Keywords: BLACAT1, colorectal cancer (CRC), tumorigenesis, invasion, survival
Introduction
Colorectal cancer (CRC) is the third-most prevalent cancer worldwide, with an estimated 0.7 million mortalities in 2012, which considerably affects human health (1,2). With the improvement and development of therapeutic approaches for CRC, substantial progress has been made in its treatment in the past decades; however, CRC-related mortality rate has not changed as expected (3). The progression and development of CRC is a multistep process and involves genetic as well as environmental factors, which cause dysregulation of many genes associated with apoptosis, proliferation, and differentiation (4-6). With the increasing popularity of molecular therapy, several studies have investigated the mechanisms underlying pathogenesis of CRC, thereby providing new insights into the progression and development of this cancer (7,8).
Long non-coding RNAs (lncRNAs) are >200 nucleotides long, lack functional open reading frames (ORFs), and rarely encode functional short peptides (9,10). Currently, lncRNAs have attracted attention for their effect on cell behavior, especially those of tumor cells. Accumulating evidence shows aberrant expression of lncRNAs in several cancers, such as cervical (11) and gastric cancers (12), non-small cell lung cancer (NSCLC) (13), and CRC (14). Thus, dysregulated lncRNAs can be used as biomarkers for diagnosis and can act as potential therapeutic targets in various cancers.
The bladder cancer associated transcript-1 (BLACAT1) on chr1q32.1 encodes a 2,616 bp long lncRNA. Emerging evidence indicates that BLACAT1 is associated with tumorigenesis of various cancers, including gastric (15), thyroid (16), and cervical cancers (17), and NSCLC (18); however, its specific role in the progression and development of CRC is poorly understood. In this study, we observed BLACAT1 overexpression in CRC tissues and cell lines. In addition, using information from Gene Expression Omnibus (GEO) and Gene Expression Profiling Interactive Analysis (GEPIA) databases, we observed that CRC patients with high BLACAT1 expression showed poor overall survival (OS). Furthermore, BLACAT1 knockdown inhibited the proliferation and invasion of CRC cells in vitro, suggesting a role of BLACAT1 in the progression and development of CRC.
Methods
Bioinformatic analysis
Microarray profiles and datasets of primary CRC were acquired from the GEO with the following entries: GSE23878 (19), GSE9348 (20), GSE22598 (21), GSE18105 (22), and GSE17538 (23) (Affymetrix Human Genome U133 Plus 2.0 platform). GSE9348 consisted of 70 primary CRC samples and 12 normal colon samples; GSE23878 consisted of 35 primary CRC samples and 24 normal colon samples; GSE22598 contained 17 pairs of CRC and adjacent non-tumor tissues; GSE18105 consisted of 44 metastatic CRC samples and 67 non-metastatic CRC samples (in the GE17538, the 44 metastatic CRC samples were from 44 cases identified as patients with metastatic CRC, and the 67 non-metastatic CRC samples were from 67 patients with non-metastatic CRC); GSE17538 was divided into the low BLACAT1 (n=146) and high BLACAT1 expression groups (n=85).
Cell culture and transfection
The human CRC cell lines (HCT116, SW480, SW620, CaCO2) and normal colon mucosal cell line NCM460 used in this study were obtained from the American Type Culture Collection (ATCC, Manassas, USA). HCT116 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Invitrogen, USA). The other cell lines (NCM460, SW480, SW620, and CaCO2) were cultured in Roswell Park memorial Institute (RPMI)-1640 medium (Invitrogen, USA) supplemented with 10% FBS. When cell densities were ~60%, 50 nM siRNAs were transfected using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions. The sequences of the BLACAT1-targeting siRNAs were: BLACAT1-si-1: 5'-AGGCUGGUUUCUGCCCUCAUCCUUU-3'; BLACAT1-si-2: 5'-GCCCAGCUUCUAGUCCUCUCCUUAU-3'; Sequences of non-target scrambled controls were provided by RiboBio (Guangzhou, China).
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
RNA was isolated from tissue and amplified as described previously (24). Total RNA from cells was isolated from using the Trizol reagent (Invitrogen, The Netherlands). For qRT-PCR, RNA was reverse transcribed to cDNA using a Revert Aid first strand cDNA synthesis kit (Fermentas, Canada). qRT-PCR was performed using a SYBR_Premix ExTaq II kit (Takara, China) and a CFX96 real-time PCR detection system (Bio-Rad, USA) to determine the relative expression levels of target genes. The sequences of the qRT-PCR primers are as follows: BLACAT1 forward 5'-GTCTCTGCCCTTTTGAGCCT-3', reverse 5'-GTGGCTGCAGTGTCATACCT-3'; human GAPDH forward 5'-TGCACCACCAACTGCTTAGC-3', reverse 5'-GGCATGGACTGTGGTCATGAG-3'; human β-actin forward 5'-ATCATGTTTGAGACCTTCAA-3', reverse 5'-CATCTCTTGCTCGAAGTCCA-3'; human U6 (nuclear) forward 5'-CTCGCTTCGGCAGCACA-3', reverse 5'-AACGCTTCACGAATTTGCGT-3'.
Cell proliferation assay
Cell proliferation was assayed using the cell counting kit-8 (CCK-8) (Dojin Laboratories, Japan), and the operating steps were performed as described previously (25).
Tumor cell clone formation assay
The tumor cell clone formation assay was performed as described previously (26). Briefly, 24 h after transfection of the si-Control and si-BLACAT1, 1×103 HCT116 cells were seeded per well in a 6-well plate and cultured for 14 days. Colonies were stained with hematoxylin. The number of clones formed per well was calculated. The histogram represents data from three independent experiments.
Transwell matrigel assays
Colorectal cell invasiveness was determined using a 24-well transwell plate (8 µM pore size; Costar), as described previously (27). Briefly, 5×104 cells were placed on the upper chamber of each insert coated with 200 mg/mL Matrigel (BD Biosciences, CA, USA). After 48 h, the invaded cells were stained with hematoxylin and counted. The number of invasive tumor cells was calculated from the total number of cells from three randomly selected 20× fields for each experiment. The histogram represents data from three independent experiments.
Statistical analysis
All statistical analyses were performed using SPSS version 18.0 and presented using the GraphPad Prism software. Data were expressed as mean ± standard error of mean (SEM). Differences between two independent groups were tested using Student’s t-test. OS was calculated using the Kaplan-Meier method, and the results of the analysis were considered significant in a log-rank test if P<0.05.
Results
BLACAT1 is upregulated in CRC tissues and cell lines
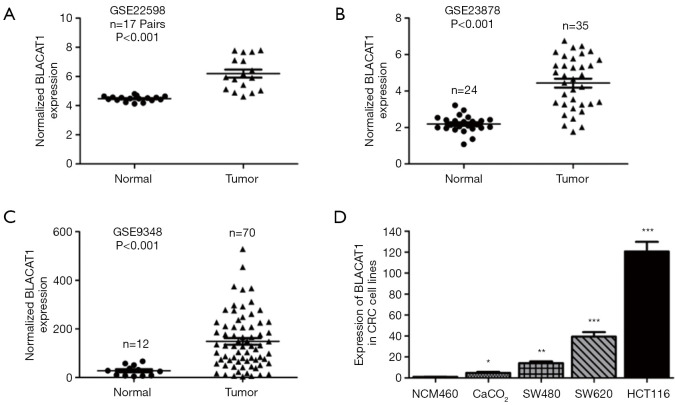
We analyzed three previously published primary CRC datasets, namely, GSE9348, GSE23878, and GSE22598, generated using the Affymetrix HG_U133 plus 2 arrays to identify lncRNAs dysregulated in CRC. We observed significant upregulation of BLACAT1 in all the analyzed datasets (P<0.001, Figure 1A,B,C). Subsequently, BLACAT1 levels in five CRC cell lines (SW480, CaCO2, HCT116, and SW620) and the normal colon mucosal cell line NCM460 were determined using qRT-PCR. We observed that BLACAT1 expression was higher in CRC cell lines than in NCM460 (P<0.05, Figure 1D). In addition, the highest BLACAT1 expression was observed in HCT116 cells.
Figure 1.
BLACAT1 is highly expressed in CRC tissues and cell lines. BLACAT1 expression, as measured using Affymetrix microarray, was higher in CRC tissues than in normal colon mucosal tissues in (A) GSE22598 (consisting of 17 pairs of CRC tissues and corresponding normal colorectal tissues) samples from the GEO database, (B) GSE23878 (consisting of 24 normal colorectal tissues and 35 CRC tissue biopsies), and (C) GSE9348 (consisting of 12 normal colorectal tissues and 70 CRC tissue biopsies). (D) BLACAT1 expression increased significantly in CRC cell lines (SW480, SW620, HCT116, and CaCO2) compared to in NCM460, a normal colon mucosal cell line. Data show mean ± SEM. *, P<0.05; **, P<0.01; ***, P<0.001 compared to control. CRC, colorectal cancer.
Upregulation of BLACAT1 predicts poor prognosis and is an independent predictor of OS in CRC
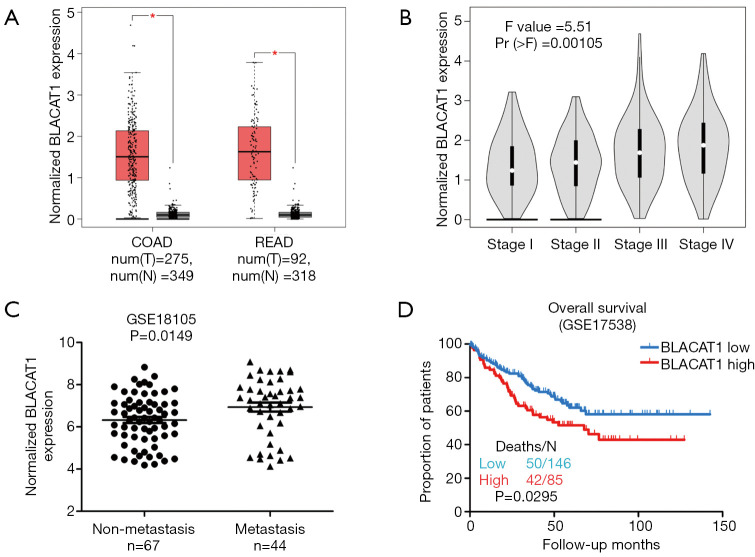
Next, we assessed the correlation of BLACAT1 expression with the pathological stages of CRC using the GEPIA database. We observed that BLACAT1 was significantly upregulated in colon (COAD) and rectum adenocarcinoma (READ) (Figure 2A). Furthermore, high BLACAT1 expression correlated significantly with the pathological stages of CRC (P=0.00105, Figure 2B). Next, we assessed the role of BLACAT1 in CRC metastasis; we used samples from GSE18105 and the Affymetrix HG_U133 plus 2 arrays to identify lncRNAs dysregulated in CRC. We observed that high BLACAT1 level correlated significantly with distant CRC metastasis (P=0.0149, Figure 2C).
Figure 2.
Relationship between BLACAT1 expression and clinicopathological features. (A) The GEPIA database was used to analyze the expression of BLACAT1 in colorectal cancer tissues; COAD stands for colon adenocarcinoma and READ for rectal adenocarcinoma; (B) the GEPIA database was used to analyze the relationship between BLACAT1 expression and pathological stages of CRC; F value is representative of the F-test; (C) relative expression of BLACAT1 in non-metastatic and metastatic human CRC tissues was obtained from the GEO database (#GSE18105, non-metastasis, n=67; metastasis, n=44); (D) the GEO database was used to analyze the clinical effect of BLACAT1 expression on CRC patient survival in a colorectal cancer specimen expression profile dataset [#GSE17538, the specimen was divided into two groups: group 1, low expression of BLACAT1, n=146, deaths (N) =50; group 2, high expression of BLACAT1, n=85, deaths (N) =42]. Data show mean ± SEM. *, P<0.05 compared to control. CRC, colorectal cancer.
To assess the prognostic value of BLACAT1 expression in patients with CRC, we investigated the association between BLACAT1 expression and OS using Kaplan-Meier analysis with the log-rank test. A previously published GEO dataset, GSE17538, was used for this analysis. Results revealed correlation of BLACAT1 expression with OS in patients with CRC (P=0.0295, Figure 2D). Patients with high BLACAT1 expression displayed lower OS. Taken together, these data indicate that high BLACAT1 level is an independent risk factor for patients with CRC.
BLACAT1 knockdown inhibits cell proliferation and invasion of CRC
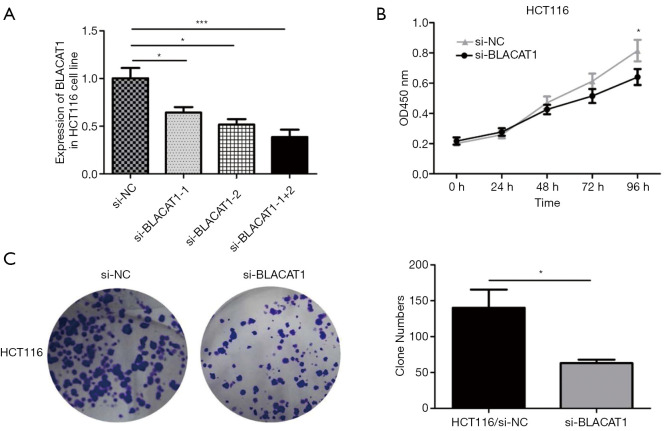
To understand BLACAT1 function in colon cancer cells, we first determined the silencing efficiency of si-BLACAT1. Highest reduction in BLACAT1 expression was observed in cells co-transfected with si-BLACAT1-1 and si-BLACAT1-2 compared to either si-BLACAT1-1 or si-BLACAT1-2 alone (Figure 3A). Therefore, co-transfection of si-BLACAT1-1 and si-BLACAT1-2 in HCT116 cells was used in further knockdown studies. Effect of BLACAT1 knockdown on CRC cell proliferation was determined using the CCK-8 proliferation assay. BLACAT1 knockdown significantly inhibited HCT116 cell proliferation compared to control cells (P<0.05, Figure 3B and Figure S1). In addition, compared to control cells, suppression of BLACAT1 inhibited the clone-forming ability of HCT116 cells (P<0.05, Figure 3C).
Figure 3.
BLACAT1 knockdown inhibits proliferation of CRC cells. The interference efficiency of si-BLACAT1 was verified in HCT116 cells. HCT116 cells were transfected with either si-NC or si-BLACAT1 (1#, 2#, 1+2#) for 48 h, following which BLACAT1 expression was analyzed using qRT-PCR (A) and CCK-8 assay (B). (C) The clonogenic assay was used to detect the proliferative ability after transfecting HCT116 cells with si-NC or si-BLACAT1 for 48 h. Data show mean ± SEM. *, P<0.05; ***, P<0.001 compared to the control group. CRC, colorectal cancer; qRT-PCR, quantitative reverse transcription-polymerase chain reaction.
Figure S1.
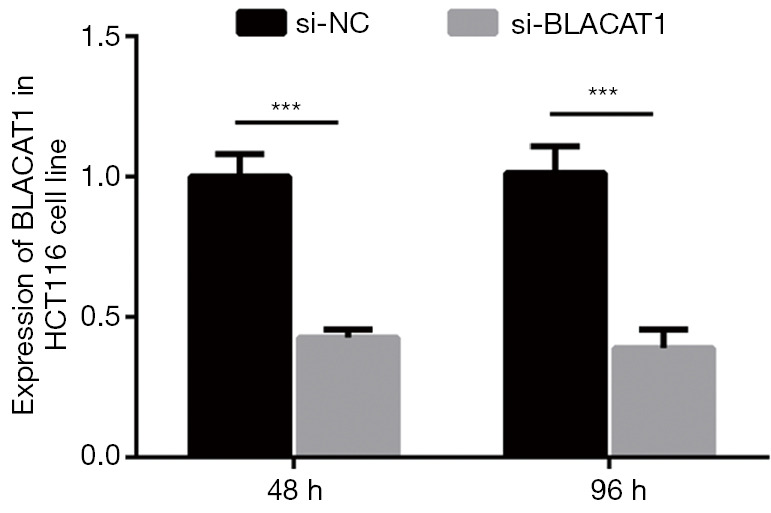
The interference efficiency of si-BLACAT1 was verified in HCT116 cells. HCT116 cells were transfected with either si-NC or si-BLACAT1 for 48 and 96 h, following which BLACAT1 expression was analyzed using qRT-PCR. ***, P<0.001. qRT-PCR, quantitative reverse transcription-polymerase chain reaction.
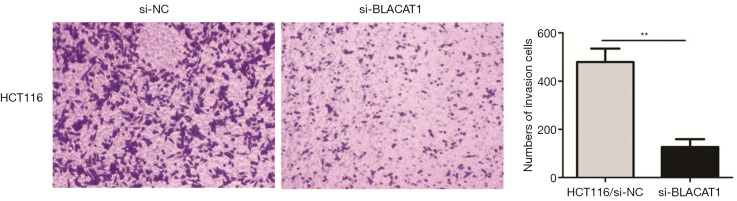
Subsequently, we also investigated the effect of BLACAT1 knockdown on invasiveness of CRC cells. The transwell Matrigel assay was performed to assess the effect of BLACAT1 on the invasiveness of CRC cells. Our results indicated that compared to the control group, BLACAT1 knockdown significantly inhibited invasiveness of HCT116 cells (P<0.01, Figure 4).
Figure 4.
BLACAT1 knockdown inhibits invasiveness of CRC cells. The cell invasion ability was determined using the transwell Matrigel assay after transfecting HCT116 cells with si-NC or si-BLACAT1 for 48 h. Cells that had invaded through the filter pores were stained with 5% crystal violet. The images were taken at ×200. Data show mean ± SEM. **, P<0.01 compared to the control group. CRC, colorectal cancer.
BLACAT1 knockdown suppresses proliferation and expression of epithelial-mesenchymal transition (EMT) markers in CRC
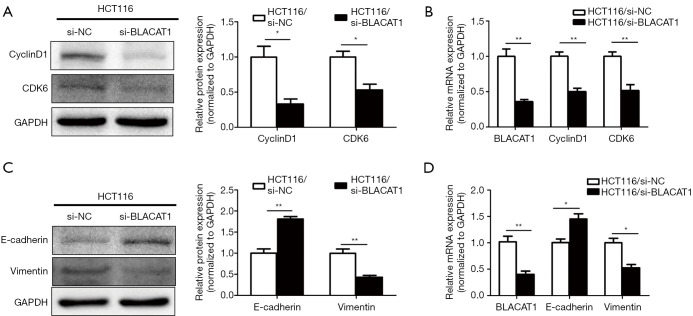
To elucidate the molecular mechanism underlying BLACAT1-mediated suppression of proliferation and invsion of CRC cells, the expression levels of proliferation and EMT markers were determined. The mRNA and protein levels of the epithelial marker E-cadherin, mesenchymal marker vimentin, and proliferation markers cyclin D1 and CDK6 in HCT116 cell line were quantified using qRT-PCR and western blotting. BLACAT1 knockdown significantly inhibited the expression of the proliferation markers cyclin D1 and CDK6 (P<0.05, Figure 5A,B). On the other hand, BLACAT1 knockdown significantly decreased the expression of the mesenchymal marker vimentin and enhanced that of the epithelial marker E-cadherin (P<0.05, Figure 5C,D), thereby inhibiting EMT. Thus, these results indicate that BLACAT1 regulates proliferation and expression of EMT markers.
Figure 5.
BLACAT1 silencing suppresses proliferation and expression of EMT markers in CRC. HCT116 cells were transfected with si-NC or si-BLACAT1-1 + si-BLACAT1-2 for 48 h, following which proliferation and expression of EMT markers were determined using western blotting and qRT-PCR. (A) Protein levels of cyclin D1 and CDK6 were determined using western blotting and (B) mRNA levels were quantitated using qRT-PCR. (C) Protein levels of E-cadherin and vimentin were analyzed using western blotting and (D) mRNA levels were determined using qRT-PCR. Data show mean ± SEM. *, P<0.05; **, P<0.01 compared to the control group. CRC, colorectal cancer; qRT-PCR, quantitative reverse transcription-polymerase chain reaction.
Discussion
CRC is one of the most common and leading causes of cancer-related mortality worldwide (28), especially in developed countries. It is estimated that in 2015, there will be approximately 0.8 million new cases and 0.4 million deaths from CRC in developed countries (29). With advancements in basic and clinical investigation, the mechanisms underlying tumorigenesis are beginning to be understood and therapeutic strategies against CRC are being considerably improved. However, the mortality and morbidity rates associated with CRC remain high, possibly owing to postsurgical recurrence and metastasis of primary tumors. Therefore, CRC treatment requires identification of novel therapeutic targets to better monitor its recurrence and metastasis.
LncRNAs act as important regulators of various biological processes by modulating gene expression at the epigenetic, transcriptional, and posttranscriptional levels (30,31), thereby regulating the fate of cellular processes, including apoptosis, proliferation, and differentiation (32). Recently, several studies have revealed that disruption or dysregulation of lncRNAs strongly correlated with the occurrence and development of malignant tumors, cancer cell proliferation, drug resistance, and EMT (33,34). As lncRNAs are easy to extract and can be detected with high specificity and sensitivity in blood and other tissues harboring steady state levels of the lncRNAs (35), they can act as novel biomarkers for predicting cancer diagnosis, recurrence, and chemo-sensitivity. Certain lncRNAs, such as MALAT1 (36), HOTAIR (37), lncARSR (38), GAS5 (39), NEAT1 (40), and TUG1 (41), have been shown to be differentially expressed in CRC and are associated with poor prognosis.
Expression of BLACAT1, a long non-coding RNA (lncRNA), is reported to correlate with the occurrence of cancers (42). BLACAT1 overexpression has recently been identified as an independent predictor of OS in various human cancers, such as gastric (15), thyroid (16), and cervical cancers (17), and NSCLC (18). However, the association of BLACAT1 expression with OS in patients with CRC has seldom been reported. In our study, we aimed to investigate whether BLACAT1 can be potentially developed into a novel biomarker for CRC diagnosis and prognosis. We reported that BLACAT1 expression in CRC tissues was significantly higher than those in matched adjacent normal tissues. Overexpression of BLACAT1 in patients with CRC resulted in poor OS, which may act as an independent poor prognostic factor for these patients.
Convincing evidence shows that BLACAT1 plays critical roles in regulating cancer cell proliferation, metastasis, cell cycle, apoptosis, stemness, and drug resistance, thereby affecting the development and progression of cancer (42). For example, Ye et al. demonstrated that BLACAT1 was overexpressed in NSCLC tissues, and knocking down BLACAT1 using small interfering RNA (siRNA) significantly suppressed NSCLC proliferation, migration, and invasion in vitro (18). Wang et al. showed that BLACAT1 upregulation promoted cervical cancer cell proliferation, invasion, and migration by activating the Wnt/β-catenin signaling pathway (17). Furthermore, Wu et al. showed that BLACAT1 sponged miR-361 to prompt the expression of ABCB1, thereby accelerating the growth and inducing the oxaliplatin-resistance of gastric cancer (15). However, the effect of BLACAT1 on CRC proliferation and invasion is not well understood. Our results demonstrated that inhibition of BLACAT1 suppressed proliferation and invasion of the HCT116 cell line.
In summary, our results indicated that BLACAT1 is upregulated in patients with CRC, and that these patients showed poor OS. In addition, multivariate analysis indicated that a high level of BLACAT1 is an independent risk factor for patients with CRC. We also demonstrated that BLACAT1 mediated the proliferation and invasion of CRC cells. BLACAT1 knockdown significantly suppressed the proliferative and invasive capacity of CRC cells. Therefore, BLACAT1 may play a crucial role in regulating the progression and development of CRC and may act as a prospective target for development of novel therapies for CRC.
Acknowledgments
Funding: This study was supported by the 5th stage “521 project” of Lianyungang research project (No. 45).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional Review Board approval was waived. Informed consent was waived due to the retrospective nature of the study.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.12.26). The authors have no conflicts of interest to declare.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin 2017;67:177-93. 10.3322/caac.21395 [DOI] [PubMed] [Google Scholar]
- 3.Sunkara V, Hebert JR. The colorectal cancer mortality-to-incidence ratio as an indicator of global cancer screening and care. Cancer 2015;121:1563-9. 10.1002/cncr.29228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ou C, Sun Z, Li S, et al. Dual roles of yes-associated protein (YAP) in colorectal cancer. Oncotarget 2017;8:75727-41. 10.18632/oncotarget.20155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peters U, Bien S, Zubair N. Genetic architecture of colorectal cancer. Gut 2015;64:1623-36. 10.1136/gutjnl-2013-306705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calvert PM, Frucht H. The genetics of colorectal cancer. Ann Intern Med 2002;137:603-12. 10.7326/0003-4819-137-7-200210010-00012 [DOI] [PubMed] [Google Scholar]
- 7.Colussi D, Brandi G, Bazzoli F, et al. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci 2013;14:16365-85. 10.3390/ijms140816365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaiopoulos AG, Athanasoula K, Papavassiliou AG. Epigenetic modifications in colorectal cancer: molecular insights and therapeutic challenges. Biochim Biophys Acta 2014;1842:971-80. 10.1016/j.bbadis.2014.02.006 [DOI] [PubMed] [Google Scholar]
- 9.He X, Ou C, Xiao Y, et al. LncRNAs: key players and novel insights into diabetes mellitus. Oncotarget 2017;8:71325-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bazzini AA, Johnstone TG, Christiano R, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. Embo J 2014;33:981-93. 10.1002/embj.201488411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang C, Zhu W, Liu T, et al. Characterization of long non-coding RNA expression profiles in lymph node metastasis of early-stage cervical cancer. Oncol Rep 2016;35:3185-97. 10.3892/or.2016.4715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song H, Sun W, Ye G, et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med 2013;11:225. 10.1186/1479-5876-11-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Li H, Hou S, et al. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One 2013;8:e65309. 10.1371/journal.pone.0065309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H, Wu R, Chen M, et al. Comprehensive analysis of differentially expressed profiles of lncRNAs and construction of miR-133b mediated ceRNA network in colorectal cancer. Oncotarget 2017;8:21095-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X, Zheng Y, Han B, et al. Long noncoding RNA BLACAT1 modulates ABCB1 to promote oxaliplatin resistance of gastric cancer via sponging miR-361. Biomed Pharmacother 2018;99:832-8. 10.1016/j.biopha.2018.01.130 [DOI] [PubMed] [Google Scholar]
- 16.Liao D, Lv G, Wang T, et al. Prognostic value of long non-coding RNA BLACAT1 in patients with papillary thyroid carcinoma. Cancer Cell Int 2018;18:47. 10.1186/s12935-018-0544-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang CH, Li YH, Tian HL, et al. Long non-coding RNA BLACAT1 promotes cell proliferation, migration and invasion in cervical cancer through activation of Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci 2018;22:3002-9. [DOI] [PubMed] [Google Scholar]
- 18.Ye JR, Liu L, Zheng F. Long Noncoding RNA Bladder Cancer Associated Transcript 1 Promotes the Proliferation, Migration, and Invasion of Nonsmall Cell Lung Cancer Through Sponging miR-144. DNA Cell Biol 2017;36:845-52. 10.1089/dna.2017.3854 [DOI] [PubMed] [Google Scholar]
- 19.Uddin S, Ahmed M, Hussain A, et al. Genome-wide expression analysis of Middle Eastern colorectal cancer reveals FOXM1 as a novel target for cancer therapy. Am J Pathol 2011;178:537-47. 10.1016/j.ajpath.2010.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong Y, Downey T, Eu KW, et al. A 'metastasis-prone' signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis 2010;27:83-90. 10.1007/s10585-010-9305-4 [DOI] [PubMed] [Google Scholar]
- 21.Okazaki S, Ishikawa T, Iida S, et al. Clinical significance of UNC5B expression in colorectal cancer. Int J Oncol 2012;40:209-16. [DOI] [PubMed] [Google Scholar]
- 22.Matsuyama T, Ishikawa T, Mogushi K, et al. MUC12 mRNA expression is an independent marker of prognosis in stage II and stage III colorectal cancer. Int J Cancer 2010;127:2292-9. 10.1002/ijc.25256 [DOI] [PubMed] [Google Scholar]
- 23.Smith JJ, Deane NG, Wu F, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology 2010;138:958-68. 10.1053/j.gastro.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ou C, Sun Z, Zhang H, et al. SPLUNC1 reduces the inflammatory response of nasopharyngeal carcinoma cells infected with the EB virus by inhibiting the TLR9/NF-kappaB pathway. Oncol Rep 2015;33:2779-88. 10.3892/or.2015.3913 [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Zhao C, Yu Z, et al. Low expression of miR-381 is a favorite prognosis factor and enhances the chemosensitivity of osteosarcoma. Oncotarget 2016;7:68585-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang W, Qin Z, Fan S, et al. miR-1207-5p suppresses lung cancer growth and metastasis by targeting CSF1. Oncotarget 2016;7:32421-32. 10.18632/oncotarget.8718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ou C, Sun Z, Li X, et al. MiR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancer. Cancer Lett 2017;399:53-63. 10.1016/j.canlet.2017.04.011 [DOI] [PubMed] [Google Scholar]
- 28.Bray F, Ren JS, Masuyer E, et al. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer 2013;132:1133-45. 10.1002/ijc.27711 [DOI] [PubMed] [Google Scholar]
- 29.Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893-917. 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 30.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41. 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 31.Peng L, Yuan X, Jiang B, et al. LncRNAs: key players and novel insights into cervical cancer. Tumour Biol 2016;37:2779-88. 10.1007/s13277-015-4663-9 [DOI] [PubMed] [Google Scholar]
- 32.Li J, Tian H, Yang J, et al. Long Noncoding RNAs Regulate Cell Growth, Proliferation, and Apoptosis. DNA Cell Biol 2016;35:459-70. 10.1089/dna.2015.3187 [DOI] [PubMed] [Google Scholar]
- 33.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011;1:391-407. 10.1158/2159-8290.CD-11-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016;29:452-63. 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Mo Y, Gong Z, et al. Circular RNAs in human cancer. Mol Cancer 2017;16:25. 10.1186/s12943-017-0598-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He X, Yan Q, Kuang G, et al. Metastasis-associated lung adenocarcinoma transcript 1 regulates tumor progression: old wine in a new bottle. J Thorac Dis 2018;10:S1088-S1091. 10.21037/jtd.2018.04.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svoboda M, Slyskova J, Schneiderova M, et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis 2014;35:1510-5. 10.1093/carcin/bgu055 [DOI] [PubMed] [Google Scholar]
- 38.Qu L, Ding J, Chen C, et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell 2016;29:653-68. 10.1016/j.ccell.2016.03.004 [DOI] [PubMed] [Google Scholar]
- 39.Yin D, He X, Zhang E, et al. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol 2014;31:253. 10.1007/s12032-014-0253-8 [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Li Y, Chen W, et al. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget 2015;6:27641-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ou C, Li G. Long non-coding RNA TUG1: a novel therapeutic target in small cell lung cancer. J Thorac Dis 2017;9:E644-E645. 10.21037/jtd.2017.06.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X, Dai M, Zhu H, et al. Evaluation on the diagnostic and prognostic values of long non-coding RNA BLACAT1 in common types of human cancer. Mol Cancer 2017;16:160. 10.1186/s12943-017-0728-2 [DOI] [PMC free article] [PubMed] [Google Scholar]