The treatment of breast cancer has progressed amazingly during the past three decades but a relatively high proportion of patients still experience resistance to pharmacotherapy (i.e., endocrine therapy, chemotherapy, and HER2-directed antibody-drugs) (1,2). Lately, two immune checkpoint inhibitors have been approved in several countries for the treatment of breast cancer. Specifically, atezolizumab (Tecentriq®, Genentech Inc, South San Francisco, CA, USA), an anti-programmed death-ligand 1 (PD-L1) drug, and pembrolizumab (Keytruda®, Merck & Co. Inc., Kenilworth, NJ, USA), that blocks PD-L1 receptor programmed cell death protein 1 (PD-1). Ongoing phase II and III trials are expected to lead to more approvals in different clinical settings.
At present, breast cancer immunotherapy is biomarker-based (3). In unresectable locally advanced or metastatic triple-negative breast cancers (TNBC), only cases in which the tumor-infiltrating immune cells express PD-L1 in ≥1% of the tumor area can be treated with atezolizumab (4). The VENTANA PD-L1 (SP142) Assay (Roche Tissue Diagnostics, Tucson, AZ, USA) comes as a companion diagnostic (CDx) test for this analysis. Results of KEYNOTE-355 recently presented at ASCO 2020 revealed progression-free survival benefits of pembrolizumab plus chemotherapy in high PD-L1 expression TNBC. However, this analysis is not yet part of the everyday in clinical practice. On the other hand, the Food and Drug Administration (FDA) approved pembrolizumab in all refractory advanced solid tumors with mismatch repair (MMR) deficiency and/or high levels of microsatellite instability (MSI) (5). Regrettably, our knowledge on the specific biology of MMR deficiency in breast cancer is limited, both in terms of identifying relative contributions of this system to patient outcomes and in understanding the role of cellular localization of MMR proteins to cancer phenotypes.
Genomic scars in the MMR system occur at relatively low frequency in breast cancer and are reported in approximately 2% of cases. However, this subject is controversial in literature given the lack of CDx and/or tumor-specific guidelines for MMR analysis (6-11). Hence, MMR data in breast cancer may vary according to the testing method employed, such as direct sequencing of microsatellite markers, next-generation sequencing (NGS), and immunohistochemistry (IHC) for the four MMR proteins (Table 1). To date, a constellation of locally developed tests can be found (12). These are generally modeled on those approved for colorectal and endometrial carcinomas, where MSI is more frequent (15% and 20–30%, respectively) than in breast cancer (13,14). It needs to be highlighted that in these types of cancer, MMR and MSI testings were not standardized to inform immunotherapy decisions but for the screening of Lynch syndrome, an inherited disorder caused by germline defects in the MMR systems (15).
Table 1. Studies on mismatch repair (MMR) status and breast cancer.
| Authors | Ref. | Year | Testing method | Nr. of patients | dMMR (%) |
|---|---|---|---|---|---|
| Cheng et al. | (6) | 2020 | IHC | 1,635 | 1.9 |
| Lopez et al. | (7) | 2020 | IHC | 608 | 13.3 |
| Fusco et al. | (8) | 2018 | IHC & MSI | 444 | 17 |
| Lee et al. | (9) | 2019 | Sanger Sequencing, IHC, MSI | 94 | 3.2 |
| Le et al. | (10) | 2017 | NGS | N/A | <2 |
| Davies et al. | (11) | 2017 | WGS | 640 | 1.7 |
dMMR, MMR deficiency; IHC, immunohistochemistry; MSI, microsatellite instability; NGS, next-generation sequencing; WGS, whole-genome sequencing.
The mechanisms underpinning the possible susceptibility of MMR-deficient breast cancers to immune-checkpoint blockade is not trivial. Dysfunction in the MMR system may result in a hypermutator state, with subsequent neo-antigen generation by the neoplastic cells and activation of the adaptative immune response (16). On the other hand, MMR defects in breast cancer induce an increase in tumor mutation burden (TMB) that is many orders of magnitude lower than that seen in archetypal Lynch syndrome spectrum cancers (17,18). Hence, there may be essential differences in how MMR functions in breast compared to other cancer sites.
Clinicopathologic features of MMR-deficient breast cancers
In this scenario, a recently published study of Cheng et al. (6) has provided important insights. The Authors profiled the expression of the four key MMR proteins [i.e., mutL homolog 1 (MLH1), PMS1 Homolog 2 (PMS2), mutS homolog 2 (MSH2), and mutS homolog 6 (MSH6)] by IHC on a large series of breast cancer patients. Their main objective was to better understand the clinical meaning of MMR deficiency in breast cancer, particularly in terms of long-term outcomes. Out of 3,992 tumors embedded in tissue microarrays, 1,635 (41%) cases were interpretable. Among them, 31 (2%) breast cancers showed the loss of nuclear staining for at least one of the MMR proteins, being identified as MMR deficient. Unlike endometrial and colorectal cancers, the majority of patients from the study of Cheng et al. presented with a single protein loss (n=25, 81%) while the remaining 6 (19%) cases had pair loss (Figure 1). Interestingly, the MutLα complex, which is composed of MLH1 and PMS2, was more targeted by alterations than MutSα (i.e., MSH2 and MSH6). The highest frequency of MMR deficiency was observed in Luminal breast cancers (n=22, 71%), as previously observed (8). The authors reported a specific correlation with high grade, low progesterone receptor (PR) expression, and high tumor-infiltrating lymphocytes (TILs) counts. However, the low proportion of MMR deficient samples precludes a conclusive understanding of the incidence rate variance between specific subclasses of patients. It could be argued that the connection between MMR deficiency and high histologic grade may be due to a higher TMB (19). However, mutations in known oncogenic drivers are not statistically enriched in MMR-defective relative to MMR-proficient breast cancers (11).
Figure 1.
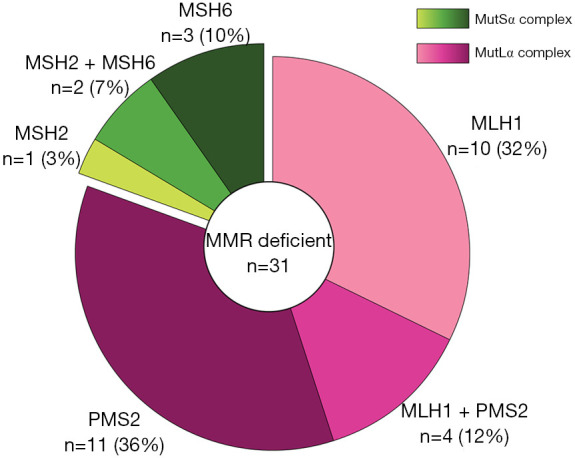
Frequency and patterns of mismatch repair protein loss across 31 mismatch repair deficient breast cancers from the British Columbia cohort.
The overall chemical integrity of the MMR system has been related to patients’ prognosis in several cancer types (20). Cheng et al. showed a non-significant decrease in the overall survival and breast cancer disease-specific survival across all breast cancer types. However, the analyses based on treatment showed that estrogen receptor (ER)-positive patients with MMR deficiency who received tamoxifen as adjuvant systemic therapy had worse survival. Of note, dysfunctions of the MutLα complex are related to resistance phenomena to all classes of endocrine therapy in breast cancer (21-23). This observation also argues against high TMB caused by MMR deficiency as the driver of poor outcomes, since MMR deficiency increases mutational load across breast cancer subtypes, and yet associates with poor outcomes only in ER+ patients (24). All these diverse observations highlight the importance of MMR testing/screening in breast cancer patients.
Real-life challenges in MMR clinical testing
Investigating MMR-deficiencies in breast cancer is not an easy task. Several issues in MMR clinical testing are related to the limitations of the existing methods and the absence of CDx and/or tumor-specific guidelines. Despite immunohistochemistry of the four MMR proteins and MSI testing being widely applied, these tests are not interchangeable in breast cancer since MMR protein loss is more commonly detected than MSI (8). Of note, not all MMR proteins equally impact either mutation load or MSI when defective. It remains to be determined how much the MMR status assessment is troubled by technical artifacts and/or intra-tumor heterogeneity phenomena. On the other hand, accumulating evidence on the significant prognostic value of MMR IHC in breast cancer seems to suggest the clinical validity of this test (6-8). To this end, specific antibody clones, CDx, and/or interpretation guidelines are warranted. Methylation-specific PCR for testing hypermethylation of MLH1 promoter akin somatic variant screening through NGS is usually performed as verification methods (12). However, in breast cancer, a tumor-specific panel of genes would be required to test any possible association of MMR deficiency with other clinically actionable genes. Finally, TMB is another emerging biomarker (25). Increased TMB was found in tumors with defects in the MMR system, thus, it could be used as a surrogate diagnostic assay. Unfortunately, TMB analysis is neither time nor cost-effective while it may prove misleading in cases of unsuitable NGS panels. Also, as in the case of MSI, TMB does not provide insight into which individual MMR protein is defective in a given patient tumor. Finally, there are no guidelines on the genes to include in the TMB count. Hence, whole-exome TMB-high tumors may not be related to MMR deficiency and/or worse prognosis (18). All these methods represent candidate tools for MMR testing in breast cancer, but they need to be profoundly tested in order to overcome existing limitations and make routine testing feasible.
The study of Cheng et al. has added great value to this topic, mostly by assessing the clinical meaning of MMR deficiency in the largest cohort of breast cancer patients thus far, while it brought to light several disadvantages of existing techniques. Due to the high degree of intratumor heterogeneity that characterizes breast cancer, sampling and analyzing different topographic areas of the same tumor could be more reliable. This intratumor heterogeneity is also specifically true for MLH1 expression, where mutational analyses demonstrate that MLH1 mutations are not founders but only occur in specific subclonal populations. It would be helpful for IHC analysis to be combined with genomic analysis in order to increase the sensitivity and specificity.
At the moment, only a few studies have tried to investigate MMR-deficiency in breast cancer, but their results are not in consensus. Considering the limitations of these studies, it is evident that locally developed methods present disadvantages and inadequate reproducibility. Therefore, tumor-specific guidelines, companion and complementary diagnostics, as well as surrogate biomarkers, are necessary for a targeted MMR status assessment in breast cancer. It is fair to conclude that improvements in MMR testing are necessary in order to enable its application in clinical practice.
Acknowledgments
Funding: This work was supported by the Italian Ministry of Health with Ricerca Corrente funds (NF) and National Cancer Institute (NCI) K22 CA229613 (SH).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1852). SH has a patent “MutL loss predicts sensitivity to CDK4/6 inhibitors in cancer” pending to Baylor College of Medicine. NF reports personal fees from Merck Sharp & Dohme (MSD) and Boehringer Ingelheim, outside the submitted work. In addition, NF has a patent “PTEN IHC as a predictor of mismatch repair status in breast cancer” pending to Italian Patent and Trademark Office. The other authors have no conflict of interest to declare.
References
- 1.Lei JT, Anurag M, Haricharan S, et al. Endocrine therapy resistance: new insights. Breast 2019;48 Suppl 1:S26-S30. 10.1016/S0960-9776(19)31118-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grizzi G, Ghidini M, Botticelli A, et al. Strategies for Increasing the Effectiveness of Aromatase Inhibitors in Locally Advanced Breast Cancer: An Evidence-Based Review on Current Options. Cancer Manag Res 2020;12:675-86. 10.2147/CMAR.S202965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emens LA. Breast Cancer Immunotherapy: Facts and Hopes. Clin Cancer Res 2018;24:511-20. 10.1158/1078-0432.CCR-16-3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmid P, Adams S, Rugo HS, et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med 2018;379:2108-21. 10.1056/NEJMoa1809615 [DOI] [PubMed] [Google Scholar]
- 5.Marcus L, Lemery SJ, Keegan P, et al. FDA Approval Summary: Pembrolizumab for the Treatment of Microsatellite Instability-High Solid Tumors. Clin Cancer Res 2019;25:3753-8. 10.1158/1078-0432.CCR-18-4070 [DOI] [PubMed] [Google Scholar]
- 6.Cheng AS, Leung SCY, Gao D, et al. Mismatch repair protein loss in breast cancer: clinicopathological associations in a large British Columbia cohort. Breast Cancer Res Treat. 2020;179:3-10. 10.1007/s10549-019-05438-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez G, Noale M, Corti C, et al. PTEN Expression as a Complementary Biomarker for Mismatch Repair Testing in Breast Cancer. International Journal of Molecular Sciences 2020;21:1461. 10.3390/ijms21041461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fusco N, Lopez G, Corti C, et al. Mismatch Repair Protein Loss as a Prognostic and Predictive Biomarker in Breast Cancers Regardless of Microsatellite Instability. JNCI Cancer Spectr 2018;2:pky056. 10.1093/jncics/pky056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SE, Lee HS, Kim KY, et al. High prevalence of the MLH1 V384D germline mutation in patients with HER2-positive luminal B breast cancer. Sci Rep 2019;9:10966. 10.1038/s41598-019-47439-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le DT, Durham JN, Smith KN, et al. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. 10.1126/science.aan6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies H, Morganella S, Purdie CA, et al. Whole-Genome Sequencing Reveals Breast Cancers with Mismatch Repair Deficiency. Cancer Res 2017;77:4755-62. 10.1158/0008-5472.CAN-17-1083 [DOI] [PubMed] [Google Scholar]
- 12.Corti C, Sajjadi E, Fusco N. Determination of Mismatch Repair Status in Human Cancer and Its Clinical Significance: Does One Size Fit All? Adv Anat Pathol 2019;26:270-9. 10.1097/PAP.0000000000000234 [DOI] [PubMed] [Google Scholar]
- 13.Stelloo E, Jansen AML, Osse EM, et al. Practical guidance for mismatch repair-deficiency testing in endometrial cancer. Ann Oncol 2017;28:96-102. 10.1093/annonc/mdw542 [DOI] [PubMed] [Google Scholar]
- 14.Sinicrope FA. DNA mismatch repair and adjuvant chemotherapy in sporadic colon cancer. Nat Rev Clin Oncol 2010;7:174-7. 10.1038/nrclinonc.2009.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mensenkamp AR, Vogelaar IP, van Zelst-Stams WA, et al. Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology 2014;146:643-6.e8. 10.1053/j.gastro.2013.12.002 [DOI] [PubMed] [Google Scholar]
- 16.Subbiah V, Kurzrock R. The Marriage Between Genomics and Immunotherapy: Mismatch Meets Its Match. Oncologist 2019;24:1-3. 10.1634/theoncologist.2017-0519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haricharan S, Bainbridge MN, Scheet P, et al. Somatic mutation load of estrogen receptor-positive breast tumors predicts overall survival: an analysis of genome sequence data. Breast Cancer Res Treat 2014;146:211-20. 10.1007/s10549-014-2991-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67-73. 10.1038/nature12113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budczies J, Bockmayr M, Denkert C, et al. Classical pathology and mutational load of breast cancer – integration of two worlds. J Pathol Clin Res 2015;1:225-38. 10.1002/cjp2.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Germano G, Amirouchene-Angelozzi N, Rospo G, et al. The Clinical Impact of the Genomic Landscape of Mismatch Repair-Deficient Cancers. Cancer Discov 2018;8:1518-28. 10.1158/2159-8290.CD-18-0150 [DOI] [PubMed] [Google Scholar]
- 21.Haricharan S, Punturi N, Singh P, et al. Loss of MutL disrupts Chk2-dependent cell cycle control through CDK4/6 to promote intrinsic endocrine therapy resistance in primary breast cancer. Cancer Discov 2017;7:1168-83. 10.1158/2159-8290.CD-16-1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anurag M, Punturi N, Hoog J, et al. Comprehensive Profiling of DNA Repair Defects in Breast Cancer Identifies a Novel Class of Endocrine Therapy Resistance Drivers. Clin Cancer Res 2018;24:4887-99. 10.1158/1078-0432.CCR-17-3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anurag M, Ellis MJ, Haricharan S. DNA damage repair defects as a new class of endocrine treatment resistance driver. Oncotarget 2018;9:36252-3. 10.18632/oncotarget.26363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez G, Costanza J, Colleoni M, et al. Molecular Insights into the Classification of Luminal Breast Cancers: The Genomic Heterogeneity of Progesterone-Negative Tumors. Int J Mol Sci 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagni F, Guerini-Rocco E, Schultheis AM, et al. Targeting Immune-Related Biological Processes in Solid Tumors: We do Need Biomarkers. Int J Mol Sci 2019;20:5452. 10.3390/ijms20215452 [DOI] [PMC free article] [PubMed] [Google Scholar]


