Abstract
Background
Small cell lung cancer (SCLC) has unique biology and chromosomal modifications; however, only a few studies have investigated the molecular map of SCLC. The present study aimed to evaluate the genomic aberrations in patients with SCLC in a Chinese cohort.
Methods
Tumor samples of SCLC were prospectively collected from Zhejiang Cancer Hospital. A total of 5 genes [EGFR (epidermal growth factor receptor) E18, EGFR E19, EGFR E20, EGFR E21, KRAS (Kirsten rat sarcoma viral oncogene homolog) E2, BRAF E15, PTEN (phosphatase and tensin homolog deleted on chromosome ten) E5, PTEN E6, PTEN E8, PIK3CA (phosphatidylinositol 3-kinase/protein kinase B) E9, PIK3CA E20] were evaluated using direct sequencing.
Results
Between November 2012 and November 2016, 30 SCLC patients were prospectively enrolled in the study. A total of 10 genomic aberrations were detected in 30 cases (33.3%): an EGFR mutation (n=6, E19, E21), a KRAS mutation (n=1, E2), PIK3CA mutations (n=1, E20), a PTEN mutation (n=2, E5, E8). No significant differences were detected in the characteristics of patients with and without genomic aberrations or patients with and without EGFR mutation.
Conclusions
The genomic aberrations of SCLC occur, offering mutational data to clinicians might be helpful for assigning patients to appropriate clinical studies, especially the anti-EFGR and PIK3CA treatment. Moreover, whether the molecular genetic profile of the SCLC patients is correlated with the effect of anti-tumor treatment, necessitating further investigation.
Keywords: Small cell lung cancer (SCLC), genetic profile, Chinese cohort
Introduction
Small cell lung cancer (SCLC) is an aggressive cancer with high recurrence rate after conventional chemotherapeutic treatment. SCLC presents unique biological characteristics, high genetic mutation load, dysregulation of tumor suppressor genes and oncogenes, and upregulated level of receptor tyrosine kinases, growth factors, and cellular markers. Currently, the analyses of genomic aberrations and searching new molecular-targeted drugs for SCLC are underway. However, only a few studies have investigated the molecular map of SCLC, and to date, none of the molecular-targeted drugs have exhibited clinical activity against SCLC. Reportedly, epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) has been successfully being used in SCLC in the event of seldom EGFR mutation seldom occurs (1,2). Although the EGFR mutations have been previously identified in SCLCs, those in the other EGFR signaling pathway genes, such as Kirsten rat sarcoma viral oncogene homolog (KRAS), human EGFR 2 (HER2), and v-raf murine sarcoma viral oncogene homolog B1 (BRAF) are yet unknown. Thus, additional studies are needed to determine the molecular characteristics of SCLC and seek other new effective targeted therapies.
The phosphoinositide 3-kinase (PI3K)/AKT pathways are vital for the proliferation, energy metabolism, cellular architecture advantages, and survival of malignant cells. Intriguingly, the suppression of the PI3K-AKT-mammalian target of rapamycin (mTOR) pathway through the lipid phosphatase activity activates phosphatase and tensin (PTEN) homolog gene that regulates a plethora of cellular processes. Moreover, the PI3K/PTEN pathway functions downstream of the receptor tyrosine kinases, including EGFR. Currently, two studies have used the targeted gene panels to identify the two common genetic aberrations in patients with SCLC: phosphatidylinositol-3-kinase catalytic subunit alpha (PI3KCA; 15%), and MET (4.4%) (3,4). Another analysis of 98 cancer patients using next-generation sequencing demonstrated that the most common aberrations occurred on the rapamycin-insensitive companion of mTOR (RICTOR; 10%), PI3KCA (6%) EGFR (5%), PTEN (5%), and KRAS (5%) (5). The identification of molecular aberrations of SCLC might facilitate the targeted therapies for the SCLC treatment.
The present study aimed to investigate the genomic aberrations in patients with SCLC in a Chinese cohort. The mutations in SCLC primarily encompass the EGFR/Ras/Raf pathway and PIK3CA/PTEN pathway genes and provide a novel potential target for SCLC. Furthermore, the identification of genomic aberrations linked to SCLC would facilitate the recognition of potential targets. A total of 5 genes (EGFR E18, EGFR E19, EGFR E20, EGFR E21, KRAS E2, BRAF E15, PTEN E5, PTEN E6, PTEN E8, PIK3CA E9, PIK3CA E20) were detected using direct sequencing in SCLC tumor samples.
Methods
Patient characteristics
A total of 30 SCLC patients treated at the Zhejiang Cancer Hospital between November 2012 and November 2016 were recruited for this study. The pathological diagnosis of SCLC in each case was based on the standard criteria defined by WHO classification. The median age of the study group was 59 (range, 35–79) years. The cohort consisting of 28 (93.3%) males and 2 (6.7%) females was further categorized into 4 non-smokers, 1 light smoker, 8 moderate smokers, and 17 heavy smokers. The stages involved were as follows: 20 were extended stage and 10 were limited stage. The study was approved by the Medical Ethics Review Committee of Zhejiang Cancer Hospital (Hangzhou, China). The patient samples of SCLC were consecutively collected, and all participants provided written informed consent.
DNA extraction and PCR amplification (direct sequencing)
The specimens from fine-needle biopsy or surgical operations were fixed using formalin and embedded in paraffin. Genomic DNA was isolated from 10 g-paraffin sections using DEXPAT® D9091 Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The target DNA fragment containing mutation loci, including EGFR exon 18, EGFR exon 20, EGFR exon21, KRAS exon2, BRAF exon15, PTEN exon5, PTEN exon6, PTEN exon8, PIK3CA exon9, PIK3CA exon20, was amplified using PCR. The amplification and sequencing primers were designed (Table 1) and synthesized by Invitrogen (Shanghai Invitrogen Biotechnology Co., Ltd, Shanghai, China). Primers with AF/AR post-fixed were the amplification primers, and those with SF post-fixed were the sequencing primers. The 25-µL PCR reaction system comprised of 12.5 µL of PrimeStar® Premix (TaKaRa), 2.5 µL of DNA sample, 1.5 µL of 10 µmol/mL AF and AR primers, and 7 µL of ddH2O. The PCR reaction was as follows: an initial denaturation step at 94 °C for 1 min, followed by 35 cycles of denaturation at 94 °C for 10 s, annealing at 58 °C for 5 s, elongation at 72 °C for 15 s, and final extension at 72 °C for 5 min; the products were held at 4 °C. The products were analyzed and identified by 2% agarose gel electrophoresis.
Table 1. Amplified primer sequences.
| Gene name | Amplified primer sequence |
|---|---|
| EGFR E18-AF | TTCCAAATGAGCTGGCAAGT |
| EGFR E 18-AR | TGGAGTTTCCCAAACACTCAG |
| EGFR E19-AF | CCCCAGCAATATCAGCCTTA |
| EGFR E19-AR | GTGGATACCAGCATGGGAGA |
| EGFR E20-AF | AGGCACAGCTTTTCCTCCAT |
| EGFR E20-AR | CATATCCCCATGGCAAACTC |
| EGFR E21-AF | ACTAACGTTCGCCAGCCATA |
| EGFR E21-AR | CGAGCTCACCCAGAATGTC |
| KRAS 1213-AF | CTTAAGCGTCGATGGAGGAG |
| KRAS 1213-AR | TTGAAACCCAAGGTACATTTCA |
| PIK3CA E9-AF | TGAAAATAAAGTCTTGCAATGAAAA |
| PIK3CA E9-AR | TTCCACAAATATCAATTTACAACCA |
| PIK3CA E20-AF | GCTCCAAACTGACCAAACTG |
| PIK3CA E20-AR | ATGCTGTTCATGGATTGTGC |
| BRAF E15-AF | TTGACTCTAAGAGGAAAGATGAAGT |
| BRAF E15-AR | AGAACACTGATTTTTGTGAATACTGG |
| PTEN E5-AF | CTCTGGAATCCAGTGTTTCTTTTA |
| PTEN E5-AR | CCTCAATAAAACTGAAGGAA |
| PTEN E6-AF | TCCTTTGTATTGATATTGCTCTTA |
| PTEN E6-AR | CAAGCTAATTAATTTGTTCAAA |
| PTEN E8-AF | TGATAGTTTATTTTGTTGACTTTTTGC |
| PTEN E8-AR | TTGTCAAGCAAGTTCTTCATCAG |
Note: AF/AR is an amplification primer.
PCR product purification
The PCR products were purified in a reaction system consisting of 25 µL PCR products, 2 µL exonuclease I, and 5 µL alkaline phosphatase (ThermoFisher Scientific Inc., Waltham, USA). The reaction mixture was incubated at 37 °C for 30 min, inactivated at 85 °C for 15 min, and stored under cold conditions until further usage.
Mutation sequencing
The purified PCR products were sequenced using BigDye® Terminator V3.1 Cycle Sequencing Kit (ABI, Foster City, USA). The 20 µL reaction system consisted of 6 µL of 10 mol/mL sequencing primer, 8 µL of 2.5× BigDye, 4 µL of ddH2O, and 2 µL of the purified PCR product. The sequencing reaction conditions were as follows: an initial pre-denaturation at 96 °C for 60 s, followed by 25 cycles of denaturation at 96 °C for 10 s, annealing at 50 °C for 5 s, and elongation at 60 °C for 4 min.
The sequencing was conducted on an ABI 3130XL genetic analyzer (ABI) after the purification of the sequencing reaction products. The sequencing data were collected using the 3130XL software and analyzed by Sequencing Analysis software v5.4.
Statistical analysis
The statistical significance of the mean values was determined by SPSS 22.0. A P value <0.05 indicated a statistically significant difference.
Results
Genomic aberrations of SCLC
We detected 10 genomic aberrations in 30 cases (33.3%): an EGFR mutation (n=6, E19, E21), a KRAS mutation (n=1, E2), PIK3CA mutations (n=1, E20), a PTEN mutation (n=2, E5, E8) (Tables 2,3). In addition, 3 cases of synonymous mutation included 2 cases of EGFR E20, 1 case of E18, and 1 case of PIK3CA E9.
Table 2. Gene mutations in 30 SCLCs.
| Gene | Mutation | |
|---|---|---|
| n | % | |
| EGFR E19 | 5 | 50 |
| EGFR E21 | 1 | 10 |
| KRAS E2 | 1 | 10 |
| PTEN E5 | 1 | 10 |
| PTEN E8 | 1 | 10 |
| PIK3CA E20 | 1 | 10 |
| Total | 10 | – |
SCLC, small cell lung cancer.
Table 3. Detail information of 10 genomic aberrations in SCLC patients.
| Case number | Gene name | Position | AA mutation | Nucleotide mutant |
|---|---|---|---|---|
| 1 | EGFR | Exon 19 | p.E746_A750del | c.2235-2249del |
| 2 | Exon 19 | p.E746_A750del | c.2235-2249del | |
| 3 | Exon 19 | p.E746_A750del | c.2235-2249del | |
| 4 | Exon 19 | p.L747_S752del | c.2239-2256del | |
| 5 | Exon 19 | p.K737_I740delinsPHWD | c.2209_2219delinsCCACACTGGGA | |
| 6 | Exon 21 | p.Q849_H850delinsHN | c.2547_2548delinsTA | |
| 7 | KRAS | G12 | p.G12D | c.35G>A |
| 8 | PIK3CA | E1034 | p.E1034Q | c.3100G>C |
| 9 | PTEN | Y315 | p.Y315H | c.943T>C |
| 10 | Exon 5 | p.F154fs*5 | c.461del |
SCLC, small cell lung cancer; AA, amino acid.
Comparison of patient characteristics classified by genomic aberrations
The patient characteristics are classified according to the status of the genomic aberrations (Table 4). No significant differences were detected in age, sex, disease extent, smoking history, and carcinoembryonic antigen (CEA) and squamous cell lung carcinoma (SCC) level between patients with and without genomic aberrations (P>0.05).
Table 4. Clinical characteristics comparison between SCLC patients with and without EGFR mutation.
| Clinical characteristics | Detected [6] | Not detected [24] | P |
|---|---|---|---|
| Gender | 0.366 | ||
| Male | 5 | 23 | |
| Female | 1 | 1 | |
| Age (median) | 0.633 | ||
| <65 | 5 | 15 | |
| ≥65 | 1 | 9 | |
| Smoking history | 0.920 | ||
| Non-smoker | 1 | 3 | |
| Light smoker | 0 | 1 | |
| Moderate smoker | 2 | 6 | |
| Heavy smoker | 3 | 14 | |
| Stage | 1.000 | ||
| Extended stage | 4 | 16 | |
| Limited stage | 2 | 8 | |
| CEA abnormal | 1 | 7 | 0.655 |
| 5 | 17 | ||
| SCC abnormal | 1 | 2 | 0.543 |
| 5 | 22 |
SCLC, small cell lung cancer; EGFR, epidermal growth factor receptor; CEA, carcinoembryonic antigen.
Furthermore, no significant differences were detected in age, sex, disease extent, smoking history, and CEA and SCC level in patients with and without EGFR mutation (P>0.05, Table 5).
Table 5. Clinical characteristics comparison between SCLC patients with and without genomic aberrations.
| Clinical characteristics | Detected [10] | Not detected [20] | P |
|---|---|---|---|
| Gender | 0.605 | ||
| Male | 9 | 19 | |
| Female | 1 | 1 | |
| Age (median) | 0.702 | ||
| <65 | 7 | 12 | |
| ≥65 | 3 | 8 | |
| Smoking history | 0.702 | ||
| Non-smoker | 1 | 3 | |
| Light smoker | 0 | 1 | |
| Moderate smoker | 4 | 4 | |
| Heavy smoker | 5 | 12 | |
| Stage | 0.231 | ||
| Extended stage | 5 | 15 | |
| Limited stage | 5 | 5 | |
| CEA abnormal | 2 | 6 | 0.682 |
| 8 | 14 | ||
| SCC abnormal | 2 | 1 | 0.532 |
| 8 | 19 |
SCLC, small cell lung cancer; CEA, carcinoembryonic antigen.
Cases of common mutations
(I). EGFR E19 c.2235–2249 del 15: EGFR codes 746 for glutamate to 750 for alanine loss
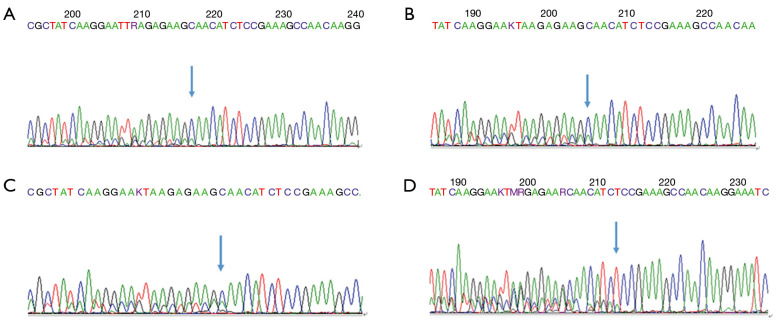
This variation, localized in the EGFR protein kinase domain, is a common variant of EGFR exon 19 (6). The E746_A750 Del mutation was reported to maintain the activity of EGFR, promote the MAPK and AKT phosphorylation downstream, and promote the tumor growth in transplanted mice (7,8) (Figure 1A,B,C).
Figure 1.
Common mutations. (A) Case 2: EGFR E19 c.2235-2249 del 15 (p.E746_A750del); (B) Case 5: EGFR E19 c.2235-2249 del 15 (p.E746_A750del); (C) Case 10: EGFR E19 c.2235-2249 del 15 (p.E746_A750del); (D) Case 7: c.2239_2256del18 (p.L747_S752del).
(II). EGFR E19 c.2239–2256 del 18 (p.L747_S752del)
EGFR 747 codes leucine to 752 codon as serine is missing. The mutation, located in the EGFR protein kinase domain, is a missing mutation on exon 19, with a frequency of 2.1% (9) (Figure 1D).
Cases of rare mutations
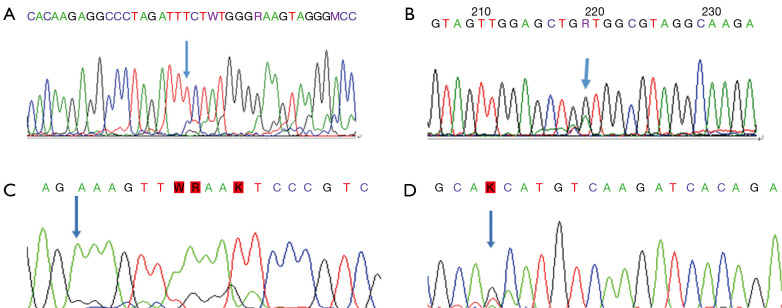
(I). PTEN E5 461delT
PTEN p.F154fs*5, PTEN 154th codon, wherein phenylalanine becomes serine, and according to the new open reading frame, 5th codon is the termination codon. The prediction analysis of this mutation led to amide-mediated mRNA degradation, resulting in the loss of protein expression. The mutation was detected in gliomas, according to the Cosmic database (Figure 2A).
Figure 2.
Rare mutations. (A) Case 6: PTEN 5 461 del T; (B) Case 12: KRAS c.35G>A (p.G12D); (C) Case 19: EGFR E19 c.2209_2219delinsCCACACTGGGA (p.K737_I740delinsPHWD); (D) Case 30: EGFR E21 c.2547_2548delinsTA (p.Q849_H850delinsHN).
(II). KRAS c.35G>A (p.G12D)
P. G12d, KRAS 12th code subglycine mutation to aspartic acid. KRAS 12D mutated GTP enzyme activity was weaker than wild-type KRAS, which enhanced the binding state of active KRAS-GTP and promoted the KRAS signaling pathway to cause cancer (10,11) (Figure 2B).
(III). EGFR E19 c.2209_2219delinsCCACACTGGGA (p.K737_I740delinsPHWD)
The 713th–740th coding positions of EGFR are deleted and inserted with proline, histidine, tryptophan, and aspartic acid. This mutation is located in exon 19 of EGFR, and no such mutation has been reported (Figure 2C).
(IV). EGFR E21 c.2547_2548delinsTA (p.Q849_H850delinsHN)
Histidine and asparagine were inserted after the deletion of the coding sequence at positions 849-950 of EGFR (Figure 2D).
Discussion
Cancer gene therapy has low toxicity than conventional chemotherapy, but no targeted drugs have been approved for the treatment of SCLC. The application of genomic technologies has begun to elucidate a large number of genetic abnormalities in SCLC. In a Japanese cohort, genomic aberrations were detected in 15% patients, including PIK3CA amplification (10%), PIK3CA mutations (5%), EGFR mutation (2%), and KRAS mutation (2%). However, in the Chinese population, the genomic aberrations in SCLC are specific. The present study demonstrated that 10 patients had genomic aberrations; EGFR mutation accounted for 60% of the patients, while PTEN mutations were found in 2 patients (20%).
The EGFR mutations found in the current study included E19 and E21, of which, E19 mutation occurred in 6 SCLC patients. Interestingly, EGFR mutations that commonly occurred in exon 19 were deletions, and those in exon 21 (L858R) were mutations. Also, in these 30 SCLC patients, rare EGFR-mutated exon 18 was found. The mutation rate of EGFR in our study was higher than in the Japanese patients, i.e., in 4% of 122 Japanese patients with SCLCs. This phenomenon might be associated with small sample size in the current research. Furthermore, patients with the p.E746_A750del variant in non-SCLC (NSCLC) are sensitive to EGFR TKI (6). Preclinical data and clinical trial results showed that exon insertion or deletion mutation 19 of EGFR was sensitive to EGFR TKI (12-14). Moreover, patients with NSCLC with L747_S752del mutation benefit from the EGFR TKI treatment (15,16).
The Ras oncogene controls several cellular functions including cell proliferation, apoptosis, migration, and differentiation, while the KRAS mutations are noted in >20% of all cancers. In this study, KRAS E2 mutation occurred in one patient (3.3% in all SCLCs), and this variation was mainly carcinogenic as identified via the PI3K-AKT, JNK, p38, and FAK signaling pathways (17,18).
PI3K might be activated by the receptor kinases and Ras, which in turn, activates AKT. However, the PI3K and EGFR signaling pathways closely interact, and PI3K signaling pathway is effectuated by additional activators and downstream targets (4). Herein, we found 1 patient with PIK3CA mutations (E20), and 2 patients harbored the PTEN mutation (E5, E8).
Furthermore, we compared the clinical characteristics between SCLC patients with and without genomic aberrations as well as between SCLC patients with and without EGFR mutation. However, no significant differences were detected in age, sex, disease extent, smoking history, and CEA and SCC level in either of the groups, which might be attributed to the small sample size as one of the underlying factors.
Nevertheless, the genomic aberrations of SCLC identified in this study offered mutational data to clinicians that might be applied for assigning patients to appropriate clinical study, especially the anti-EFGR treatment.
Acknowledgments
Funding: This work was supported by the Zhejiang Province Medical Science Project of China (2015ZHA006), the 1022 Talent Training Program of Zhejiang Cancer Hospital, and the Zhejiang Provincial National Science Foundation of China (Q18H290004).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Review Committee of Zhejiang Cancer Hospital (Hangzhou, China). The number of ethics approval is [2012]-10-20. Written informed consent was obtained from the patient for publication of this manuscript and any accompanying images.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.01.26). The authors have no conflicts of interest to declare.
References
- 1.Okamoto I, Araki J, Suto R, et al. EGFR mutation in gefitinib-responsive small-cell lung cancer. Ann Oncol 2006;17:1028-9. 10.1093/annonc/mdj114 [DOI] [PubMed] [Google Scholar]
- 2.Zakowski MF, Ladanyi M, Kris MG, et al. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med 2006;355:213-5. 10.1056/NEJMc053610 [DOI] [PubMed] [Google Scholar]
- 3.Govindan R, Page N, Morgensztern D, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol 2006;24:4539-44. 10.1200/JCO.2005.04.4859 [DOI] [PubMed] [Google Scholar]
- 4.Lu HY, Zhang G, Cheng QY, et al. Expression and mutation of the c-kit gene and correlation with prognosis of small cell lung cancer. Oncol Lett 2012;4:89-93. 10.3892/ol.2012.679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dy GK, Miller AA, Mandrekar SJ, et al. A phase II trial of imatinib (ST1571) in patients with c-kit expressing relapsed small-cell lung cancer: a CALGB and NCCTG study. Ann Oncol 2005;16:1811-6. 10.1093/annonc/mdi365 [DOI] [PubMed] [Google Scholar]
- 6.Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. 10.1038/nrc2088 [DOI] [PubMed] [Google Scholar]
- 7.Sakai K, Arao T, Shimoyama T, et al. Dimerization and the signal transduction pathway of a small in-frame deletion in the epidermal growth factor receptor. FASEB J 2006;20:311-3. 10.1096/fj.05-4034fje [DOI] [PubMed] [Google Scholar]
- 8.Carey KD, Garton AJ, Romero MS, et al. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res 2006;66:8163-71. 10.1158/0008-5472.CAN-06-0453 [DOI] [PubMed] [Google Scholar]
- 9.Furuyama K, Harada T, Iwama E, et al. Sensitivity and kinase activity of epidermal growth factor receptor (EGFR) exon 19 and others to EGFR-tyrosine kinase inhibitors. Cancer Sci 2013;104:584-9. 10.1111/cas.12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janakiraman M, Vakiani E, Zeng Z, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res 2010;70:5901-11. 10.1158/0008-5472.CAN-10-0192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith G, Bounds R, Wolf H, et al. Activating K-Ras mutations outwith 'hotspot' codons in sporadic colorectal tumours - implications for personalised cancer medicine. Br J Cancer 2010;102:693-703. 10.1038/sj.bjc.6605534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He M, Capelletti M, Nafa K, et al. EGFR exon 19 insertions: a new family of sensitizing EGFR mutations in lung adenocarcinoma. Clin Cancer Res 2012;18:1790-7. 10.1158/1078-0432.CCR-11-2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang CH, Shih JY, Chen KC, et al. Survival outcome and predictors of gefitinib antitumor activity in East Asian chemonaive patients with advanced nonsmall cell lung cancer. Cancer 2006;107:1873-82. 10.1002/cncr.22220 [DOI] [PubMed] [Google Scholar]
- 14.Asahina H, Yamazaki K, Kinoshita I, et al. A phase II trial of gefitinib as first-line therapy for advanced non-small cell lung cancer with epidermal growth factor receptor mutations. Br J Cancer 2006;95:998-1004. 10.1038/sj.bjc.6603393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung KP, Wu SG, Wu JY, et al. Clinical outcomes in non-small cell lung cancers harboring different exon 19 deletions in EGFR. Clin Cancer Res 2012;18:3470-7. 10.1158/1078-0432.CCR-11-2353 [DOI] [PubMed] [Google Scholar]
- 16.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. 10.1073/pnas.0405220101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Céspedes MV, Sancho FJ, Guerrero S, et al. K-ras Asp12 mutant neither interacts with Raf, nor signals through Erk and is less tumorigenic than K-ras Val12. Carcinogenesis 2006;27:2190-200. 10.1093/carcin/bgl063 [DOI] [PubMed] [Google Scholar]
- 18.Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst 2012;104:228-39. 10.1093/jnci/djr523 [DOI] [PMC free article] [PubMed] [Google Scholar]