Abstract
Nucleic acid sequence-based amplification (NASBA), an isothermal amplification technique, was established and evaluated for the detection of Aspergillus RNA and compared with a previously published, well-defined real-time PCR assay amplifying a region of the Aspergillus 18S rRNA gene. NASBA showed a lower detection limit of 1 CFU and detected RNA from five different clinically relevant Aspergillus species, including Aspergillus fumigatus. All 77 blood samples tested by PCR and NASBA showed identical results in both assays. Results with the NASBA technique were obtained within 6 h. Thus, the NASBA technique provided a valuable tool for sensitive, specific, fast, and reliable detection of Aspergillus RNA with potential for routine diagnosis, including the possibility to test the viability of cells.
Aspergillus species cause life-threatening acute invasive disease in immunocompromised patients. The incidence of invasive aspergillosis has been steadily increasing over the last few decades due to an increasing number of patients undergoing chemotherapy or bone marrow and solid organ transplantation and to an improved survival rate among patients with AIDS. The incidence is estimated to be up to 25% and the mortality is up to 90% in patients with acute leukemia (5). Early diagnosis is essential for appropriate and successful antifungal therapy. However, conventional tests for the detection of Aspergillus spp., such as blood culture and serology, have limited sensitivity and specificity (5). Diagnosis by biopsy or histopathology is difficult due to the severity of illness in these patients. Recently developed methods such as antigen detection and PCR-based assays may provide sensitive tools for the diagnosis of invasive aspergillosis.
Nucleic acid sequence-based amplification (NASBA) is an isothermal amplification technology which specifically amplifies RNA sequences in a DNA background by using T7 RNA polymerase (2). NASBA-based assays have been described for the detection of Candida spp. (19), Salmonella spp. (15), human immunodeficiency virus (14), cytomegalovirus (1), and hepatitis C virus (4) RNA in clinical specimens. However, to our knowledge, NASBA protocols have not yet been applied to the detection of Aspergillus RNA.
The objective of our study was to establish and to evaluate a NASBA-based assay for a sensitive and specific extraction, amplification, and detection of RNA from different Aspergillus species in blood and to compare these data with a previously published, well-defined real-time PCR assay amplifying a region of the Aspergillus 18S rRNA gene (11).
For the determination of the lower detection limit of the NASBA assay, as well as for RNA degradation experiments, blood from healthy volunteers was spiked with Aspergillus fumigatus conidia (106 to 100/ml, in serial dilution). A. fumigatus cultures (DSM 790) were obtained from the German Collection of Microorganisms (DSM) and were cultured on Sabouraud-glucose-agar for 72 h at 30°C. Serial dilutions of fungal cells were prepared with sterile saline suspensions, adjusted to a McFarland standard of 0.5 (106 CFU/ml).
In order to determine possible cross-reactions of the NASBA oligonucleotide with other filamentous fungi, RNA from defined cultures of Aspergillus flavus (DSM 818), Aspergillus niger (DSM 737), Aspergillus versicolor (DSM 1943), Aspergillus glaucus (ATCC 14567), Scopulariopsis brevicaulis (DSM 1218), Curvularia inaequalis (DSM 62462), Absidia corymbifera (DSM 1144), Fusarium solani (DSM 1164), Rhizopus oryzae (DSM 905), Acremonium chrysogenum (DSM 880), Penicillium brevicompactum (DSM 3825), Penicillium chrysogenum (DSM 844), and Alternaria alternata (DSM 1102), as well as RNA from Candida albicans (DSM 1665), cytomegalovirus, and human fibroblasts from healthy individuals, were analyzed.
Additionally, 77 blood specimens from neutropenic patients after allogeneic bone marrow transplantation (n = 20) were analyzed in parallel by real-time PCR and NASBA-based assays for the presence of Aspergillus nucleic acid.
RNA was extracted using a protocol for filamentous fungi. Briefly, 100-μl portions of fungal suspensions were prepared followed by immersion in liquid nitrogen and incubation for 3 min at 60°C. Alternatively, for RNA extraction from whole blood, 300 μl of RLT lysis buffer (Qiagen, Hilden, Germany) was added to 100 μl of blood, followed by immersion in liquid nitrogen. Then, 4.5 μl (20 μl when RNA was extracted from blood samples) of RNA Secure (Ambion, Austin, Tex.) was added, and the mixture was incubated for 20 min at 60°C. Isolation and purification of the RNA were performed according to the manufacturer's protocol by using the RNeasy Minikit and QiaShredder spin columns (Qiagen). Next, 80 μl of eluate was obtained and stored at −80°C until further use.
The design of NASBA oligonucleotide was based on comparison of the sequence of 18S rRNA genes of Aspergillus species and other fungi in the GenBank database. Primers 2.1 (5′-GCCGCGGTAATTCCAGCTCCAATA) and 1.2 (5′-AATTCTAATACGACTCACTATAGGGGAGCAAAGGCCTGCTTTGAACA) (0.4 mM each, with the T7 promoter sequence in italics) bind to a highly conserved region of the 18S rRNA gene. A biotinylated oligonucleotide probe (5′-GGTCCGCCTCACCGCGAGTACTG) was chosen that binds to clinically relevant Aspergillus species (A. fumigatus, A. versicolor, A. flavus, A. niger, and A. glaucus). Next, 5 μl of target RNA was added to a prereaction mixture according to the protocol of the Basic Kit Amplification Module (Organon Teknika, Boxtel, The Netherlands), followed by an incubation at 65°C for 5 min and at 41°C for another 5 min. Then, 5 μl of enzyme solution (AMV-RT, RNase H, T7 RNA polymerase, bovine serum albumin, and sorbitol) was added, and the reaction mixture was incubated for 90 min at 41°C for isothermal amplification of RNA. After amplification, detection reagents were prepared by vortexing a bead-oligo suspension (biotinylated Aspergillus oligonucleotide bound to streptavidin-coated paramagnetic beads) until an opaque solution was formed. Bead-oligo suspension and a generic ruthenium-labeled electrochemiluminescence (ECL) probe were mixed. Then, 20 μl of this mixture was added to 5 μl of the diluted NASBA product (1:10) and incubated for 30 min at 41°C. After that, 300 μl of Assay Buffer (Organon Teknika) was added to the hybridization tube, and the ECL readings were performed in the NucliSens Reader as described elsewhere (6). To evaluate whether genomic Aspergillus DNA present in the extract was detectable by NASBA, aliquots of extracted Aspergillus nucleic acid were incubated with DNase-free RNase (Roche Molecular Biochemicals, Mannheim, Germany) for 1 h at 37°C.
DNA was extracted as described previously (9) using recombinant lyticase (Sigma, Deissenhofen, Germany) and the QIAmp Tissue Kit (Qiagen). DNA amplification was performed with primers (5′-ATT GGA GGG CAA GTC TGG TG, 5′-CCG ATC CCT AGT CGG CAT AG; Roth, Karlsruhe, Germany) binding to conserved regions of the fungal 18S rRNA gene in a real-time PCR format using the LightCycler instrument (11). The detection system is based on fluorescence resonance energy transfer with two different specific oligonucleotides. Hybridization probe 1 (5′-GTT CCC CCC ACA GCC AGT GAA GGC) was labeled with fluorescein; hybridization probe 2 (5′-TGA GGT TCC CCA GAA GGA AAG GTC CAG C) was labeled with Light Cycler Red 640. Both probes are Aspergillus genus-specific and can hybridize in a head-to-tail arrangement which brings the two fluorescent dyes into close proximity. A transfer of energy between the two probes results in the emission of red fluorescent light, which is measured by photohybrids.
To minimize the risk of carryover contaminations, RNA and DNA extraction, NASBA-based amplification and detection, and the LightCycler PCR were performed in separate rooms with equipment (pipettes, tips, and glassware) exclusively used for these purposes. Workers performing NASBA and PCR assays were single-use gowns, sterile gloves, and face masks and were not allowed to move from the detection into the extraction area on the same day.
In order to control the presence of RNA in extracts, 23 samples were analyzed with the Titan One Tube RT-PCR Kit (Roche Molecular Biochemicals). The assay is a reverse transcription-PCR (RT-PCR) technique using avian myeloblastosis virus for first-strand synthesis and Taq polymerase, together with the proofreading Pwo polymerase for the PCR part (12). All samples were treated with DNase I (RNase-free; Roche Molecular Biochemicals) for 1 h at 37°C, and PCR was performed as described before (9). Amplicons were detected by gel electrophoresis (2% agarose, in TAE) and visualized by GelStar DNA staining (FMC Bioproducts, Hessisch Oldendorf, Germany).
For determination of the lower detection limit of the RNA assay, NASBA-based amplification of RNA was compared to the detection of Aspergillus DNA by PCR. NASBA-based assays showed a lower detection limit of 1 CFU, whereas the lower detection limit of the PCR was 10 CFU (11). The Aspergillus genus-specific NASBA probe detected RNA extracted from cultures of A. fumigatus, A. flavus, A. versicolor, A. glaucus, and A. niger. Additionally, the probe cross-reacted with P. brevicompactum, P. chrysogenum, and A. alternata RNA. No signal was obtained with RNA from six filamentous fungi, C. albicans, as well as with RNA from cytomegalovirus and human fibroblasts (Table 1). To determine whether inhibitors that might be present in the blood influenced the amplification performance, 28 series of diluted A. fumigatus conidia (106 to 100 CFU) were analyzed. Of these reactions, none turned out to be invalid (no amplification process), demonstrating the robustness of the assay.
TABLE 1.
ECL counts obtained by NASBA assay and CFU counts obtained by quantitative LightCycler-PCR (LC-PCR) for A. fumigatus in defined dilutions for different filamentous fungi and for C. albicans, cytomegalovirus, and human RNA
| Inoculum (CFU)a | Mean ECL count ± SD as determined by NASBA assay | Mean CFU/ml ± SD as determined by LC-PCRb |
|---|---|---|
| Aspergillus fumigatus (105) | 82,841 ± 52,282 | 100,650 ± 38,112 |
| Aspergillus fumigatus (104) | 61,770 ± 46,673 | 11,230 ± 5,416 |
| Aspergillus fumigatus (103) | 32,901 ± 18,073 | 990 ± 523 |
| Aspergillus fumigatus (102) | 7,201 ± 11,412 | 125 ± 68 |
| Aspergillus fumigatus (101) | 3,133 ± 3,067 | 9.3 ± 0.1 |
| Aspergillus fumigatus (1) | 2,891 ± 3,450 | Neg |
| Aspergillus flavus∗ | 230,117 | ND |
| Aspergillus niger∗ | 293,710 | ND |
| Aspergillus versicolor∗ | 138,254 | ND |
| Aspergillus glaucus∗ | 123,072 | ND |
| Scopulariopsis brevicaulis∗ | 325 | Neg |
| Curvularia inaequalis∗ | 134 | Neg |
| Absidia corymbifera∗ | 134 | Neg |
| Fusarium solani∗ | 146 | Neg |
| Rhizopus oryzae∗ | 180 | Neg |
| Acremonium chrysogenum∗ | 130 | Neg |
| Penicillium brevicompactum∗ | 17,519 | Pos |
| Penicillium chrysogenum∗ | 44,226 | Pos |
| Alternaria alternata∗ | 844 | Pos |
| Candida albicans∗ | 196 | Neg |
| Cytomegalovirus | 134 | Neg |
| Human fibroblast | 209 | Neg |
| Double-distilled water | 90 ± 66 | Neg |
Organisms tested in an undiluted suspension are indicated with an asterisk.
Neg, negative; Pos, positive; ND, not done.
To evaluate whether coinfections with two or more fungal species in a patient influence the performance of the NASBA assay, blood samples were spiked with A. fumigatus and C. inaequalis, a fungus which is not detected by this assay (103 CFU). A minimal reduction in the ECL counts was observed (18,760 counts) compared to samples spiked with A. fumigatus alone (29,575 counts). If blood samples were spiked with A. fumigatus and A. niger (103 CFU), an increase in the ECL counts of 15,097 was observed.
Additionally, 4 blood samples which were Aspergillus PCR positive and 73 samples which were PCR negative from patients after allogeneic bone marrow transplantation (n = 20) were analyzed by PCR and NASBA. All PCR-negative samples (n = 73) were also NASBA negative, and the four PCR-positive samples were also NASBA positive. These preliminary data indicate the possibility of using the NASBA technique for analyzing RNA in clinical specimens.
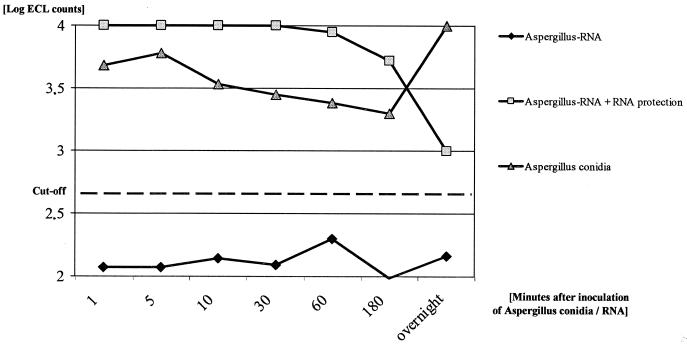
In order to control the degradation of RNA, blood samples were spiked with A. fumigatus conidia or RNA. ECL counts were measured in the presence or absence of RNA protection buffer (RNA Secure), respectively. In the presence of the RNA protection buffer, Aspergillus conidia and RNA were detectable 1, 5, 10, 30, 60, and 180 min and 24 h postspiking, whereas without buffer only the conidia were detectable (24 h postinoculation). No ECL counts were detectable with spiked RNA, a result most likely due to rapid RNA degradation (Fig. 1). NASBA performed on RNase-treated culture extracts of A. fumigatus yielded ECL signals which were not different from the assay negative control signals. In contrast, positive results were obtained in all 23 spiked samples if cDNA was amplified by RT-PCR performed on DNase-treated nucleic acid extracts.
FIG. 1.
Influence of time between sampling and extraction on detectable RNA level.
Invasive aspergillosis has been reported with an increasing frequency in bone marrow and solid organ transplant recipients, in patients receiving intense chemotherapy, in AIDS patients, and in patients with cystic fibrosis. For the early diagnosis of disseminated disease, rapid, sensitive, specific, and reproducible detection methods are mandatory (8). In selected groups of patients, PCR-based assays demonstrated a potential value for the early diagnosis of invasive fungal infections (7, 16). We recently showed that prospective screening of patients after allogeneic stem cell transplantation by PCR revealed a very high negative predictive value of 100% (7). However, as also found for other sensitive assays, the positive predictive value in an unselected cohort of patients was found to be rather low. In contrast, RT-PCR (13) and NASBA-based assays (1) proved to be highly specific methods for the detection of viral infection as it directly reflects transcriptional activity. Widjojoatmodjo et al. demonstrated that NASBA is a good alternative to PCR for the detection of candidemia (19). Candida RNA was extracted according to a modified protocol from van Deventer et al. (18), which allows RNA isolation within 3 h. However, for Aspergillus RNA extraction, a protocol including immersion of the fungal cells in liquid nitrogen and incorporation with β1,3-glucanase is mandatory because of the complexity of its cell wall. The assay described offers a very sensitive tool for the detection of Aspergillus RNA in blood. The NASBA process requires fewer cycles than PCR to obtain similar sensitivity since 10 to 100 copies of RNA are generated in each transcription step (2). Thus, the total incubation time and the error frequency are reduced with NASBA. PCR-based assays followed by specific hybridization require a minimum of 9 h, whereas the total incubation time of the NASBA assay is less than 4 h. Therefore, NASBA-based assays (extraction, amplification, and detection) can be performed within 1 working day. However, the use of a buffer (RNA Secure) which protects RNA against degradation by RNase is strongly recommended, since all untreated samples remained negative, whereas in samples containing buffer, spiked RNA was detectable at >3 h postspiking.
The primers described are binding to 18S rRNA. In bacteria, 16S rRNA is present in approximately 1,000 copies per cell, whereas in fungal cells, 100 to 300 copies of 18S rRNA can be found. This further enhances the sensitivity of the assay compared to the amplification of nucleic acids from single-copy genes with a detection limit of 100 CFU (3). The NASBA probe showed specificity for Aspergillus and Penicillium species with no cross-reaction to yeast, viral, or human RNA. We demonstrated previously (10) that lyticase or commercially available PCR buffers contained DNA from Saccharomyces cerevisiae and other yeasts. Thus, a potential risk of contamination with RNA extracted from fungi present in enzymes and buffers (10) could be minimized.
The NASBA assay offers practical advantages compared to PCR or RT-PCR. PCR requires rapid temperature changes for which thermal cyclers are required, whereas NASBA is an isothermal amplification process. Unlike RT-PCR, the NASBA assay allows detection of unspliced mRNA and, since thermal denaturation is absent, a contaminating background of genomic DNA is not a concern. Specimens can be stored in NASBA lysis buffer at −80°C until further processing. In addition, the NASBA technique requires only 100 μl of whole blood compared to 5 ml of blood for PCR. By both techniques, we obtained an identical sensitivity of 1 to 10 CFU.
In conclusion, NASBA-based assays are valuable tools for sensitive, specific, fast, and reliable detection of various pathogens. They have a potential value for routine diagnosis, including the possibility to test viability of cells. Further studies will be performed for a prospective comparison of PCR, NASBA, and the detection of fungal cell wall components for an early and sensitive detection of Aspergillus spp. and to clarify the value of these assays in preemptive antifungal therapy.
Acknowledgments
We thank Organon Teknika, Boxtel, The Netherlands for supplying the NASBA Basic Kits.
This project has been supported by the Deutsche Krebshilfe grant 70-2199-Ka1.
REFERENCES
- 1.Blok M J, Goossens V J, Vanherle S J, Top B, Tacken N, Middeldorp J A, Christiaans H, van Hooff J P, Bruggeman C A. Diagnostic value of monitoring cytomegalovirus late pp67 mRNA expression in renal allograft recipients by nucleic acid sequence based amplification. J Clin Microbiol. 1998;36:1341–1346. doi: 10.1128/jcm.36.5.1341-1346.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 3.Crampin A C, Matthews R C. Application of the polymerase chain reaction to the diagnosis of candidosis by amplification of an HSP90 gene fragment. J Med Microbiol. 1993;39:233–238. doi: 10.1099/00222615-39-3-233. [DOI] [PubMed] [Google Scholar]
- 4.Damen M, Sillekens P, Cuypers H T, Frantzen I, Melsert R. Characterization of the quantitative HCV NASBA assay. J Virol Methods. 1999;82:45–54. doi: 10.1016/s0166-0934(99)00079-8. [DOI] [PubMed] [Google Scholar]
- 5.Denning D W. Invasive aspergillosis. Clin Infect Dis. 1998;26:781–805. doi: 10.1086/513943. [DOI] [PubMed] [Google Scholar]
- 6.Gudibande S R, Kenten J H, Link J, Friedman K, Massey R J. Rapid, non-separation electrochemiluminescent DNA hybridization assays for PCR products, using 3′-labeled oligonucleotide probes. Mol Cell Probe. 1992;6:495–503. doi: 10.1016/0890-8508(92)90046-z. [DOI] [PubMed] [Google Scholar]
- 7.Hebart H, Loeffler J, Meisner C, Serey F, Schmidt D, Böhme A, Martin H, Engel A, Kern W V, Schumacher U, Kanz L, Einsele H. Early detection of Aspergillus infection after allogeneic stem cell transplantation by polymerase chain reaction screening. J Infect Dis. 2000;181:1713–1719. doi: 10.1086/315435. [DOI] [PubMed] [Google Scholar]
- 8.Latgé J P. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeffler J, Hebart H, Sepe S, Schumacher U, Einsele H. Detection of PCR-amplified fungal DNA by using a PCR-ELISA system. Med Mycol. 1998;36:275–279. doi: 10.1080/02681219880000441. [DOI] [PubMed] [Google Scholar]
- 10.Loeffler J, Hebart H, Bialek R, Hagmeyer L, Schmidt D, Serey F, Hartmann M, Eucker J, Einsele H. Contaminations occuring in fungal PCR assay. J Clin Microbiol. 1999;37:1200–1202. doi: 10.1128/jcm.37.4.1200-1202.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeffler J, Henke N, Hebart H, Schmidt D, Hagmeyer L, Schumacher U, Einsele H. Quantification of fungal DNA by using fluorescence resonance energy transfer and the LightCycler system. J Clin Microbiol. 2000;38:586–590. doi: 10.1128/jcm.38.2.586-590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallet F, Oriol G, Mary C, Verrier B, Mandrand B. Continuous RT-PCR using AMV-RT and Taq DNA polymerase: characterization and comparison to uncoupled procedures. BioTechniques. 1995;18:678–687. [PubMed] [Google Scholar]
- 13.Randhawa P S, Manez R, Frye B, Ehrlich G D. Circulating immediate-early mRNA in patients with cytomegalovirus infection after solid organ transplantation. J Infect Dis. 1994;170:1264–1267. doi: 10.1093/infdis/170.5.1264. [DOI] [PubMed] [Google Scholar]
- 14.Shepard R N, Schock J, Robertson K, Shugars D C, Dyer J, Vernazza P, Hall C, Cohen M S, Fiscus S A. Quantitation of human immunodeficiency virus type 1 RNA in different biological compartments. J Clin Microbiol. 2000;38:1414–1418. doi: 10.1128/jcm.38.4.1414-1418.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpkins S A, Chan A B, Hays J, Pöpping B, Cook N. An RNA transcription based amplification technique (NASBA) for the detection of viable Salmonella enterica. Lett Appl Microbiol. 2000;30:75–79. doi: 10.1046/j.1472-765x.2000.00670.x. [DOI] [PubMed] [Google Scholar]
- 16.van Burik J-A, Myerson D, Schreckhise R W, Bowden R A. Panfungal PCR assay for detection of fungal infection in human blood specimens. J Clin Microbiol. 1998;36:1169–1175. doi: 10.1128/jcm.36.5.1169-1175.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Deursen P B, Gunther A W, van Riel C C, van der Eijnden M M, Vos H L, van Gemen B, van Strijp D A, Tackent N M, Bertina R M. A novel quantitative multiplex NASBA method: application to measuring tissue factor and CD14 mRNA levels in human monocytes. Nucleic Acids Res. 1999;27:e15. doi: 10.1093/nar/27.17.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Deventer A J, Goessens W H, van Belkum A, van Vliet H J, van Etten E W, Verbrugh H A. Improved detection of Candida albicans by PCR in blood of neutropenic mice with systemic candidiasis. J Clin Microbiol. 1995;33:625–628. doi: 10.1128/jcm.33.3.625-628.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Widjojoatmodjo M K, Borst A, Schukkink R A, Box A T, Tacken N M, van Gemen B, Verhoef J, Top B, Fluit A C. Nucleic acid sequence-based amplification (NASBA) detection of medically important Candida species. J Microbiol Methods. 1999;38:81–90. doi: 10.1016/s0167-7012(99)00079-2. [DOI] [PubMed] [Google Scholar]