Abstract
Objective
To describe and discuss the progression of the non-coding RNA as biomarkers in early esophageal cancer.
Background
Esophageal cancer without obvious symptoms during early stages is one of the most common cancers, the current clinical treatments offer possibilities of a cure, but the survival rates and the prognoses remain poor, it is a serious threat to human life and health. Most patients are usually diagnosed during terminal stages due to low sensitivity of esophageal cancer’s early detection techniques. With the development of molecular biology, an increasing number of non-coding RNAs are found to be associated with the occurrence, development, and prognosis of esophageal cancer. Some of these have begun to be used in clinics and laboratories for diagnosis, treatment, and prognosis, with the goal of reducing mortality.
Methods
The information for this paper was collected from a variety of sources, including a search of the keynote’s references, a search for texts in college libraries, and discussions with experts in the field of esophageal cancer clinical treatment.
Conclusions
Non-coding RNA does play a regulatory role in the development of esophageal cancer, which can predict the occurrence or prognosis of tumors, and become a new class of tumor markers and therapeutic targets in clinical applications. In this review, we survey the recent developments in the incidence, diagnosis, and treatment of esophageal cancer, especially with new research progresses on non-coding RNA biomarkers in detail, and discuss its potential clinical applications.
Keywords: Esophageal cancer, clinical treatment, diagnosis, non-coding RNA
Introduction
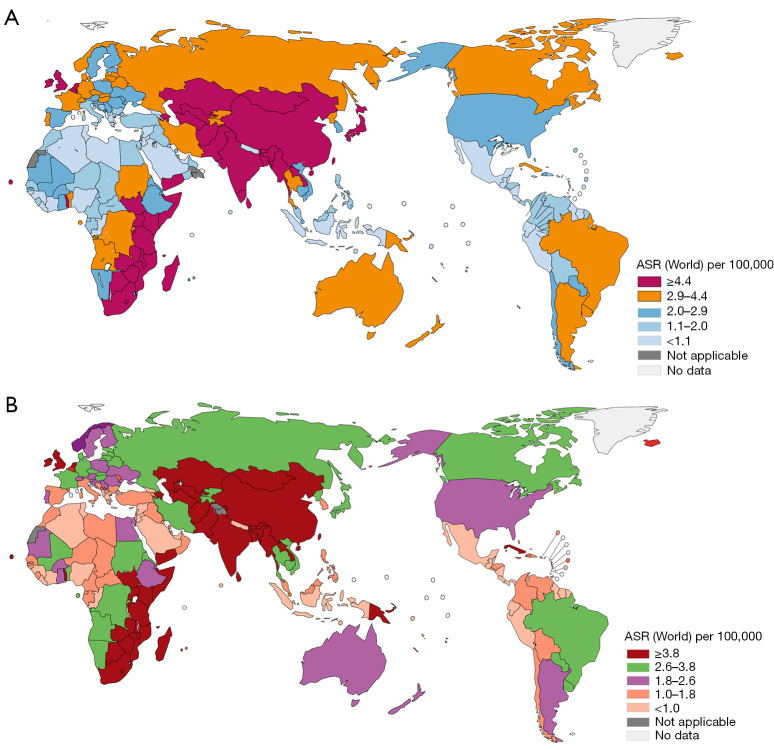
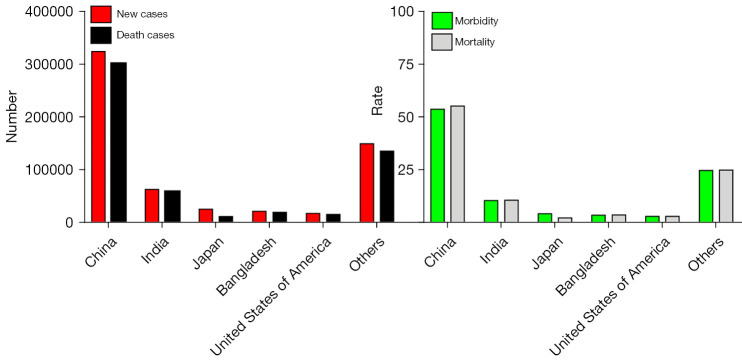
As one of the most common gastrointestinal tumors, esophageal cancer is a primary malignant tumor that can develop anywhere along the esophagus, originating from the esophageal epithelial cells. With the improvement in human living standards and hence life expectancy, esophageal cancer has shown an alarming upward trend in both morbidity and mortality and has even become endemic in many parts of the world, especially in developing countries. EC often has two pathologic types: squamous carcinoma (ESCC) and adenocarcinoma (EAC), with mainly common adenocarcinoma in the west, while China is mainly squamous carcinoma. According to the GLOBOCAN2020 database (http://gco.iarc.fr/today), 604,100 new cases of esophageal cancer were reported worldwide, accounting for 3.1% of all cancer cases and ranking the eighth, and the number of deaths reached 544,076, accounting for 5.5%, ranking the sixth in 2020 alone. Also, the ratio of deaths caused by esophageal cancer to all cancer death cases is 1.7 times that of esophageal cancer cases to all cancer new cases, indicating its poor treatment effect. Esophageal cancer is one of the malignant tumors with the highest incidence variation among all cancers, and the prevalence ratio in high-risk areas is 37 times higher than that in low-risk areas, in China in the year 2020 (Figure 1). The incidence and death rate of oesophageal cancer was 13.8 per 100,000 person-years and 12.7 per 100,000 person-years, and the 5-year prevalence was 24.04 per 100,000. While in Europe, the incidence and death rate is less than in China, 3.3 and 2.7 respectively; The number of new cases of esophageal cancer reached 324,000, and the global burden rate reached 54 percent; the number of death cases reached 301,000, accounting for half of worldwide, so the highest incidence of esophageal cancer is in China (Figure 2), and researches show that it is highly regionalized, especially high in north China regions, and higher in rural areas than urban areas, forming a typical epidemiological characteristic (1). The morbidity and mortality rates of males are higher than those of females, and the standardized mortality rates of both sexes are higher than the global level in recent years (2).
Figure 1.
Age-standardized incidence and mortality rates (world) in 2020. (A) Age-standardized incidence rates (world) in 2020, esophagus, both sexes, all age, ASR (World) per100,000; (B) Age-standardized mortality rates (world) in 2020, esophagus, both sexes, all age, ASR (World) per100,000. Note: all data cite from GLOBOCAN2020 database (http://gco.iarc.fr/today).
Figure 2.
Estimated number of new and death cases of high-incidence countries in 2020, both sexes and all ages. Note: data cite from GLOBOCAN2020 database (IARC website, http://gco.iarc.fr/today).
The sharp increase in the incidence and mortality of esophageal cancer indicates that the prevention, diagnosis, treatment, and prognosis of esophageal cancer are unsatisfactory. Different pathogenic sites lead to a big difference in incidence, therefore the onset of esophageal cancer is regarded as a complex process of multi-factor actions, multi-gene controls, and gradual changes (3). Nowadays, it is universally acknowledged that esophageal cancer is related to poor dietary habits, such as eating hot food (4), smoking (5), and ingesting N-nitrosamine compounds (6), which gradually lead to changes in an organism’s genetic materials and metabolism, and the cells and tissues grow out of control consequently, resulting in the initiation and progression of esophageal cancer. Recently it also has been reported that esophageal cancer has the possibility of inheritance (7), and the increase of aging population is a main driving factor for the increase in esophageal cancer cases, implying weakened immunity with aging may be the underlying cause of esophageal cancer.
In this review, we begin by discussing the current state of clinical treatment of esophageal cancer, followed by an overview of the recent developments in the incidence, diagnosis, and treatment of esophageal cancer, especially with new research progresses on non-coding RNA biomarkers in detail.
We present the following article in accordance with the Narrative Review reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-687).
Methods
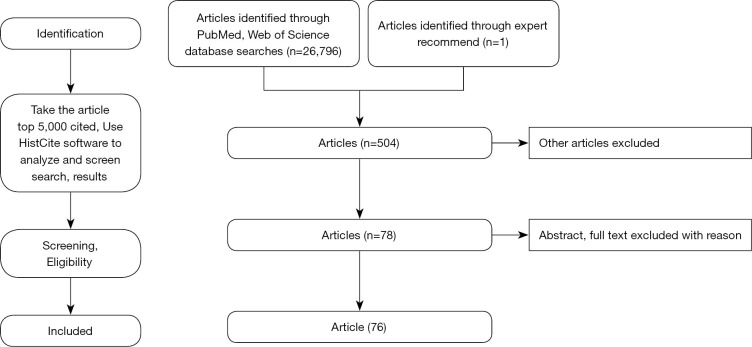
Information used to write this paper was collected by handing searches of the references of retrieved literature, using personal and college libraries to search for texts on keywords, and discussing with experts in the field of clinical and research of EC (Figure 3). For example, using PubMed and Web of Science, we performed relative articles, searching terms: ((((Esophageal cancer) AND (LncRNA)) OR (micro RNA)) OR (circle RNA)) OR (Non-coding RNA). Articles selected were required to be original articles published in English between 2021 and 2011.The article examines studies that contain the latest clinical practices for treating EC in the past 10 years. It should also be noted that the focus of this article is only on research articles, the publication types (in vitro, in vivo and, in situ) is not limited, and news articles, editorials, and unpublished works were excluded.
Figure 3.
Flowchart of article selection process.
Clinical treatment
Esophageal cancer is relatively obscure and lacks specific clinical features in its early stage, therefore when most patients suffer from the symptoms of gradual dysphagia and unconscious weight loss and seek medical help, they are usually in the middle or late stage, missing the best treatment period. At that time, it is harder to get curative treatment, even if they wanted to. And it is of a high probability of recurrences, so they are often advised to choose alternative therapies such as palliative care to control tumor-related clinical symptoms and prolong survival (8-10). As a result, when patients are diagnosed with esophageal cancer, they usually already missed the best chance of cure, which is the fundamental reason for the low overall survival rate and high mortality rate of esophageal cancer patients. Now the treatment strategies for EC are based on TNM-related tumor staging (10).
In the 1960s, Professor Qiong Shen invented the esophageal trawl method and established the diagnosis of esophageal balloon cytology to identify respectable early cancers and precursor lesions, a deflated balloon covered with silk net or cotton is swallowed in the stomach, inflated, and when collected scraping the surface of the esophagus endometrium, smearing on a microscope slide, then fixing the spiders with ethanol, staining with Pap staining, and the cells are read Abnormal signs, which is one of the historic events in the early diagnosis of esophageal cancer (11,12). In the 1980s, endoscopy and iodine staining techniques were widely used in the clinical diagnosis of esophageal cancer, which improved the diagnostic efficiency of early-stage esophageal cancer (13,14). Nowadays, with the continuous development of medical science and technology, more diversified technologies are applied in the early diagnosis of esophageal cancer, such as image logical examinations, pathological examinations, biomarkers, etc. but each has its shortcomings. Imageological examination mainly includes X-ray and CT examination. Studies have shown that CT examination has a higher diagnosis rate than X-rays in the diagnosis of esophageal cancer, avoiding misdiagnosis (15), though, in early-stage esophageal cancer, CT examination cannot detect the situation of the nidus. For the middle or late-stage esophageal cancer, CT examination can not only show the relationship between the nidus and the surrounding structure, reveal enlarged lymph nodes, but also find the metastasis of distant organs (16). Now imaging diagnosis is usually confirmed by endoscopy and confirmed histopathologically by biopsy, as the “gold standard” for the diagnosis of esophageal cancer. However, the “gold standard” is not 100% accurate due to the requirements of pathological examination on the skill and experience of pathologists and surgeons, and the development of endoscopic technology, especially whether the lesion can be accurately obtained when the lesions are not typical (10). With the rapid development of biomarkers, a new inspection technology emerged (17,18), using the joint detection of tumor markers in the diagnosis of esophageal cancer has high diagnostic efficiency, such as carcinoembryonic antigen (CEA), alpha-fetoprotein (AFP), carbohydrate antigen 199 (CA-199) and carbohydrate antigen 72-4 (CA72-4). But due to the low specificity and sensitivity of tumor markers, it cannot satisfy the diagnosis needs of all kinds of early esophageal cancers (19,20). It is now found that the diagnosis rate of gastroscopy for middle and terminal stage esophageal cancer is as high as 100% and China has included gastroscopy as one of the routine physical examination items, but the diagnosis rate of gastroscopy for early-stage esophageal cancer is not as precise as that of terminal stage esophageal cancer., The accuracy rate is only about 50% to 60%, thus how to reduce mortality by early-stage esophageal cancer screening is still the focus of future research directions (21).
Currently, the clinical treatments for patients with middle and terminal esophageal cancer mainly are surgical treatment, radiotherapy, and chemotherapy, among which surgical treatment is the main treatment, and radiotherapy and chemotherapy are adjuvant treatments. In 1913, the first esophagectomy for esophageal cancer was successfully performed in the world, which laid the foundation for surgical treatment of esophageal cancer. Nowadays for early and terminal stage esophageal cancer with distant metastasis, surgery is still the first choice; for stage I and II esophageal cancer, surgical resection is the only possible way to remove the tumor (22,23), but it may have side effects (23,24), such as intraoperative trauma, complication (pulmonary complications and anastomotic fistula, etc. (24), survival problem (such as difficulty in feeding and nutrition, and the long-term survival rate is low) as well as tumor recurrence and metastasis (25). Theoretically, chemo-radiotherapy and other adjuvant therapies can eliminate residual lesion tissues and lesion tissues caused by tumor cell metastasis in vivo, thus reducing local recurrence and improving overall survival rate (OS), but there are still problems in radiotherapy sensitivity and resistance to chemotherapy drugs (26). Along with the continuous renewal of medical pieces of equipment and the improvement of medical technology, minimally invasive surgery (27), photodynamic therapy (28), thoracoscope technology (29) are also applied in the treatment of esophageal cancer, but still exist problems, such as low survival rate and high recurrence rate. Because esophageal cancer is relatively obscure and lacks specific clinical features in its early stage, most patients who experience gradual dysphagia and unconscious weight loss seek medical help when they are usually in the middle or late stages, missing out on the best treatment options. At that time, it is harder to get curative treatment, even if they wanted to. In addition, because there is a high probability of recurrence, patients are often advised to choose alternative therapies such as palliative care to control tumor-related clinical symptoms and prolong survival (8-10). As a result, when patients are diagnosed with esophageal cancer, they usually already missed the best chance of cure, which is the fundamental reason for the low overall survival rate and high mortality rate of esophageal cancer patients.
The current clinical treatment for patients with effect is not ideal enough, it still needs further development and progress (Table 1).
Table 1. The Potential algorithm of therapy for extended locally advanced esophageal cancer and salvage treatment.
| Early esophageal cancer | Extended esophageal cancer | Locally advanced esophageal cancer | Salvage treatment for local failure of esophageal cancer | |
|---|---|---|---|---|
| Algorithm of therapy | esophagectomy (10) | nimotuzumab combined with chemotherapy (30) | DCF therapy with prophylactic administration of pegfilgrastim (31) | Salvage surgery (32) |
| ESD (miR-205 levels may serve as a marker) (33,34) | HiPorfin Photodynamic Therapy (35) | targeted therapy: Gefitinib (36): lapatinib combined with paclitaxel (37) | ER (38) | |
| EMR (remove lesions only smaller than 2 cm en bloc) (39,40) | combined immunotherapy and chemotherapy (41) | CRT (37) | repeated tPDT (42) |
DCF, Docetaxel, cisplatin, and 5-FU; ER, endoscopic resection; CRT, Chemoradiotherapy; tPDT, talaporfin sodium photodynamic therapy; ESD, endoscopic submucosal dissection; EMR, endoscopic mucosal resection.
Previous studies on the genomics of esophageal cancer
Cancer is the disease with the highest mortality rate. It originates from the disorder of related genes. The main mechanisms include insertion of enhancers or promoters, reduction of GTPase activity, structural mutations or deletions of key genes, etc. (43-49). However, this may not necessarily cause malignant transformation of cells during part of the tumor formation process. It must be caused by the combination of genes or other factors that affect cell proliferation and apoptosis (48,50). Esophageal cancer-related studies have proved that esophageal cancer is a complex disease. Its occurrence, development, metastasis, and other processes are caused by multifactorial abnormalities such as carcinogen metabolism activation and inactivation, cell proliferation, and apoptosis, and are accompanied by damage repair Multi-gene mutations such as related genes and regulatory genes provide a strategy for studying the genetics and epigenetics of different cancers (51-53). In the past semi century, researchers worldwide on the pathogenesis of esophageal cancer have made a series of achievements in genes, protein, and metabolic levels (54), but the exact pathogenesis of esophageal cancer is still unknown and still under active research and exploration. At the genomics level, the International Tumor Genome Collaboration Consortium (ICGC) was formally established and launched a global cancer genome research work in 2007, which conducts tumor molecular-level researches from the variation of DNA sequences and maps somatic gene mutation profiles. Till now, many research teams from different countries have summarized data on specific cancers and reported their research results including esophageal cancer. There are researches on the gene-level systematic analysis of esophageal cancer, such as genetic alterations, over-expression of molecules thought to cause malignant transformation, related single nucleotide variations (SNP) and other genetic variations, and exploring driver genes and corresponding signal regulatory networks, Genomics studies of esophageal cancer have found that gene expression disorders are expected to serve as a marker for early diagnosis of esophageal cancer (55,56), and DNA methylation may also indicate the occurrence of tumors, such as EPB41L3 (57), STK3 (58) promoter AKAP12 (59) and so on, which have been proved to be used as a biomarker for the early diagnosis of esophageal cancer.
Non-coding RNA and esophageal cancer
After the completion of the Human Genome Sequencing Project, it is confirmed that only about 2% of the genes in the entire transcriptome can be transcribed and translated into proteins, more than 90% of the remaining cannot be used to encode proteins but can be transcribed into RNAs. However, then the ENCODE research project has discovered that 75% of the genome sequence could be transcribed into RNA, among which nearly 74% of the transcripts are non-coding RNAs (ncRNA, non-coding RNA). In follow-up researches, non-coding RNAs have been gradually confirmed to play a pivotal role in the life activities of humans and many other species, regulating different levels of gene expression (60-62). Till now, the types of non-coding RNAs with regulatory effects mainly include long non-coding RNA (lncRNA), microRNA (miRNA) (63), and novel endogenous non-coding RNA - circular RNA (circRNA) (64).
There conclude some methods for the detection and verification of non-coding RNA biomarkers for early diagnosis of esophageal cancer (Figure 4) (65-71) such as genome-wide profiling, RNA-seq. After rigorous sequencing technology, the differential expression between the groups is compared, and then the bioinformatics correlation analysis is performed to obtain the effective and desired target RNA, and the experimental verification and value evaluation at different levels are carried out. RNA-seq and microarray are the two popular methods employed for genome-wide transcript profiling. Among the several methods, RNA-seq and microarray stand out as the two widely used genome-wide gene expression quantification methods (72-75), RNA-Seq is the direct sequencing of transcripts by high-throughput sequencing technologies, microarray is using the principle of molecular hybridization, the compared specimen (labeled) is hybridized with the microarray, and the intensity of the hybridization signal and data processing are detected to convert it into a specific gene abundance so that the difference in gene expression level of the different specimens is comprehensively compared, they have their own advantages and disadvantages (Table 2) (76-83). Leveraging RNA-seq for daily diagnostic activities is no longer a dream but a consolidated reality, it is a powerful technology that is predicted to replace microarrays for transcriptome profiling (84), although established best practices exist. Managing RNA-seq data is not easy, such as plan library preparation, budget optimization, accuracy of downstream sequencing, data assembly, and comparison, etc. it still faces many challenges (85-87).
Figure 4.
Common methods for the detection of non-coding RNA biomarkers.
Table 2. Comparison of the advantages and disadvantages of RNA-seq and microarray.
| RNA-seq | Microarray | |
|---|---|---|
| Afford | Company | Laboratory, company |
| Technology limitation | Short sequence reads, not map uniquely to a single gene or isoform (mapping uncertainty), | Background hybridization, cross-hybridization, specific hybridization, probe design |
| Testing dynamic range | Wider (low to extremely abundant, more sensitive, accurate) | Cannot distinguish ‘‘no’’ from ‘‘low’’ expression. |
| Data analysis | Far from mature, vulnerable to general biases and errors, the absence of a gold standard for analysis | Relatively simple, faster |
| Cost | Expensive | Cheap |
| Advantage | Can discover unknown transcripts and rare transcripts, higher detection throughput, highly reproducible, with relatively little technical variation | Can provide highly consistent data, the analysis software is quite mature, and give clinical diagnostic value |
With the development of high-throughput sequencing technology, researches on non-coding RNAs of esophageal cancer have been deepened, and researchers have found several biomarkers for the diagnosis, therapeutic effect evaluation, and prognosis judgment of esophageal cancer.
LncRNA and esophageal cancer
LncRNA is a class of non-coding RNA that regulates gene expression with a length of more than 200 nucleotides and is characterized by its huge amount, various action patterns, conservation in evolution, and several broad categories, including sense, antisense, bidirectional, intronic, and intergenic, all of which have a certain association with cancer (88,89). Researches of recent years have found that lncRNAs not only participates in a variety of biological activities of gene level, chromosome level, and material transportation aspect but also play a more extensive, elaborate, and complex regulatory role than microRNAs in cell differentiation and development, gene expression, heredity and so on (90-93). Various lncRNAs have been proved to promote or inhibit tumor formation and transfer (94,95). Also, a variety of researches confirmed that many lncRNAs in esophageal cancer are differentially expressed and performing important regulation functions (96-99). These differentially expressed lncRNAs have been studied to further reveal their biological functions and molecular mechanisms, and have shown their potential to be biomarkers of esophageal cancer or potential therapeutic targets for esophageal cancer (Table 3): Using the method of real-time fluorescent quantitative PCR (RT-qPCR), it was found that LINC00152 is highly expressed in esophageal cancer, and LINC00152/EZH2/ZEB1 axis could regulate the Epithelial-to-mesenchymal transition (EMT) of esophageal cancer cells and resistance of EC cells to oxaliplatin (L-OHP), thus could present a potential diagnostic marker and therapeutic target for the treatment of esophageal cancer (100,101). Another lncRNA HOTTIP, which was first discovered in a case of gastric cancer, recently has been found to be expressed significantly higher in esophageal cancer areas than in normal tissues, which upregulated snail1 by competitively binding miR-30b and subsequently promoted epithelial-mesenchymal transformation (EMT). In addition, the overall survival rate of esophageal cancer patients with high expression of lncRNA HOTTIP is significantly lower than that with low expression, thus Hottip-miR-30b-HOXA13 axis may be a potential diagnostic marker or drug target for ESCC therapy and HOTTIP could be used as a molecular marker for the prognosis of esophageal cancer patients (102). Another highly expressed lncRNA FOXD2-AS1 participates in the regulation of miR-145-5p/CDK6 axis, and significantly inhibits proliferation and invasion of esophageal cancer, thus has functions of clinically determining markers and therapeutic targets for lymph node metastasis (103,104). LncRNA HOTAIR, first discovered in breast cancer and highly expressed in esophageal squamous cell carcinoma, has the ability to inhibit tumor cell apoptosis and promote tumor metastasis because it finds that HOTAIR directly decreased WIF-1 expression by promoting its histone H3K27 methylation in the promoter region and then activated the Wnt / β-catenin signaling pathway, and now HOTAIR has been reported as an indicator of survival and a prognostic tumor marker for patients (97,105-107). PVT1, whose expression is significantly higher in esophageal squamous cell carcinoma tissues than in paracancerous tissues, promotes tumor progression by competitively binding with miRNA-203 to regulate the expression of human recombinant LIM and SH3 protein1 (LASP1) and is often used in clinical practice to predict the overall prognosis of ESCC patients (108). LncNAAK001796, which regulates the MDM2/p53 signaling pathway, not only is highly expressed in esophageal squamous cell carcinoma but also could inhibit cell cycle and tumor growth after being knocked out, therefore can be used as an adverse prognostic factor to determine the prognostic quality (109). MEG3, lowly expressed in esophageal cancer, shown in studies that its mechanism of action is as a sponge of miRNA-4261 to inhibit recombinant human Dickkopf-related protein 2 (DKK2) thus block the Wnt/β-catenin (β-catenin) signaling pathway (110), has its role of predicting tumor progression and as a prognostic marker, LncRNA-TUSC7, significantly down-regulated in ESCC tissues and could significantly inhibit the migration, chemotherapy resistance and invasion ability of esophageal cancer cells through the expression of related proteins, can be used as a molecular marker for the diagnosis and treatment of esophageal squamous cell carcinoma and metastasis monitoring (111). In addition to the above signaling pathways, the involvement of lncRNAs has also been found in EGFR/SATA1, RAS/MARK pathways, and so on (112). Among them, the highly expressed LINC00152 discovered in the study was collected from 76 patients with disease tissues, and RT-PCR was used to prove that its expression was also up-regulated. Then, through related experiments on cell lines and mouse models, the molecular marker function was further confirmed, so it is speculated that it is possible to further advance clinical research. Therefore, the role of lncRNA in esophageal cancer is incomparable, and it is of great importance to study it.
Table 3. List of common lncRNA biomarkers for esophageal cancer.
| Name | Expression | Mechanism | Biomarker potency | Site of detection | Detection method | Reference |
|---|---|---|---|---|---|---|
| LINC00152 | ↑ (100) | Interacts with EZH2, positively regulate the expression of ZEB1, enhance the EMT of EC cells (101) | (i), (iii) | Whole blood (100), Animal (100,101) (a, b) | RT-qPCR, Tumor formation study, | (100,101) |
| HOTTIP | ↑ | Up-regulated snail1 by competitively binding miR-30b, regulate the Hottip-miR-30b-HOXA13 axis in ESCC | (i), (ii), (iii) | Tumor, cell line (a) | RT-qPCR, Dual-luciferase reporter assay | (102) |
| FOXD2-AS1 | ↑ (103) | Regulate miR-145-5p/CDK6 axis, significantly inhibit the proliferation and invasion of esophageal cancer cells (104) | (i), (iii), (iv) (103,104) | Tumor (a) | RT-qPCR, WB | (103,104) |
| HOTAIR | ↑ (97) | HOTAIR decreased WIF-1 expression, regulate HOTAIR/WIF-1 axis (105) | (ii) (106,107) | Tumor, serum (a) | Microarray, RT-qPCR | (97,105-107) |
| PVT1 | ↑ | Competitively bind miRNA- 203, regulate the expression of human recombinant LIM and SH3 protein 1 in ESCC | (ii) | tumor, mice (a, b) | RT-qPCR, Immunohistochemical, Tumor formation study, | (108) |
| AK001796 | ↑ | Regulate the MDM2/P53 signal pathway in ESCC | (ii) | Tumor, mice (a, b) | RT-qPCR, RT-PCR, Tumor formation study, | (109) |
| MEG3 | ↓ | Interact with MI R-4261, inhabit recombinant DKK2 and block Wnt/β-catenin signaling, inhibit the occurrence and development of ESCC | (i), (ii) | Tumor, mice (a, b) | RT-qPCR, Luciferase reporter assay, WB, Tumor formation study, | (110) |
| LncRNA-TUSC7 | ↓ | bound to miR-224, miR-224 specifically bound to DESC1, DESC1 inhibited chemotherapy resistance of ESCC cells via EGFR/AKT | (i), (iii), (iv) | Tumor (a) | RT-qPCR, bioinformatics | (111) |
↑, represents up-regulated gene expression in tumor tissue; ↓, represents down-regulated gene expression in tumor tissue; (i), diagnostic; (ii), prognostic; (iii), therapeutic; (iv), metastasis monitoring. RT-qPCR, real-time quantitative PCR; WB, western blot; RT-PCR, reverse transcription PCR; ESCC, esophageal squamous cell cancer.
MicroRNA and esophageal cancer
In recent years, cancer has become one of the most serious threats to human life and health, and miRNAs play an important role in cancer. It was first confirmed by Calin et al. in 2004 using human precursor cells and the microarray of mature miRNA oligonucleotides probes, reporting the genome-wide expression profile of miRNA in human B-cell chronic lymphocytic leukemia (CLL) and confirming that the differential expression of genes is related to Zap-70, and these data indicate that miRNA expression patterns will affect leukemia biology and clinical behavior (113). Since then, along with the rapid development of molecular biology, studies on the relationship between miRNAs and tumors have gradually become clearer, and more miRNAs have been proved to play an important role in the forming of tumors. Due to these studies, the biological functions of the miRNAs have gradually become clearer. Multiple data have proved that some specific miRNA expression will get increased or decreased between the tissues getting cervical cancer (114), prostate cancer (115), gastric cancer (116), esophageal cancer (117), adjacent tissues, and abnormal expression also occurs when genetic or epigenetic changes occur (118). After in-depth research on the relationship between miRNA and tumors, it is found that these miRNAs have different functions, which can be divided into two categories: miRNAs up-regulated in tumor cells and miRNAs down-regulated in tumor cells. MicroRNAs highly expressed in tumor tissues stimulate the over-expression of oncogenes while those whose expression is down-regulated are reduced or even silenced in tumor cells (119), thereby affecting the occurrence, development, invasion, and metastasis of cancer. Some scholars even found that the expression level of some miRNAs also affects the overall survival rate of patients with esophageal cancer when studying the relationship between esophageal cancer and miRNA (120-122), such as miRNA221 (123), mi RNA23a (124), miRNA193-3p (125), etc. This also shows that miRNAs can be used to assess the overall survival of the esophageal cancer patients. Compared with the traditional tumor markers, with low specificity and weak sensitivity, miRNAs are evolutionarily conserved in the human body, widely distributed in easily accessible body fluids, and have high stability. Therefore, miRNAs are a new type of diagnostic marker with great potential. As diagnostic markers, miRNAs are also used in various aspects of clinical treatments of esophageal cancer (Table 4): For example, the most classic small-molecule RNA miRNA21 used in the treatment of various tumors has also been found with higher expression in esophageal cancer tissues than normal tissues. It has been experimentally verified that miRNA-21 in esophageal cancer will target the key protein of the PTEN /PI3K/AKT signaling pathway, therefore, affects the proliferation and apoptosis of tumor cells and other life activities, thus miRNA-21 can be used at the diagnosis, treatment, and prognosis of esophageal cancer (126-128); Highly expressed miRNA miR-18a, similar to miR-21, regulates the PTEN-PI3K-AKT-mTOR signal axis, increases the expression of cyclin D1 and promotes the proliferation of esophageal squamous cell carcinoma cells, and shown in studies that it can be used not only as a prognostic indicator of esophageal cancer but also as a diagnostic biomarker (collaborate with miR-21), will get higher sensitivity and better effect (129-131); Jing et al. by getting microRNA chip via RT-q PCR assay demonstrated that miR-17/20a overexpression can inhibit Tβ-β / integrin β6 subunit pathway by targeting TβR2 and Smad anchor receptor activation and degradation. Therefore, miR-17/20a can be used as diagnostic and prognostic markers (132). MAP3K1 was a direct target of miR-203, which was first discovered in breast cancer and whose expression level is much higher in esophageal cancer tissues than in adjacent tissues. Overexpression of MAP3K1 can eliminate the inhibition of miR-203 on cell proliferation and invasion, MiR-203 is clinically used to determine the TNM stage, pathological differentiation, and lymph nodes of esophageal cancer metastasis and is used as a prognostic marker (133-135). MiR-1290, which is highly expressed in esophageal cancer and has been demonstrated to operate as a tumor oncogene in the progression of ESCC by targeting NFIX, has also been used as a prognostic marker (136-138). Except for the fact that up-regulated miRNAs play an important regulatory role in esophageal cancer, some down-regulated miRNAs, such as the recently discovered miR-145, also play a role in cell proliferation, migration, or invasion, making it a biomarker, treatment target, and prognostic indicator for esophageal cancer (139-141); miR-143 and miR-122, both found differentially expressed in cancer cells and normal tissues, have also been found to be used as therapeutic targets for esophageal cancer (142-144), and miR-133b, with low expression in ESCC, can inhibit the MAPK/ERK and PI3K/AKT pathways (common tumor signaling pathways) by down-regulating the expression of EGFR to resist tumor regulatory role, so can be used for the detection and prognosis of early esophageal cancer (145,146). Using SYBR green real-time quantitative reverse transcription-polymerase chain reaction, the high expression of miR-18a in 105 surgical specimens from ESCC patients was detected, and The miR-18a expression positively correlated with tumor stage, then through further experiments, it was found that comparing traditional esophageal tumor markers, serum miR-18a, and miR-21 are more sensitive to the diagnosis of esophageal cancer, and the combination of traditional esophageal tumor markers can further improve the sensitivity and accuracy of esophageal cancer diagnosis. Therefore, it is speculated that the new miR biomarkers can be combined with each other or even combined with traditional tumor markers to achieve better results.
Table 4. List of common miRNA biomarkers for esophageal cancer.
| Name | Expression | Mechanism | Biomarker potency | Study setting | Detection method | Reference |
|---|---|---|---|---|---|---|
| miR-21 | ↑ (126,127) | Target the key proteins of the PTEN/PI3K/AKT signal pathway (128) | (i), (ii), (iii) (126-128) | Serum (126), tumor (127,128) (a) | RT-qPCR (128), Affymetrix GeneChip miRNA 1.0 Array (127) | (126-128) |
| miR-18a | ↑ (129) | Regulate the PTEN-PI3K-AKT-mTOR signal axis, incerase the expression of cylin D1 in ESCC (130) | (i), (ii) (collaborate with miR-21 will get higher sensitivity and better effect) (129,131) | Serum (129), tumor (129-131) (a, b) | RT-PCR (129), RT-qPCR (130,131), IHC (131), WB (131), Tumor formation study | (129-131) |
| miR-17/20a | ↑ | Inhibit Tβ-β / integrin β6 subunit pathway by targeting TβR2 and Smad anchor receptor activation and degradation in ESCC | (i), (ii) | cell lines, mice (a, b) | qPCR, IHC, Tumor formation study, | (132) |
| mi R-203 | ↑ (133) | MiR-203 targets MAP3K1, thereby inhibiting cell proliferation and invasion (134) | (ii), (iv) (135) | Tumor (a) | RT-qPCR (133), WB (134), Dual-luciferase reporter assay (134) | (133-135) |
| miR-1290 | ↑ (136) | Target NFIX (137) | (iv) (138) | Serum (a) | miRNA microarray | (136-138) |
| mi R-145 | ↓ (139) | Target SMAD5, promote cell proliferation and migration/invasion (140) | (ii), (iii) (141) | Tumor, cell, mice (a, b) | RT-qPCR, WB, IHC, Luciferase reporter assay | (139-141) |
| miR-143 | ↓ (142) | MAPK7 pathway (143) | (iii) | Tumor (a) | RT-qPCR | (142,143) |
| miR-122 | ↓ | Target and down-regulate PKM2 to inhibit the proliferation of esophageal carcinoma cells | (iii) | Cell (a) | Bioinformatics analysis, RT-qPCR, WB | (144) |
| miR-133b | ↓ (145) | Down-regulate the expression of EGFR in ESCC to inhibit MAPK/ERK and PI3K/AKT pathway (146) | (i), (ii) | Tumor, cell, mice (a, b) | RT-qPCR (145), WB (146), Tumor formation study, | (145,146) |
↑, represents up-regulated gene expression in tumor tissue; ↓, represents down-regulated gene expression in tumor tissue; (i), diagnostic; (ii), prognostic; (iii), therapeutic; (iv), metastasis monitoring. RT-qPCR, real-time quantitative PCR; WB, western blot; RT-PCR, reverse transcription PCR; ESCC, esophageal squamous cell cancer.
Although the number of mi RNAs discovered already is not small, the current research on miRNAs is still at a relatively early stage, and the number of experimentally proven microRNAs with clear functions is only a drop in the ocean compared to the number of predicted microRNAs found by bioinformatics methods. Therefore, it is necessary to go deeper in future researches to screen out highly sensitive and specific microRNAs and their corresponding type of esophageal cancer to ensure that more accurate diagnosis and more valuable evaluation results can be provided in future clinical tests.
CircRNA and esophageal cancer
With the continuous development of Next-Generation Sequencing (NGS) technology, low cost, high speed, and high throughput, and the progress of corresponding matching algorithms, the application value of circular RNA (circRNA) in the clinical field of tumors has gradually emerged. Circular RNA, as a type of evolutionarily conservative ancient RNA that has the function of regulating gene expression in eukaryotic creatures, is a new kind of endogenous non-coding circular RNA (147). It forms a special circular double-stranded closed structure by a covalent bond with variable length and does not have the “cap” and “tail” characteristics of chain RNAs (148,149). Several studies have proved that circRNAs are widely present in a variety of biological cells (150-152). For example, in 1976, circRNA was first discovered in a viroid in a plant and then was found in animal cells and fungal yeast; so far, tens of thousands of of circRNAs have been discovered, and they can be divided into three types according to their constituent sequences and sources: circular intronic RNA (ciRNA) (153) and exonic circRNA (ecircRNA) (154) and exon-intron circRNA (ElciRNA) (155). And these three types of circular RNA molecules are universally acknowledged to be formed by two kinds of synthetic mechanisms: direct reverse splicing and lasso-driven cyclization (156,157). CircRNA is a kind of particular RNA molecule with special structures, which is not comparatively easy to be recognized and degraded by enzymes, and it is not completely dependent on the linear mRNA expression of parent genes. CircRNA is specifically expressed and plays an important role in cells, tissues, and ontogenetic stages, with strong stability and Spatio-temporal specificity (148,158,159). After the in-depth study of circular RNAs, it has been found that circRNAs are mainly involved in the regulation of biological processes at the transcriptional and post-transcriptional levels, and nowadays its universally acknowledged that the main mechanisms of circRNAs are involved in alternative splicing, the transcription process, adjusting the parent gene expression, acting as microRNA molecular “sponge”, regulating RNA-binding protein, participating in protein translation and so on (97,160-164), among which, molecular sponge action is the most classic mechanism of circRNAs. Molecular sponge action refers to the biological regulation process of miRNA inhibition and up-regulation of target gene expression when a circRNA has a specific site that could bind to a miRNA. Till now the mechanism has also been proved by a large number of experiments (154,165,166).
With the in-depth research on the structure, mechanism, and function of circRNAs, it has been found that circRNAs not only play an important biological function in physiological processes but also plays a crucial role in the occurrence and development of diseases, such as viral hepatitis (167), vascular diseases (168), central nervous system degenerative diseases (169,170), diabetes (171), various cancers (172-174), etc. With the progress of testing technologies and the improvement of medical care, and cancer is gradually found to be the disease with the highest mortality rate, people are paying more and more attention to the diagnose and treatments of cancer. Therefore, as circRNAs have been found in various cancers and have certain regulatory effects, the researches on RNAs in tumors has gradually been focused on the distribution of circRNAs in different kinds of tumors and their roles in tumor genesis and development, especially the molecular mechanism of how circRNAs regulate tumor cells proliferation, apoptosis, invasive movement, and angiogenesis. So far, various studies have identified the abnormal expression of a variety of circRNAs in different tumors, including esophageal cancer (Table 5), which preliminarily reveals the unusual clinical significance of circRNAs. For example, the expression of classical tumor-associated ciRS-7 in esophageal squamous cell carcinoma tumor tissues is significantly higher than that in normal tissues, in which ciRS-7 is supposed to act as a molecular sponge of miR-7 and miR-876-5p to enhance the antigen MAGE-A family expression. Therefore, ciRs-7 can be used as a potential target for diagnosis and treatment of esophageal cancer (175,176); CircHIPK3, one of the most thoroughly studied circRNA, is up-expressed in esophageal cancer tissues and can be used as a potential therapeutic target for esophageal cancer by regulating P53-Akt-Mdm2 signaling pathway to promote cell proliferation and inhibit apoptosis (177,178). So does circ-0006168, as a potential diagnostic biomarker and therapeutic target, and its mechanism of action is to target and regulate miR-100/ mTOR and miR-339-5p/CDC25A pathways to promote the progression of esophageal cancer (179); 20 cancer tissues cases and 105 control cases were detected by RNA sequencing technology and RT-PCR, getting the result that the expression of hsa_circ_0004771 was up-regulated, and the reason why it was of great value for the diagnosis and prognosis of malignant tumors is that it acted on the miR-339-5p/CDC25A pathway (180). When screening the differentially expressed genes of esophageal cancer patients and normal humans, it was found that there were also down-regulated circRNAs in the esophageal cancer group, which play an important role in the occurrence, development, invasion, and metastasis of esophageal cancer. circVRK1, as potential biomarkers and therapeutic targets for human malignant tumors, and its mechanism of action is that CircVRK1 inactivated PI3K/AKT signaling pathway by up-regulating PTEN and CircVRK1-miR-624-3p-PTEN pathway regulated ESCC progression (181). Circ-ITCH is not only poorly expressed in esophageal cancer, but also compete with the 3’untranslated region (3’UTR) of the messenger RNA of the homologous protein ITCH to bind with the miRNAs related to tumors such as miR-7, miR-17, miR-214, etc., thus reducing the inhibition and enhancing the expression of ITCH, then it plays a regulatory role through ITCH/DVL2/Wnt pathway, so Circ-ITCH is related to cancer formation and could be used to predict tumor progression (182,183); Circ-0043898, discovered in esophageal cancer and significantly down-regulated in esophageal cancer, plays a role by participating in the mechanism of protein synthesis. It has been found that circ-0043898 is involved in the synthesis of tumor proteoglycan, RAP1, and mTOR signaling pathways and has the potential to become a therapeutic target and one of the biomarkers for diagnosis of esophageal cancer (164,184); Also At the same time, high-throughput sequencing technology is commonly used to construct circRNA expression profiles in esophageal squamous cell carcinoma to analyze the differentially expressed circRNA, the highly differentially expressed circRNA obtained seem to be potential biomarkers for esophageal squamous cell carcinoma, such as hsa_circ_0007541, whose expression level in esophageal squamous cell carcinoma tissues significantly differs from adjacent tissues and has a regulatory effect on the progression of the tumor (185). Through the discovery and induction of known biomarkers of esophageal cancer circle RNA, we can see that circle RNA generally regulates cell signaling pathways by acting on miR. This mechanism of action has been obtained in both in vivo and in vitro experiments. It has been confirmed that, for example, up-regulated ciRS-7 and down-regulated circ-ITCH, their regulation mechanism is to up-regulate or down-regulate the expression of related proteins as the corresponding miR molecular sponges, thereby regulating cell proliferation or decay signaling pathways.
Table 5. List of common circRNA biomarkers for esophageal cancer.
| Name | Expression | Mechanism | Biomarker potency | Study setting | Detection method | Reference |
|---|---|---|---|---|---|---|
| ciRS-7 | ↑ (175) | As miR-876-5p and miR-7sponge, inhibit them to regulate antigen MAGE-A family in ESCC (175,176) | (i), (iii) (175) | Tumor (a) | RT-PCR | (175,176) |
| CircHIPK3 | ↑ | Regulate P53-Akt-Mdm2 signal pathway in ESCC | (iii) | Tumor (a) | RT-qPCR, Bioinformatics analysis, WB | (177,178) |
| circ-0006168 | ↑ | Target regulate miR-100/mTOR and miR-339-5p/CDC25A pathway | (i), (iii) | Tumor, cell (a) | RT-qPCR, Dual-Luciferase reporter system | (179) |
| hsa_circ0004771 | ↑ | Regulate miR-339-5p/CDC25A pathway in ESCC | (i), (ii) (low invasive biomarker) | plasma (a) | RT-qPCR | (180) |
| circVRK1 | ↓ | Regulate miR-624-3p/PTEN/PI3K/AKT signaling pathway | (iii) | Tumor, cell (a) | RT-qPCR | (181) |
| circ-ITCH | ↓ | Make miR-7, miR-17, miR-214 molecular sponges, enhancing the expression of ITCH to ubiquitin degrade phosphorylated DVL2 and thus inhibit the development of the Wnt pathway in ESCC | (iv) | Tumor, cell, mice (a, b) | RT-qPCR, Bioinformatics analysis, WB, Tumor formation study | (182,183) |
| circ-0043898 | ↓ | Participate in the synthesis of tumor proteoglycan and RAP1 and mTOR signaling pathways | (i), (iii) | Tumor, cell, mice (a, b) | RNA-seq, RT-PCR, GO, KEGG, Tumor formation study | (164,184) |
↑, represents up-regulated gene expression in tumor tissue, ↓, represents down-regulated gene expression in tumor tissue, (i) diagnostic, (ii) prognostic and (iii) therapeutic (iv) metastasis monitoring, a in-vitro, b, in-vivo, c, in-situ, RT-qPCR, real-time quantitative PCR, WB, western blot, RT-PCR, reverse transcription PCR, ESCC, esophageal squamous cell cancer.
Based on all the above, circular RNAs play an important role in the process of life and could be used as a biomarker for many diseases. However, compared with lncRNAs and miRNAs, the studies of the circular RNAs whose functions are experimentally identified in the researches are still in the preliminary stage. Identifying the specific mechanisms and functions of the circular RNAs is only the tip of an iceberg, it takes a lot of effort to further explore the mysteries of circular RNAs in future researches.
Conclusions
In recent years, with the development of medical technology and the deepening of tumor molecular biology research, tumor markers are helpful to the diagnosis, treatment, and prognosis of patients with esophageal cancer. We found that compared with mRNA, lncRNA and circRNA have higher tissue and Spatio-temporal specificity, and can play a regulatory role at the transcription and post-transcriptional levels in a variety of ways. They are the best candidates for early diagnosis and prognostic biomarkers of the disease. However, the current mechanism of esophageal cancer is not very clear. Some biomarkers are common markers of multiple cancers and have similar abnormal expressions, which are unique to non-esophageal cancer or even early esophageal cancer. Furthermore, many markers are still in experiments. At this stage, although it has been further verified on animal models and has certain reference value, it cannot be fully applied or suitable for human entity disease. It has certain limitations. Therefore, finding effective and specific markers is faced in the diagnosis of esophageal cancer. The main challenge is also an urgent and necessary task. In the future, the best marker profile of non-coding RNA is mainly based on RNA-seq technology, combined with large sample data, to screen and identify RNA molecules specifically expressed in disease tissues or patients, and guide early diagnosis and prognosis of diseases, such as Bioinformatics (186), microarray (187,188) and other methods (189). In addition, different factors such as esophageal cancer pathology, stage of development, age, and other factors can also lead to differential expression of non-coding RNA. Therefore, when developing and evaluating the best marker profile of non-coding RNA, it is necessary to increase the sample size as much as possible and carry out clinical classification. And from the above results, it can be seen that the use of a single marker for early diagnosis of esophageal cancer still has certain limitations. Therefore, in future studies, researchers not only need to pay more attention to the sensitivity and specificity of tumor markers but also explore the application of the combined detection of multiple markers to ensure the provision of more valuable diagnosis and evaluation results. It is also possible to use a combination of multi-omics techniques to study the mechanism of esophageal cancer occurrence, development, invasion and metastasis, so as to better guide the patients with esophageal cancer diagnosis, treatment, and prognosis!
Acknowledgments
We would like to thank Vivian and Feifei Zhu for their help in polishing our paper.
Funding: This work was supported by the National Natural Science Foundation of China [grant numbers [31861143051, 31872425].
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-687
Peer Review File: Available at https://dx.doi.org/10.21037/tcr-21-687
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-687). The authors have no conflicts of interest to declare.
References
- 1.Zuo TT, Zheng RS, Zeng HM, et al. Incidence and trend analysis of esophageal cancer in China. Zhonghua Zhong Liu Za Zhi 2016;38:703-8. [DOI] [PubMed] [Google Scholar]
- 2.Cai Z, Liu Q. Understanding the Global Cancer Statistics 2018: implications for cancer control. Sci China Life Sci 2021;64:1017-20. 10.1007/s11427-019-9816-1 [DOI] [PubMed] [Google Scholar]
- 3.Lu SH. Alterations of oncogenes and tumor suppressor genes in esophageal cancer in China. Mutat Res 2000;462:343-53. 10.1016/S1383-5742(00)00023-5 [DOI] [PubMed] [Google Scholar]
- 4.Mao WM, Zheng WH, Ling ZQ. Epidemiologic risk factors for esophageal cancer development. Asian Pac J Cancer Prev 2011;12:2461-6. [PubMed] [Google Scholar]
- 5.Gao YT, McLaughlin JK, Gridley G, et al. Risk factors for esophageal cancer in Shanghai, China. II. Role of diet and nutrients. Int J Cancer 1994;58:197-202. 10.1002/ijc.2910580209 [DOI] [PubMed] [Google Scholar]
- 6.Lu SH, Montesano R, Zhang MS, et al. Relevance of N-nitrosamines to esophageal cancer in China. J Cell Physiol Suppl 1986;4:51-8. [DOI] [PubMed] [Google Scholar]
- 7.Torre LA, Siegel RL, Ward EM, et al. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol Biomarkers Prev 2016;25:16-27. 10.1158/1055-9965.EPI-15-0578 [DOI] [PubMed] [Google Scholar]
- 8.Allum WH, Stenning SP, Bancewicz J, et al. Long-term results of a randomized trial of surgery with or without preoperative chemotherapy in esophageal cancer. J Clin Oncol 2009;27:5062-7. 10.1200/JCO.2009.22.2083 [DOI] [PubMed] [Google Scholar]
- 9.Shapiro J, van Lanschot JJB, Hulshof MCCM, et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): long-term results of a randomised controlled trial. Lancet Oncol 2015;16:1090-8. 10.1016/S1470-2045(15)00040-6 [DOI] [PubMed] [Google Scholar]
- 10.Lagergren J, Smyth E, Cunningham D, et al. Oesophageal cancer. Lancet 2017;390:2383-96. 10.1016/S0140-6736(17)31462-9 [DOI] [PubMed] [Google Scholar]
- 11.Shen Q. Establishment of esophageal phagocytic cell diagnosis and on-site prevention and treatment of esophageal cancer. Bulletin of Chinese Cancer 2000;000:215-7. [Google Scholar]
- 12.Dawsey SM, Shen Q, Nieberg RK, et al. Studies of esophageal balloon cytology in Linxian, China. Cancer Epidemiol Biomarkers Prev 1997;6:121-30. [PubMed] [Google Scholar]
- 13.Domper Arnal MJ, Ferrández Arenas Á, Lanas Arbeloa Á. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol 2015;21:7933-43. 10.3748/wjg.v21.i26.7933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimizu Y, Tukagoshi H, Fujita M, et al. Endoscopic screening for early esophageal cancer by iodine staining in patients with other current or prior primary cancers. Gastrointest Endosc 2001;53:1-5. 10.1067/mge.2001.111387 [DOI] [PubMed] [Google Scholar]
- 15.Yu J, Yang M. The Comparison of Clinical Diagnosis Value about Esophageal Cancer with CT and X-ray. Guide of China Medicine 2010;8:29. [Google Scholar]
- 16.Gong C, Shao K. CT Role in Staging Intermediate and Advanced Esophageal Carcinoma. Journal of Clinical Radiology 2000;19:619-21. [Google Scholar]
- 17.Kunizaki M, Hamasaki K, Wakata K, et al. Clinical Value of Serum p53 Antibody in the Diagnosis and Prognosis of Esophageal Squamous Cell Carcinoma. Anticancer Res 2018;38:1807-13. [DOI] [PubMed] [Google Scholar]
- 18.Gion M, Tremolada C, Mione R, et al. Tumor markers in serum of patients with primary squamous cell carcinoma of the esophagus. Tumori 1989;75:489-93. 10.1177/030089168907500519 [DOI] [PubMed] [Google Scholar]
- 19.Zhu N, Peng J, Yuan J, et al. Diagnosis value of CEA, AFP, CA-199, CA72-4 tumor markers joint inspection on esophageal cancer. Shaanxi Medical Journal 2018;47:120-2. [Google Scholar]
- 20.Hou JJ. To evaluate the combined detection of multiple tumor markers in early diagnosis of esophageal cancer. China Medical Engineering 2012;20:16-7. [Google Scholar]
- 21.Zhou SZ. Value Analysis of gastroscopy examination in the detection rate of esophageal cancer. Journal of Anhui Health Vocational & Technical College 2019;18:48-9. [Google Scholar]
- 22.Hofstetter W. Evolving Therapies in Esophageal Carcinoma, An Issue of Thoracic Surgery Clinics, E-Book. Elsevier Health Sciences, 2013. [DOI] [PubMed] [Google Scholar]
- 23.Rizk N. Surgery for esophageal cancer: goals of resection and optimizing outcomes. Thorac Surg Clin 2013;23:491-8. 10.1016/j.thorsurg.2013.07.009 [DOI] [PubMed] [Google Scholar]
- 24.Zhao X, Huai M, Zhang Y, et al. Analysis of Early Complications and Causes of Death After Thoracoscopic Resection of Esophageal Cancer. China Health Standard Management 2017;8:51-3. [Google Scholar]
- 25.Kong Y, Gao H, Li Y, et al. Efficacy and Prognosis of Chemoradiotherapy for 501 Patients With Postoperative Recurrence of Esophageal Cancer. 2020. doi: 10.21203/rs.3.rs-94545/v1 [DOI]
- 26.Berger B, Stahlberg K, Lemminger A, et al. Impact of radiotherapy, chemotherapy and surgery in multimodal treatment of locally advanced esophageal cancer. Oncology 2011;81:387-94. 10.1159/000335263 [DOI] [PubMed] [Google Scholar]
- 27.Miyasaka D, Okushiba S, Sasaki T, et al. Clinical evaluation of the feasibility of minimally invasive surgery in esophageal cancer. Asian J Endosc Surg 2013;6:26-32. 10.1111/j.1758-5910.2012.00158.x [DOI] [PubMed] [Google Scholar]
- 28.Moghissi K. Endoscopic photodynamic therapy (PDT) for oesophageal cancer. Photodiagnosis Photodyn Ther 2006;3:93-5. 10.1016/j.pdpdt.2006.03.005 [DOI] [PubMed] [Google Scholar]
- 29.Booka E, Takeuchi H, Kikuchi H, et al. Recent advances in thoracoscopic esophagectomy for esophageal cancer. Asian J Endosc Surg 2019;12:19-29. 10.1111/ases.12681 [DOI] [PubMed] [Google Scholar]
- 30.Han X, Lu N, Pan Y, et al. Nimotuzumab Combined with Chemotherapy is a Promising Treatment for Locally Advanced and Metastatic Esophageal Cancer. Med Sci Monit 2017;23:412-8. 10.12659/MSM.902645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunjigake K, Okamoto K, Gabata R, et al. Successful Treatment of Advanced Esophageal Cancer with DCF Chemotherapy and Pegfilgrastim. Gan To Kagaku Ryoho 2019;46:61-3. [PubMed] [Google Scholar]
- 32.Tachimori Y, Kanamori N, Uemura N, et al. Salvage esophagectomy after high-dose chemoradiotherapy for esophageal squamous cell carcinoma. J Thorac Cardiovasc Surg 2009;137:49-54. 10.1016/j.jtcvs.2008.05.016 [DOI] [PubMed] [Google Scholar]
- 33.Neureiter D, Mayr C, Winkelmann P, et al. Expression of the microRNA-200 Family, microRNA-205, and Markers of Epithelial-Mesenchymal Transition as Predictors for Endoscopic Submucosal Dissection over Esophagectomy in Esophageal Adenocarcinoma: A Single-Center Experience. Cells 2020;9:486. 10.3390/cells9020486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das A, Singh V, Fleischer DE, et al. A comparison of endoscopic treatment and surgery in early esophageal cancer: an analysis of surveillance epidemiology and end results data. Am J Gastroenterol 2008;103:1340-5. 10.1111/j.1572-0241.2008.01889.x [DOI] [PubMed] [Google Scholar]
- 35.Zeng R, Liu C, Li L, et al. Clinical Efficacy of HiPorfin Photodynamic Therapy for Advanced Obstructive Esophageal Cancer. Technol Cancer Res Treat 2020;19:1533033820930335. 10.1177/1533033820930335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Xie Z, Shi Y, et al. Gefitinib single drug in treatment of advanced esophageal cancer. J Cancer Res Ther 2016;12:C295-7. 10.4103/0973-1482.200760 [DOI] [PubMed] [Google Scholar]
- 37.Hecht JR, Bang YJ, Qin SK, et al. Lapatinib in Combination With Capecitabine Plus Oxaliplatin in Human Epidermal Growth Factor Receptor 2-Positive Advanced or Metastatic Gastric, Esophageal, or Gastroesophageal Adenocarcinoma: TRIO-013/LOGiC--A Randomized Phase III Trial. J Clin Oncol 2016;34:443-51. 10.1200/JCO.2015.62.6598 [DOI] [PubMed] [Google Scholar]
- 38.Makazu M, Kato K, Takisawa H, et al. Feasibility of endoscopic mucosal resection as salvage treatment for patients with local failure after definitive chemoradiotherapy for stage IB, II, and III esophageal squamous cell cancer. Dis Esophagus 2014;27:42-9. 10.1111/dote.12037 [DOI] [PubMed] [Google Scholar]
- 39.Ning B, Abdelfatah MM, Othman MO. Endoscopic submucosal dissection and endoscopic mucosal resection for early stage esophageal cancer. Ann Cardiothorac Surg 2017;6:88-98. 10.21037/acs.2017.03.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo HM, Zhang XQ, Chen M, et al. Endoscopic submucosal dissection vs endoscopic mucosal resection for superficial esophageal cancer. World J Gastroenterol 2014;20:5540-7. 10.3748/wjg.v20.i18.5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Xuan TT, Chen Y, et al. Investigative therapy for advanced esophageal cancer using the option for combined immunotherapy and chemotherapy. Immunotherapy 2020;12:697-703. 10.2217/imt-2020-0063 [DOI] [PubMed] [Google Scholar]
- 42.Tamaoki M, Yokoyama A, Horimatsu T, et al. Repeated talaporfin sodium photodynamic therapy for esophageal cancer: safety and efficacy. Esophagus 2021. [Epub ahead of print]. doi: . 10.1007/s10388-021-00853-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo K, Wang WP, Jiang T, et al. Assessment of epidermal growth factor receptor mutation/copy number and K-ras mutation in esophageal cancer. J Thorac Dis 2016;8:1753-63. 10.21037/jtd.2016.06.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park HY, Oh HJ, Kim KH, et al. Quantification of epidermal growth factor receptor (EGFR) mutation may be a predictor of EGFR-tyrosine kinase inhibitor treatment response. Thorac Cancer 2016;7:639-47. 10.1111/1759-7714.12378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adil Butt M, Pye H, Haidry RJ, et al. Upregulation of mucin glycoprotein MUC1 in the progression to esophageal adenocarcinoma and therapeutic potential with a targeted photoactive antibody-drug conjugate. Oncotarget 2017;8:25080-96. 10.18632/oncotarget.15340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ueda M, Iguchi T, Masuda T, et al. Somatic mutations in plasma cell-free DNA are diagnostic markers for esophageal squamous cell carcinoma recurrence. Oncotarget 2016;7:62280-91. 10.18632/oncotarget.11409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Chen X, Sun Q, et al. Overexpression of A613T and G462T variants of DNA polymerase β weakens chemotherapy sensitivity in esophageal cancer cell lines. Cancer Cell Int 2016;16:85. 10.1186/s12935-016-0362-x [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Peng F, Hu D, Lin X, et al. Analysis of Preoperative Metabolic Risk Factors Affecting the Prognosis of Patients with Esophageal Squamous Cell Carcinoma: The Fujian Prospective Investigation of Cancer (FIESTA) Study. EBioMedicine 2017;16:115-23. 10.1016/j.ebiom.2017.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin HD, Liao XY, Chen YB, et al. Genomic Characterization of Esophageal Squamous Cell Carcinoma Reveals Critical Genes Underlying Tumorigenesis and Poor Prognosis. Am J Hum Genet 2016;98:709-27. 10.1016/j.ajhg.2016.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Luo Z, Yang L, et al. The association between four SNPs of X-ray repair cross complementing protein 1 and the sensitivity to radiotherapy in patients with esophageal squamous cell carcinoma. Oncol Lett 2016;11:3508-14. 10.3892/ol.2016.4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bird-Lieberman EL, Fitzgerald RC. Early diagnosis of oesophageal cancer. Br J Cancer 2009;101:1-6. 10.1038/sj.bjc.6605126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bennett WP, Hollstein MC, He A, et al. Archival analysis of p53 genetic and protein alterations in Chinese esophageal cancer. Oncogene 1991;6:1779-84. [PubMed] [Google Scholar]
- 53.Zhao R, Casson AG. Epigenetic aberrations and targeted epigenetic therapy of esophageal cancer. Curr Cancer Drug Targets 2008;8:509-21. 10.2174/156800908785699306 [DOI] [PubMed] [Google Scholar]
- 54.Spizzo R, Almeida MI, Colombatti A, et al. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene 2012;31:4577-87. 10.1038/onc.2011.621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuwano H, Kato H, Miyazaki T, et al. Genetic alterations in esophageal cancer. Surg Today 2005;35:7-18. 10.1007/s00595-004-2885-3 [DOI] [PubMed] [Google Scholar]
- 56.Karagoz K, Lehman HL, Stairs DB, et al. Proteomic and Metabolic Signatures of Esophageal Squamous Cell Carcinoma. Curr Cancer Drug Targets 2016. [Epub ahead of print]. 10.2174/1568009616666160203113721 [DOI] [PubMed] [Google Scholar]
- 57.Li X, Zhou F, Jiang C, et al. Identification of a DNA methylome profile of esophageal squamous cell carcinoma and potential plasma epigenetic biomarkers for early diagnosis. PLoS One 2014;9:e103162. 10.1371/journal.pone.0103162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pu W, Wang C, Chen S, et al. Targeted bisulfite sequencing identified a panel of DNA methylation-based biomarkers for esophageal squamous cell carcinoma (ESCC). Clin Epigenetics 2017;9:129. 10.1186/s13148-017-0430-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin Z, Hamilton JP, Yang J, et al. Hypermethylation of the AKAP12 promoter is a biomarker of Barrett's-associated esophageal neoplastic progression. Cancer Epidemiol Biomarkers Prev 2008;17:111-7. 10.1158/1055-9965.EPI-07-0407 [DOI] [PubMed] [Google Scholar]
- 60.Dou C, Cao Z, Yang B, et al. Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci Rep 2016;6:21499. 10.1038/srep21499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010;142:409-19. 10.1016/j.cell.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li W, Zhao W, Lu Z, et al. Long Noncoding RNA GAS5 Promotes Proliferation, Migration, and Invasion by Regulation of miR-301a in Esophageal Cancer. Oncol Res 2018;26:1285-94. 10.3727/096504018X15166193231711 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Gerstein MB, Lu ZJ, Van Nostrand EL, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 2010;330:1775-87. 10.1126/science.1196914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733. 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang WT, Sun YM, Huang W, et al. Genome-wide Long Non-coding RNA Analysis Identified Circulating LncRNAs as Novel Non-invasive Diagnostic Biomarkers for Gynecological Disease. Sci Rep 2016;6:23343. 10.1038/srep23343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamada A, Okugawa Y, Boland CR, et al. Sa1916 RNA-Sequencing Approach for the Identification of Novel Long Non-Coding RNA Biomarkers in Colorectal Cancer. Gastroenterology 2015;148:S-354. 10.1016/S0016-5085(15)31190-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu J, Zhou R, Deng M, et al. Long non-coding RNA DIO3OS binds to microRNA-130b to restore radiosensitivity in esophageal squamous cell carcinoma by upregulating PAX9. Cancer Gene Ther 2021. [Epub ahead of print]. doi: . 10.1038/s41417-021-00344-2 [DOI] [PubMed] [Google Scholar]
- 68.Yoffe L, Gilam A, Yaron O, et al. Early Detection of Preeclampsia Using Circulating Small non-coding RNA. Sci Rep 2018;8:3401. 10.1038/s41598-018-21604-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elias-Rizk T, El Hajj J, Segal-Bendirdjian E, et al. The long non coding RNA H19 as a biomarker for breast cancer diagnosis in Lebanese women. Sci Rep 2020;10:22228. 10.1038/s41598-020-79285-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bouckenheimer J, Fauque P, Lecellier CH, et al. Differential long non-coding RNA expression profiles in human oocytes and cumulus cells. Sci Rep 2018;8:2202. 10.1038/s41598-018-20727-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu Z, Zhang QC, Lee B, et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 2016;165:1267-79. 10.1016/j.cell.2016.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marguerat S, Bähler J. RNA-seq: from technology to biology. Cell Mol Life Sci 2010;67:569-79. 10.1007/s00018-009-0180-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nagalakshmi U, Wang Z, Waern K, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008;320:1344-9. 10.1126/science.1158441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilhelm BT, Marguerat S, Watt S, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 2008;453:1239-43. 10.1038/nature07002 [DOI] [PubMed] [Google Scholar]
- 75.Schena M, Heller RA, Theriault TP, et al. Microarrays: biotechnology's discovery platform for functional genomics. Trends Biotechnol 1998;16:301-6. 10.1016/S0167-7799(98)01219-0 [DOI] [PubMed] [Google Scholar]
- 76.Kuwabara PE. DNA Microarrays and Gene Expression: From Experiments to Data Analysis and Modeling. Brief Funct Genomic Proteomic 2003;2:80-1. 10.1093/bfgp/2.1.80 [DOI] [Google Scholar]
- 77.Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008;5:621-8. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 78.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 2010;464:773-77. 10.1038/nature08903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garber M, Grabherr MG, Guttman M, et al. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods 2011;8:469-77. 10.1038/nmeth.1613 [DOI] [PubMed] [Google Scholar]
- 80.Sîrbu A, Kerr G, Crane M, et al. RNA-Seq vs dual- and single-channel microarray data: sensitivity analysis for differential expression and clustering. PLoS One 2012;7:e50986. 10.1371/journal.pone.0050986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fu X, Fu N, Guo S, et al. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics 2009;10:161. 10.1186/1471-2164-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kerr G, Ruskin HJ, Crane M, et al. Techniques for clustering gene expression data. Comput Biol Med 2008;38:283-93. 10.1016/j.compbiomed.2007.11.001 [DOI] [PubMed] [Google Scholar]
- 83.Nazarov PV, Muller A, Kaoma T, et al. RNA sequencing and transcriptome arrays analyses show opposing results for alternative splicing in patient derived samples. BMC Genomics 2017;18:443. 10.1186/s12864-017-3819-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mutz KO, Heilkenbrinker A, Lönne M, et al. Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol 2013;24:22-30. 10.1016/j.copbio.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 85.Geraci F, Saha I, Bianchini M. Editorial: RNA-Seq Analysis: Methods, Applications and Challenges. Front Genet 2020;11:220. 10.3389/fgene.2020.00220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei L, Feng J, Tao J. editors. Workshop: Transcriptome assembly from RNA-Seq data: Objectives, algorithms and challenges. IEEE 1st International Conference on Computational Advances in Bio and Medical Sciences, ICCABS 2011, Orlando, FL, USA, February 3-5, 2011; 2011. [Google Scholar]
- 87.Esteve-Codina A. RNA-Seq Data Analysis, Applications and Challenges. Compr Anal Chem 2018;82:71-106. 10.1016/bs.coac.2018.06.001 [DOI] [Google Scholar]
- 88.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41. 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 89.He TY, Li SH, Huang J, et al. Prognostic value of long non-coding RNA CRNDE in gastrointestinal cancers: a meta-analysis. Cancer Manag Res 2019;11:5629-42. 10.2147/CMAR.S201646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clark MB, Johnston RL, Inostroza-Ponta M, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res 2012;22:885-98. 10.1101/gr.131037.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature 2011;470:284-8. 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sarma K, Levasseur P, Aristarkhov A, et al. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci U S A 2010;107:22196-201. 10.1073/pnas.1009785107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gutschner T, Baas M, Diederichs S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res 2011;21:1944-54. 10.1101/gr.122358.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yuan SX, Yang F, Yang Y, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231-41. 10.1002/hep.25895 [DOI] [PubMed] [Google Scholar]
- 95.Shoshani O, Massalha H, Shani N, et al. Polyploidization of murine mesenchymal cells is associated with suppression of the long noncoding RNA H19 and reduced tumorigenicity. Cancer Res. 2012;72:6403-13. 10.1158/0008-5472.CAN-12-1155 [DOI] [PubMed] [Google Scholar]
- 96.Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010;465:1033-8. 10.1038/nature09144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen FJ, Sun M, Li SQ, et al. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog 2013;52:908-15. 10.1002/mc.21944 [DOI] [PubMed] [Google Scholar]
- 98.Cui Z, Ren S, Lu J, et al. The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urol Oncol 2013;31:1117-23. 10.1016/j.urolonc.2011.11.030 [DOI] [PubMed] [Google Scholar]
- 99.Khaitan D, Dinger ME, Mazar J, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res 2011;71:3852-62. 10.1158/0008-5472.CAN-10-4460 [DOI] [PubMed] [Google Scholar]
- 100.Yang Y. Expression and clinical significance of long non-coding RNA Linc00152,H19 and ROR in patients with esophageal cancer. International Journal of Laboratory Medicine 2019:226-229,233. [Google Scholar]
- 101.Zhang S, Liao W, Wu Q, et al. LINC00152 upregulates ZEB1 expression and enhances epithelial-mesenchymal transition and oxaliplatin resistance in esophageal cancer by interacting with EZH2. Cancer Cell Int 2020;20:569. 10.1186/s12935-020-01620-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lin C, Wang Y, Wang Y, et al. Transcriptional and posttranscriptional regulation of HOXA13 by lncRNA HOTTIP facilitates tumorigenesis and metastasis in esophageal squamous carcinoma cells. Oncogene 2017;36:5392-406. 10.1038/onc.2017.133 [DOI] [PubMed] [Google Scholar]
- 103.Bao J, Zhou C, Zhang J, et al. Upregulation of the long noncoding RNA FOXD2-AS1 predicts poor prognosis in esophageal squamous cell carcinoma. Cancer Biomark 2018;21:527-33. 10.3233/CBM-170260 [DOI] [PubMed] [Google Scholar]
- 104.Shi W, Gao Z, Song J, et al. Silence of FOXD2-AS1 inhibited the proliferation and invasion of esophagus cells by regulating miR-145-5p/CDK6 axis. Histol Histopathol 2020;35:1013-21. [DOI] [PubMed] [Google Scholar]
- 105.Ge XS, Ma HJ, Zheng XH, et al. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci 2013;104:1675-82. 10.1111/cas.12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Song W, Zou SB. Prognostic role of lncRNA HOTAIR in esophageal squamous cell carcinoma. Clin Chim Acta 2016;463:169-73. 10.1016/j.cca.2016.10.035 [DOI] [PubMed] [Google Scholar]
- 107.Wang W, He X, Zheng Z, et al. Serum HOTAIR as a novel diagnostic biomarker for esophageal squamous cell carcinoma. Mol Cancer 2017;16:75. 10.1186/s12943-017-0643-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li PD, Hu JL, Ma C, et al. Upregulation of the long non-coding RNA PVT1 promotes esophageal squamous cell carcinoma progression by acting as a molecular sponge of miR-203 and LASP1. Oncotarget 2017;8:34164-76. 10.18632/oncotarget.15878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu B, Pan CF, Yao GL, et al. The long non-coding RNA AK001796 contributes to tumor growth via regulating expression of p53 in esophageal squamous cell carcinoma. Cancer Cell Int 2018;18:38. 10.1186/s12935-018-0537-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ma J, Li TF, Han XW, et al. Downregulated MEG3 contributes to tumour progression and poor prognosis in oesophagal squamous cell carcinoma by interacting with miR-4261, downregulating DKK2 and activating the Wnt/β-catenin signalling. Artif Cells Nanomed Biotechnol 2019;47:1513-23. 10.1080/21691401.2019.1602538 [DOI] [PubMed] [Google Scholar]
- 111.Chang ZW, Jia YX, Zhang WJ, et al. LncRNA-TUSC7/miR-224 affected chemotherapy resistance of esophageal squamous cell carcinoma by competitively regulating DESC1. J Exp Clin Cancer Res 2018;37:56. 10.1186/s13046-018-0724-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chu C, Qu K, Zhong FL, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 2011;44:667-78. 10.1016/j.molcel.2011.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Calin GA, Liu CG, Sevignani C, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A 2004;101:11755-60. 10.1073/pnas.0404432101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tan Y, Wang H, Zhang C. MicroRNA-381 targets G protein-Coupled receptor 34 (GPR34) to regulate the growth, migration and invasion of human cervical cancer cells. Environ Toxicol Pharmacol 2021;81:103514. 10.1016/j.etap.2020.103514 [DOI] [PubMed] [Google Scholar]
- 115.Sharma N, Baruah MM. The microRNA signatures: aberrantly expressed miRNAs in prostate cancer. Clin Transl Oncol 2019;21:126-44. 10.1007/s12094-018-1910-8 [DOI] [PubMed] [Google Scholar]
- 116.Liu D, Hu X, Zhou H, Shi G, Wu J. Identification of Aberrantly Expressed miRNAs in Gastric Cancer. Gastroenterol Res Pract. 2014;2014:473817. 10.1155/2014/473817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pisanu A, Lecca D, Mulas G, et al. Dynamic changes in pro- and anti-inflammatory cytokines in microglia after PPAR-γ agonist neuroprotective treatment in the MPTPp mouse model of progressive Parkinson's disease. Neurobiol Dis 2014;71:280-91. 10.1016/j.nbd.2014.08.011 [DOI] [PubMed] [Google Scholar]
- 118.Guo L, Cai X, Hu W, et al. Expression and clinical significance of miRNA-145 and miRNA-218 in laryngeal cancer. Oncol Lett 2019;18:764-70. 10.3892/ol.2019.10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bobryshev YV, Orekhov AN, Chistiakov DA. MicroRNAs in Esophageal Adenocarcinoma: Functional Significance and Potential for the Development of New Molecular Disease Markers. Curr Pharm Des 2015;21:3402-16. 10.2174/1381612821666150311124418 [DOI] [PubMed] [Google Scholar]
- 120.Meng F, Qian L, Lv L, et al. miR-193a-3p regulation of chemoradiation resistance in oesophageal cancer cells via the PSEN1 gene. Gene 2016;579:139-45. 10.1016/j.gene.2015.12.060 [DOI] [PubMed] [Google Scholar]
- 121.Tian J, Shang M, Shi SB, et al. Cetuximab plus pemetrexed as second-line therapy for fluorouracil-based pre-treated metastatic esophageal squamous cell carcinoma. Cancer Chemother Pharmacol 2015;76:829-34. 10.1007/s00280-015-2854-0 [DOI] [PubMed] [Google Scholar]
- 122.Huang Y, Zhang R, Fang Y. Overexpression of miR-218 enhances the chemosensitivity of esophageal squamous cell carcinoma to cisplatin. Acta Universitatis Medicinalis Anhui 2018;53:923-7. [Google Scholar]
- 123.Wang Y, Zhao Y, Herbst A, et al. miR-221 Mediates Chemoresistance of Esophageal Adenocarcinoma by Direct Targeting of DKK2 Expression. Ann Surg 2016;264:804-14. 10.1097/SLA.0000000000001928 [DOI] [PubMed] [Google Scholar]
- 124.Xing Y, Zha WJ, Li XM, et al. Circular RNA circ-Foxo3 inhibits esophageal squamous cell cancer progression via the miR-23a/PTEN axis. J Cell Biochem 2020;121:2595-605. 10.1002/jcb.29481 [DOI] [PubMed] [Google Scholar]
- 125.Bibby B, Cawthorne C, Reynolds J, et al. MicroRNA-330-5p downregulation in oesophageal adenocarcinoma is a potential therapeutic target for enhancing chemoradiation sensitivity in patients. Eur J Cancer 2016;61:S158. 10.1016/S0959-8049(16)61559-9 [DOI] [Google Scholar]
- 126.Kurashige J, Watanabe M, Kamohara H, et al. Abstract 1155: Serum microRNA-21 is a novel biomarker in patients with esophageal squamous cell carcinoma. In: Proceedings of the 102nd Annual Meeting of the American Association for Cancer Research; 2011 Apr 2-6; Orlando, FL. Philadelphia (PA): AACR; Cancer Res 2011;71:Abstract nr 1155. doi: 10.1158/1538-7445.AM2011-1155. 10.1158/1538-7445.AM2011-1155 [DOI] [Google Scholar]
- 127.Winther M, Alsner J, Tramm T, et al. Evaluation of miR-21 and miR-375 as prognostic biomarkers in esophageal cancer. Acta Oncol 2015;54:1582-91. 10.3109/0284186X.2015.1064161 [DOI] [PubMed] [Google Scholar]
- 128.Wu YR, Qi HJ, Deng DF, et al. MicroRNA-21 promotes cell proliferation, migration, and resistance to apoptosis through PTEN/PI3K/AKT signaling pathway in esophageal cancer. Tumour Biol 2016;37:12061-70. 10.1007/s13277-016-5074-2 [DOI] [PubMed] [Google Scholar]
- 129.Yao LH, Wang GR, Cai Y, et al. The expressions and diagnostic values of miR-18a and miR-21 in esophageal cancer. Zhonghua Zhong Liu Za Zhi 2019;41:107-11. [DOI] [PubMed] [Google Scholar]
- 130.Wang F, Liu S, Liu J, et al. SP promotes cell proliferation in esophageal squamous cell carcinoma through the NK1R/Hes1 axis. Biochem Biophys Res Commun 2019;514:1210-6. 10.1016/j.bbrc.2019.05.092 [DOI] [PubMed] [Google Scholar]
- 131.Xu XL, Jiang YH, Feng JG, et al. MicroRNA-17, microRNA-18a, and microRNA-19a are prognostic indicators in esophageal squamous cell carcinoma. Ann Thorac Surg 2014;97:1037-45. 10.1016/j.athoracsur.2013.10.042 [DOI] [PubMed] [Google Scholar]
- 132.Jing C, Ma G, Li X, et al. MicroRNA-17/20a impedes migration and invasion via TGF-β/ITGB6 pathway in esophageal squamous cell carcinoma. Am J Cancer Res 2016;6:1549-62. [PMC free article] [PubMed] [Google Scholar]
- 133.Hezova R, Kovarikova A, Srovnal J, et al. Diagnostic and prognostic potential of miR-21, miR-29c, miR-148 and miR-203 in adenocarcinoma and squamous cell carcinoma of esophagus. Diagn Pathol 2015;10:42. 10.1186/s13000-015-0280-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zong M, Feng W, Wan L, et al. miR-203 affects esophageal cancer cell proliferation, apoptosis and invasion by targeting MAP3K1. Oncol Lett 2020;20:751-7. 10.3892/ol.2020.11610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu Y, Dong Z, Liang J, et al. Methylation-mediated repression of potential tumor suppressor miR-203a and miR-203b contributes to esophageal squamous cell carcinoma development. Tumour Biol 2016;37:5621-32. 10.1007/s13277-015-4432-9 [DOI] [PubMed] [Google Scholar]
- 136.Ding Y, Ma Q, Guo X, et al. A preliminary screening of differentially expressed miRNAs in serum of patients with esophageal squamous cell carcinoma. Shandong Medical Journal 2018;58:1-4. [Google Scholar]
- 137.Mao Y, Liu J, Zhang D, et al. MiR-1290 promotes cancer progression by targeting nuclear factor I/X (NFIX) in esophageal squamous cell carcinoma (ESCC). Biomed Pharmacother 2015;76:82-93. 10.1016/j.biopha.2015.10.005 [DOI] [PubMed] [Google Scholar]
- 138.Sun H, Wang L, Zhao Q, et al. Diagnostic and prognostic value of serum miRNA-1290 in human esophageal squamous cell carcinoma. Cancer Biomark 2019;25:381-7. 10.3233/CBM-190007 [DOI] [PubMed] [Google Scholar]
- 139.Zheng TL, Li DP, He ZF, et al. miR-145 sensitizes esophageal squamous cell carcinoma to cisplatin through directly inhibiting PI3K/AKT signaling pathway. Cancer Cell Int 2019;19:250. 10.1186/s12935-019-0943-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang Q, Gan H, Song W, et al. MicroRNA-145 promotes esophageal cancer cells proliferation and metastasis by targeting SMAD5. Scand J Gastroenterol 2018;53:769-76. 10.1080/00365521.2018.1476913 [DOI] [PubMed] [Google Scholar]
- 141.Jin W, Luo W, Fang W, et al. miR-145 expression level in tissue predicts prognosis of patients with esophageal squamous cell carcinoma. Pathol Res Pract 2019;215:152401. 10.1016/j.prp.2019.03.029 [DOI] [PubMed] [Google Scholar]
- 142.Ansari MH, Irani S, Edalat H, et al. Deregulation of miR-93 and miR-143 in human esophageal cancer. Tumour Biol 2016;37:3097-103. 10.1007/s13277-015-3987-9 [DOI] [PubMed] [Google Scholar]
- 143.Xia C, Yang Y, Kong F, et al. MiR-143-3p inhibits the proliferation, cell migration and invasion of human breast cancer cells by modulating the expression of MAPK7. Biochimie 2018;147:98-104. 10.1016/j.biochi.2018.01.003 [DOI] [PubMed] [Google Scholar]
- 144.Zhang HS, Zhang FJ, Li H, et al. Tanshinone IIA inhibits human esophageal cancer cell growth through miR-122-mediated PKM2 down-regulation. Arch Biochem Biophys 2016;598:50-6. 10.1016/j.abb.2016.03.031 [DOI] [PubMed] [Google Scholar]
- 145.Zeng W, Zhu JF, Liu JY, et al. miR-133b inhibits cell proliferation, migration and invasion of esophageal squamous cell carcinoma by targeting EGFR. Biomed Pharmacother 2019;111:476-84. 10.1016/j.biopha.2018.12.057 [DOI] [PubMed] [Google Scholar]
- 146.Zhu JF, Liu Y, Huang H, et al. MicroRNA-133b/EGFR axis regulates esophageal squamous cell carcinoma metastases by suppressing anoikis resistance and anchorage-independent growth. Cancer Cell Int 2018;18:193. 10.1186/s12935-018-0684-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Heitzer E, Auer M, Gasch C, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res 2013;73:2965-75. 10.1158/0008-5472.CAN-12-4140 [DOI] [PubMed] [Google Scholar]
- 148.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol 2015;12:381-8. 10.1080/15476286.2015.1020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell 1976;8:547-55. 10.1016/0092-8674(76)90223-3 [DOI] [PubMed] [Google Scholar]
- 151.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979;280:339-40. 10.1038/280339a0 [DOI] [PubMed] [Google Scholar]
- 152.Matsumoto Y, Fishel R, Wickner RB. Circular single-stranded RNA replicon in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 1990;87:7628-32. 10.1073/pnas.87.19.7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792-806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 154.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 155.Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777. 10.1371/journal.pgen.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA 2015;6:563-79. 10.1002/wrna.1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A 1996;93:6536-41. 10.1073/pnas.93.13.6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Werfel S, Nothjunge S, Schwarzmayr T, et al. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol 2016;98:103-7. 10.1016/j.yjmcc.2016.07.007 [DOI] [PubMed] [Google Scholar]
- 159.Szabo L, Morey R, Palpant NJ, et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 2015;16:126. 10.1186/s13059-015-0690-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA Biogenesis Competes with Pre-mRNA Splicing. Molecular cell 2014;56:55-66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 161.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011;30:4414-22. 10.1038/emboj.2011.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kos A, Dijkema R, Arnberg AC, et al. The hepatitis delta (delta) virus possesses a circular RNA. Nature 1986;323:558-60. 10.1038/323558a0 [DOI] [PubMed] [Google Scholar]
- 165.Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet 2014;5:8. 10.3389/fgene.2014.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA 2010;16:2043-50. 10.1261/rna.2414110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Taylor JM. Hepatitis delta virus. Virology 2006;344:71-6. 10.1016/j.virol.2005.09.033 [DOI] [PubMed] [Google Scholar]
- 168.Burd CE, Jeck WR, Liu Y, et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010;6:e1001233. 10.1371/journal.pgen.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Correa-Medina M, Bravo-Egana V, Rosero S, et al. MicroRNA miR-7 is preferentially expressed in endocrine cells of the developing and adult human pancreas. Gene Expr Patterns 2009;9:193-9. 10.1016/j.gep.2008.12.003 [DOI] [PubMed] [Google Scholar]
- 170.Junn E, Lee KW, Jeong BS, et al. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A 2009;106:13052-7. 10.1073/pnas.0906277106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhao Z, Li X, Jian D, et al. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol 2017;54:237-45. 10.1007/s00592-016-0943-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Qu S, Song W, Yang X, et al. Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genom Data 2015;5:385-7. 10.1016/j.gdata.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark 2016;16:161-9. 10.3233/CBM-150552 [DOI] [PubMed] [Google Scholar]
- 174.Liu J, Zheng X, Liu H. Hsa_circ_0102272 serves as a prognostic biomarker and regulates proliferation, migration and apoptosis in thyroid cancer. J Gene Med 2020;22:e3209. 10.1002/jgm.3209 [DOI] [PubMed] [Google Scholar]
- 175.Sang M, Meng L, Sang Y, et al. Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett 2018;426:37-46. 10.1016/j.canlet.2018.03.049 [DOI] [PubMed] [Google Scholar]
- 176.Chen J, Yang J, Fei X, et al. CircRNA ciRS-7: a Novel Oncogene in Multiple Cancers. Int J Biol Sci 2021;17:379-89. 10.7150/ijbs.54292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wen J, Liao J, Liang J, et al. Circular RNA HIPK3: A Key Circular RNA in a Variety of Human Cancers. Front Oncol 2020;10:773. 10.3389/fonc.2020.00773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gou Y, Ma J, Han S, et al. CircHIPK3 Affects Malignant Phenotypes in Esophageal Squamous Cell Carcinoma by Regulating p53-Akt-Mdm2 Signaling Pathways. Cancer Research on Prevention and Treatment 2019;46:987-93. [Google Scholar]
- 179.Xie ZF, Li HT, Xie SH, et al. Circular RNA hsa_circ_0006168 contributes to cell proliferation, migration and invasion in esophageal cancer by regulating miR-384/RBBP7 axis via activation of S6K/S6 pathway. Eur Rev Med Pharmacol Sci 2020;24:151-63. [DOI] [PubMed] [Google Scholar]
- 180.Huang E, Fu J, Yu Q, et al. CircRNA hsa_circ_0004771 promotes esophageal squamous cell cancer progression via miR-339-5p/CDC25A axis. Epigenomics 2020;12:587-603. 10.2217/epi-2019-0404 [DOI] [PubMed] [Google Scholar]
- 181.He Y, Mingyan E, Wang C, et al. CircVRK1 regulates tumor progression and radioresistance in esophageal squamous cell carcinoma by regulating miR-624-3p/PTEN/PI3K/AKT signaling pathway. Int J Biol Macromol 2019;125:116-23. 10.1016/j.ijbiomac.2018.11.273 [DOI] [PubMed] [Google Scholar]
- 182.Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget 2015;6:6001-13. 10.18632/oncotarget.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li H, Lan M, Liao X, et al. Circular RNA cir-ITCH Promotes Osteosarcoma Migration and Invasion through cir-ITCH/miR-7/EGFR Pathway. Technol Cancer Res Treat 2020;19:1533033819898728. 10.1177/1533033819898728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang W, Ma J, Lu J, et al. Circ0043898 acts as a tumor inhibitor and performs regulatory effect on the inhibition of esophageal carcinoma. Cancer Biol Ther 2018;19:1117-27. 10.1080/15384047.2018.1480889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang W, Zhu D, Zhao Z, et al. RNA sequencing reveals the expression profiles of circRNA and identifies a four-circRNA signature acts as a prognostic marker in esophageal squamous cell carcinoma. Cancer Cell Int 2021;21:151. 10.1186/s12935-021-01852-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Li JS, Xie YF, Song LT, et al. Expression profile of long non-coding RNA in gingival tissues from the patients with aggressive periodontitis. Zhonghua Kou Qiang Yi Xue Za Zhi 2018;53:635-9. [DOI] [PubMed] [Google Scholar]
- 187.Song H, Sun W, Ye G, et al. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med 2013;11:225. 10.1186/1479-5876-11-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Huang Y, Liu HM, Wu LL, et al. Long non-coding RNA and mRNA profile analysis in the parotid gland of mouse with type 2 diabetes. Life Sci 2021;268:119009. 10.1016/j.lfs.2020.119009 [DOI] [PubMed] [Google Scholar]
- 189.Patentstar. Method for detecting ncRNA. Available online: https://cprs.patentstar.com.cn/Search/Detail?ANE=8CEA4DBA3ADA6ABAFHHA9DIC5BEA5EAA9FAFHHIA5EAA9HEC