Abstract
Background
Denosumab is an inhibitor of receptor activator of NF-κB ligand (RANKL), which inhibits bone metastasis (BM) in breast cancer (BC), but does not completely control cancer cell BM in some BC patients. This study was designed to study whether denosumab inhibits human BC cells (MCF-7) cell line-induced spontaneous osteoclastogenesis via RANKL/metastasis-associated lung adenocarcinoma transcript 1 (MALAT1)/miR-124 axis.
Methods
We established a co-culture system of MCF-7-induced spontaneous osteoclastogenesis in RAW 264.7 cells, and denosumab is added into the co-culture system to inhibit RAW 264.7 cell differentiation into osteoclasts. Real-time PCR (RT-PCR), immunofluorescence and western blotting analysis were used to detect gene expression, while tartrate-resistant acid phosphatase (TRAP) staining was used to assess osteoclast formation.
Results
Denosumab inhibits MCF-7 cell line-induced spontaneous osteoclastogenesis, and the inhibition of denosumab was found to be more pronounced after MALAT1 downregulation and miR-124 overexpression. However, MALAT1 knockdown or miR-124 overexpression did not alter RANKL protein expression. Moreover, the dual luciferase gene reporter system showed that miR-124 targeted the inhibition of MALAT1, while si-MALAT1 upregulated miR-124 expression. miR-124-mimics were able to decrease the expression of Rab27a, IL-11, activated T-cell nuclear factor 1 (NFATc1) and TARP protein.
Conclusions
Denosumab inhibits MALAT1 expression by inhibiting RANKL, thereby upregulating miR-124 expression, which ultimately inhibits MCF-7 cell line-induced pseudo osteoclastogenesis.
Keywords: Denosumab, osteoclastogenesis, receptor activator of NF-κB ligand (RANKL), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), miR-124
Introduction
Breast cancer (BC) is a common malignant tumor in women, accounting for most malignant tumors in women. Approximately 1.2 million new BC patients are diagnosed and around 500,000 people die of BC each year worldwide (1,2). Clinical studies have shown that the incidence of bone metastases (BMs) in BC is highest in two common tumor metastatic organs, the lung and liver (3,4). Since the BM mechanism of BC has not been fully elucidated, BM cannot be cured, resulting in a high mortality rate (5,6).
Denosumab targets the inhibition of receptor activator of NF-κB ligand (RANKL), which inhibits the activation of osteoclasts and decreases bone absorption and destruction (7). It has been widely used to treat or prevent BM of solid tumors, such as BC, kidney cancer and urinary system tumors, and has been proven to exert a therapeutic effect (8). However, for some BM patients, the effect of denosumab treatment on the prevention of BM is extremely low (9,10), but the specific cause is unknown, and we suspect that this may be related to the expression of effector molecules downstream of RNAKL. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was found to be highly expressed in BC tissues and promoted the invasion and migration of BC cells (11,12). In addition, previous studies have found that RANKL can promote the expression of MALAT1 in human osteoblasts, thereby regulating the biological characteristics of human osteoblasts (13). miR-124 has been shown to be downregulated in BC tissues and inhibits EMT and metastasis in BC (14,15), and regulates MALAT1 expression (16).
This study aimed to determine whether the downstream molecular mechanism of denosumab inhibition of BM in BC proceeds by inhibiting RANKL and if it is related with MALAT1 and miR-124 expression. It was also determined whether MALAT1 and miR-124 exert a feedback effect on denosumab. In this study, we constructed a BM model by establishing a MCF-7 and RAW 264.7 non-contact co-culture system, and altered the expression of MALAT1 and miR-124 in RAW 264.7 cells to explore the effect of MALAT1 and miR-124 on the inhibition of MCF-7-induced osteoclastogenesis in vitro caused by denosumab. We found that denosumab inhibits MALAT1 expression by inhibiting RANKL, thereby upregulating miR-124 expression, and ultimately inhibiting MCF-7 cell line-induced pseudo osteoclastogenesis.
Methods
Cell culture and treatment
RAW 264.7 (SC-6003, ATCC, USA) and MCF-7 cells (CRL-10781) were all cultured in Dulbecco’s Modified Eagle Medium (DMEM) solutions (31600091, Gibco, USA), into which 10% fetal bovine serum (10099-141, Gibco, USA) was added, while RAW 264.7 cells were co-cultured with MCF-7 cells in a non-contact system (Figure 1A). Denosumab (Amgen, USA) was added into the co-culture system for 5 days at a concentration of 0.1 mg/mL.
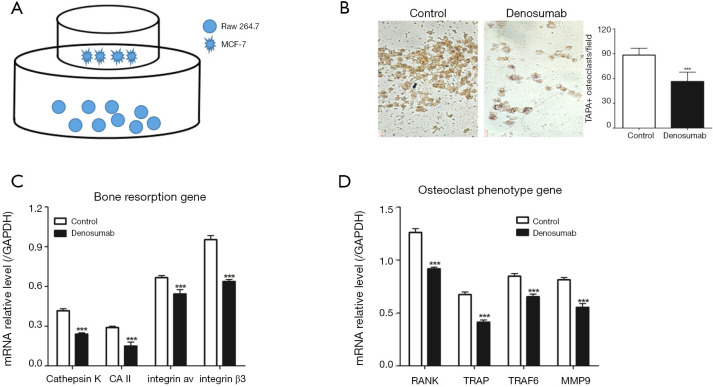
Figure 1.
Denosumab inhibits MCF-7-induced spontaneous osteoclastogenesis of RAW 264.7 cells. (A) Co-culture system for spontaneous osteoclastogenesis of RAW 264.7 cells induced by MCF-7 cells; (B) representative field of co-culture after TRAP staining and statistics of TRAP+ osteoclastogenesis; (C,D) the mRNA expression of bone resorption gene (C) and osteoclast phenotype gene (D). Three times for each experiment and ***, P<0.001 vs. control group. Scale bar =20 mm. TRAP, tartrate-resistant acid phosphatase.
Tartrate-resistant acid phosphatase (TRAP) staining
Cells in cell culture suspension and urine were collected through centrifugation, and phosphate buffer saline (PBS) was used to resuspend the sediment. Then, the cells were cytospun onto slides, and TRAP staining was performed using a TRAP staining solution kit (Solarbio, G1942, China).
Cell transfection
The miR-negative control (miR-NC) and mimic of miR-124, and si-NC and si-MALAT1 were purchased from Sangon Biotech (Shanghai, China), and were directly transferred into cells using LipofectamineTM 2000 transfection reagent (11668019, Invitrogen, CA, USA). The wild type and mutated mRNA 3'-UTR of MALAT1 were first connected to pisCHECK2 (Promega, WI, USA) before being transfected into cells as miRNAs.
Real-time quantitative PCR (RT-qPCR)
RT-qPCR was used to detect mRNA, miRNA and lncRNA expression, as previously described (17), with the PCR primers used given in Table 1.
Table 1. Primers for RT-qPCR.
| Gene | Primer sequence (5'–3') |
|---|---|
| MALAT1 | Forward: AAAGCAAGGTCTCCCCACAAG |
| Reverse: GGTCTGTGCTAGATCAAAAGGCA | |
| miR-124 | Forward: ACACTCCAGCTGGGCGTGTTCACAGCGGAC |
| Reverse: TGGTGTCGTGGAGTCG | |
| Cathepsin K | Forward: CTGAAGATGCTTCCCATATGTGGG |
| Reverse: GCAGGCGTTGTTCTTATTCCGAGC | |
| CAII | Forward: CTTCAGGACAATGCAGTGC |
| Reverse: ATCCAGGTCACACATTCCAGC | |
| Integrin av | Forward: GCCAGCCCATTGAGTTTGATT |
| Reverse: GCTACCAGGACCACCGAGAAG | |
| Integrin β3 | Forward: TTACCCCGTGGACATCTACTA |
| Reverse: AGTCTTCCATCCAGGGCAATA | |
| RANK | Forward: ACCTCCAGTCAGCAAAGAAGT |
| Reverse: TCACAGCCCTCAGAATCCAC | |
| TRAP | Forward: ACACAGTGATGCTGTGTGGCAACTC |
| Reverse: CCAGAGGCTTCCACATCTATGCTGG | |
| TRAF6 | Forward: AGCCCACGAAAGCCAGAAGAA |
| Reverse: CCCTTATGGATTTGATGATGC | |
| MMP-9 | Forward: CGAGTGGACGCGACCGTAGTTGG |
| Reverse: CAGGCTTAGAGCCACGACCATACAG | |
| U6 | Forward: CTCATAGGGTTGTTCGCTCGG |
| Reverse: AACGCTTCACGAATTTGCGT | |
| GAPDH | Forward: TAGCGGCTAGCGGTAT |
| Reverse: CGGGGCTATGGCTAGCTAGCTTTC |
RT-qPCR, real-time quantitative PCR.
Western blotting analysis
Levels of Rab27a, IL-11, activated T-cell nuclear factor 1 (NFATc1), TRAP and GAPDH protein were analyzed using western blotting analysis, as previously described (17). The primary antibodies used were Rab27a (ab55667, 1:500), IL-11 (ab187167, 1:1,000), NFATc1 (ab25916, 1:1,000), TRAP (ab65854, 1:1,500) and GAPDH (ab8245, 1:3,000), which were all purchased from Abcam.
Cellular immunofluorescence
Cells in cell culture suspension and urine were collected through centrifugation, and PBS was used to resuspend the sediment. Then, the cells were cytospun onto slides, and fixed using 4% paraformaldehyde for 0.5 hours at room temperature. Thereafter, they were incubated with a primary body, RANKL (ab45039, 1:100, ABCAM, UK), at 4 °C and left overnight. Goat anti-rabbit IgG (H+L) cross-adsorbed secondary antibody (A11008, 1:500, Invitrogen, USA) was used as the secondary antibody at room temperature for 1 hour, followed by incubation with 5 ug/mL 4',6-diamidino-2-phenylindole (DAPI) (D8417, Sigma, USA) at room temperature for 5 minutes.
Statistical analysis
Data were analyzed using SPSS 20.0 software. Data between two groups were compared using student’s t-test, and data between multiple groups were compared using one-way ANOVA, with Duncan test as the post hoc test. A P value of <0.05 was considered to indicate a significant difference.
Results
Denosumab inhibits spontaneous osteoclastogenesis of RAW 264.7 cells
First, we established a non-contact co-culture system (Figure 1A), in which MCF-7 cells were cultured in the upper chamber and RAW 264.7 cells were cultured in the lower chamber. As shown in Figure 1B, the TRAP+ osteoclast of the control group was significantly higher than that of the denosumab group, which was added with denosumab to the co-culture system to prevent osteoclast differentiation of RAW 264.7 cells, induced by MCF-7 cells. Moreover, we also measured the expression of bone resorption genes, such as cathepsin K, CAII, integrin av and integrin β3, and the expression of osteoclast phenotype genes, such as RANK, TRAP, TRAF6 and MMP-9. We found that the mRNA expression levels of bone resorption genes (Figure 1C) and osteoclast phenotype genes (Figure 1D) in control group were all significantly higher than that of the denosumab group.
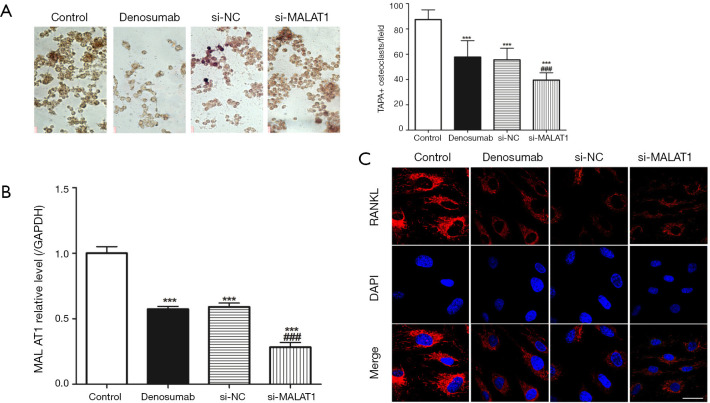
MALAT1 knockdown enhances denosumab function
We transferred si-MALAT1 into RAW 264.7 cells to knockdown MALAT1, and then the MALAT1 knockdown cells were placed in the lower chamber of the co-culture system to be cultured. TARP staining was used to measure TARP+ osteoclast. We found that (Figure 2A) the TRAP+ osteoclast in control group was highest, while the TRAP+ osteoclast in si-MALAT1 group was the lowest, and the results of RT-qPCR confirmed that (Figure 2B) the level of MALAT1 in the si-MALAT1 group was the lowest. However, immunofluorescence detection of RANKL in the si-MALAT1 group found that its expression was not significantly different from that of the denosumab group (Figure 2C).
Figure 2.
MALAT1 regulates spontaneous osteoclastogenesis of RAW 264.7 cells inhibited by denosumab. (A) Representative field of co-culture after TRAP staining and statistics of TRAP+ osteoclastogenesis; (B) MALAT expressed in each group; (C) immunofluorescence staining of RANKL in RAW 264.7 cells of each group. Three times for each experiment; ***, P<0.001 vs. control group, and ###, P<0.001 vs. denosumab group. Scale bar =50 µm. TRAP, tartrate-resistant acid phosphatase; RANKL, receptor activator of NF-κB ligand.
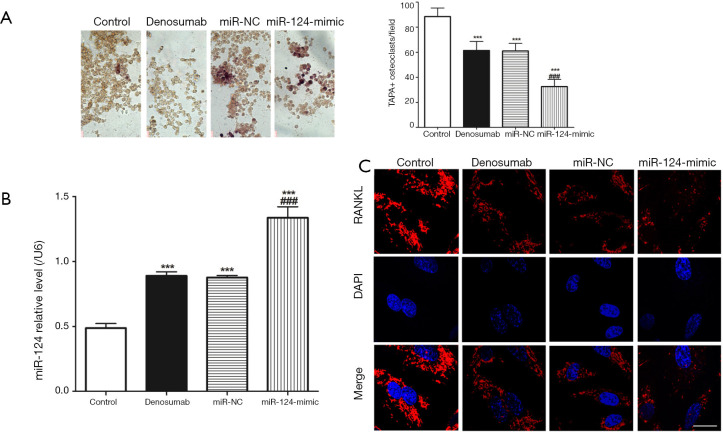
miR-124 overexpression enhances denosumab function
We transfected the miR-124-mimic into RAW 264.7 cells to upregulate miR-124, and then the resulting miR-124 overexpressing cells were placed in the lower chamber of the co-culture system to be cultured. TARP staining was used to measure TARP+ osteoclast. We found that the TRAP+ osteoclast in control group was the highest (Figure 3A), and that the TRAP+ osteoclast in miR-124-mimic group was the lowest, and the results of RT-qPCR confirmed that (Figure 3B) the level of miR-124 in the miR-124-mimic group was the lowest. However, immunofluorescence detection of RANKL in the miR-124-mimic group found that its expression was not significantly different from that of the denosumab group (Figure 3C).
Figure 3.
miR-124 regulates spontaneous osteoclastogenesis of RAW 264.7 cells inhibited by denosumab. (A) Representative field of co-culture after TRAP staining and statistics of TRAP+ osteoclastogenesis; (B) miR-124 expressed in each group; (C) immunofluorescence staining of RANKL in RAW 264.7 cells of each group. Three times for each experiment, ***, P<0.001 vs. control group, and ###, P<0.001 vs. denosumab group. Scale bar =50 µm. TRAP, tartrate-resistant acid phosphatase; RANKL, receptor activator of NF-κB ligand.
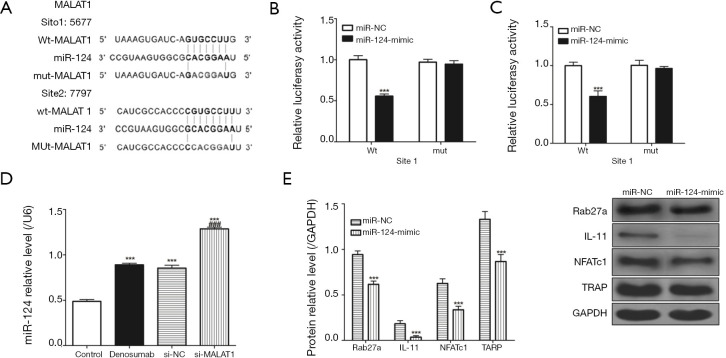
miR-124 negatively regulates osteoclast differentiation and mutual inhibition along with MALAT1
After analyzing the sequences of MALAT1 and miR-124, we found that MALAT1 and miR-124 have complementary sequences (Figure 4A). We validated the results using the luciferase gene reporter system and found that transfection with the miR-124-mimic significantly increased WT type 3'-UTR luciferase activity of RAW 264.7 cells, but did not have the same effect on that of MUT (Figure 4B,C). In addition, we also found that MALAT1 knockdown by si-MALAT1 could increase miR-124 expression, and that miR-124 overexpression could decrease the expression of Rab27a, IL-11, NFATc1 and TARP proteins (Figure 4D,E).
Figure 4.
miR-124 and MALAT1 inhibit each other in RAW 264.7 cells. (A) Sequence in which MALAT1 and miR-124 bind to each other; (B,C) dual luciferase gene reporter system validates sequence of direct binding of MALAT1 to miR-124; (D) miR-124 expressed in each group; (E) levels of Rab27a, IL-11, NFATc1 and TARP protein in RAW 264.7 cells after transferring to miR-NC or miR-124-mimic. Three times for each experiment, ***, P<0.001 vs. control group, and ###, P<0.001 vs. denosumab group. MALAT1, metastasis-associated lung adenocarcinoma transcript 1; NFATc1, activated T-cell nuclear factor 1; miR-NC, miR-negative control; TRAP, tartrate-resistant acid phosphatase.
Discussion
There are four basic elements required for the development of BM in malignant tumor cells: cancer cells, osteoblasts, osteoclasts and bone matrix (18). Metastatic cancer cells cannot directly destroy bone, and its metastases must first activate osteoclasts to differentiate and mature, and then only can osteoclasts mediate bone resorption to cause tumorous bone destruction for further local growth, which involves an important signal transduction pathway, the RANK/RANKL/OPG pathway (19). Normal bone metabolism maintains a dynamic balance between the osteogenic effects of osteoblasts and the bone resorption effect of osteoclasts, with the RANKL/RANK/OPG pathway being one of the main mechanisms of regulating bone homeostasis. OPG, RANKL and RANK are members of the tumor necrosis factor family, and OPG and RANKL are expressed in osteoblasts and bone stromal cells, while RANK is expressed on the surface of osteoclast precursors. During the process of bone destruction, RANKL acts as an activating factor that induces the maturation and activation of osteoclasts by binding to RANK on the surface of osteoclast precursors, which ultimately leads to bone resorption. However, OPG is a soluble RANKL inhibitor that binds to RANKL and inhibits the binding of RANKL to RANK, thereby inhibiting the differentiation and maturation of osteoclast precursors (20).
In this study, we found that MCF-7 could induce the spontaneous osteoclast differentiation of RAW 264.7 cells in the non-contact co-culture system of MCF-7 and RAW 264.7 cells, and that denosumab not only significantly inhibited the number of TARP osteoclasts, but also decreased the expression of bone resorption genes and osteoclast phenotype genes. This indicates that: (I) the non-contact co-culture system of MCF-7 and RAW 264.7 cells in this study can be used to demonstrate BM mechanism of BC cells in vitro; (II) Denosumab can inhibit the spontaneous osteoclast differentiation of RAW 264.7 cells induced by MCF-7. As an inhibitor of RNAKL, denosumab has been widely used in clinical settings to treat BM of cancer and has been shown to produce a good effect. Theoretically, denosumab can block the pathway of BM by interfering with the RNAKL/RANK axis of bone, delay the progression of BM, decrease tumor burden and prolong patient survival time, and this theory has been confirmed in animal models (21). Canon and Gonzalez-Suarez found that denosumab can decrease tumor burden of bone in advanced BM animal models, hinder tumor progression, prolong the formation in mice and decrease the development of spontaneous lung metastases (22,23). However, denosumab did not prolong survival and BM time of patients with malignant tumors in clinical trials (24). Two theories that may explain the difference in these outcomes have been accepted. On the one hand, activation of osteoclasts not only involves OPG, RANKL and RANK, but involves many other related factors involved in the regulation of activation of bone cells, such as M-CSF and sRANKL (25,26). On the other hand, the OPG/RANKL/RANK pathway regulated osteoclast activation also requires downstream effector molecules, which undergo various changes in BM.
In this study, we found that inhibition of MALAT1 expression or upregulation of miR-124 expression in RAW 264.7 cells did not alter RANKL expression, but significantly increased denosumab induced inhibition of spontaneous osteoclast differentiation caused by MCF-7 cells, while denosumab could also inhibit MALAT1 expression and increase miR-124 expression.
MALAT1 is also named nuclear-enriched autosomal transcript 2, and is an important member of the lncRNA family that was discovered in NSCLC tissues in 2003 (27). Many studies have found that MALAT1 is abnormally expressed in multiple tumor tissues (28,29). Previous research has found that MALAT1 can promote the proliferation, metastasis and invasion of tumor cells (30,31) through the recruitment of specific SR protein family members (32,33), and is involved in epigenetic regulation (34,35) and cell cycle regulation (36). In BC, MALAT1 is considered as an oncogene and was found to be highly expressed in BM tumor tissues, and the high expression of MALAT1 was found to promote the proliferation, migration and invasion of BC cells. In addition, MALAT1 has been shown to promote cancer cell BM in non-small cell lung cancer (37). In human mature bone cells, RNAKL promotes the expression of MALAT1. Combined with the results of the present study, MALAT1 may be considered as a downstream effector molecule of RANKL, and high expression of MALAT1 may play a role in promoting BM of BC cells.
miR-124 is a miRNA that is closely involved with malignant tumors, and many studies have shown that miR-124 is downregulated in a variety of malignant tumor tissues, including BC, and in vitro and in vivo experiments have confirmed that miR-124 is a tumor suppressor gene that inhibits the proliferation, invasion and migration of many malignant tumor cells, including BC cells. Moreover, previous research has also found that miR-124 can inhibit osteoclast differentiation (38). In this study, we found that miR-124 targeted the inhibition of MALAT1 expression, but its expression was also inhibited by MALAT1, and miR-124 could significantly inhibit the expression of Rab27a, IL-11, NFATc1 and TARP proteins. Previous studies have found that miR-124 could inhibit the osteoclastic differentiation of RAW 264.7 cells, induced in BC cell lines through the targeted inhibition of IL-11 (39), Rab27a (40) and NFATc1 (41).
Conclusions
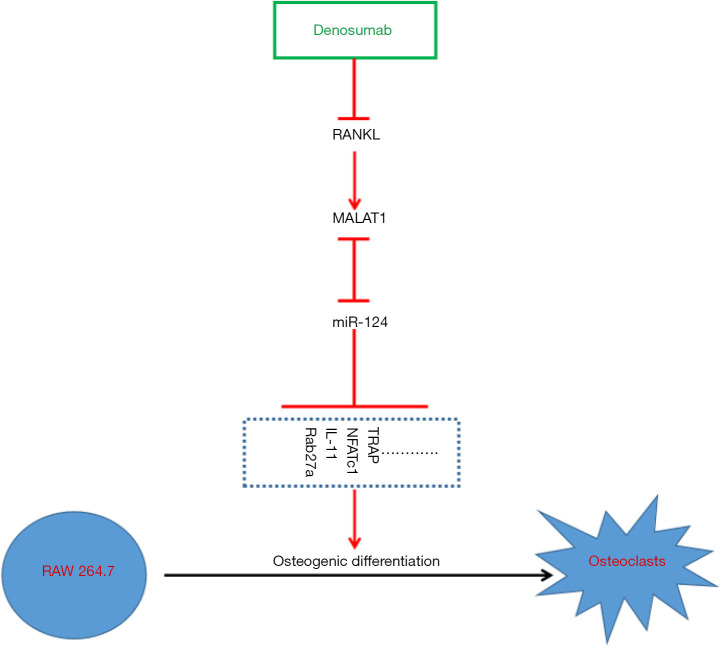
Taken together, the results of this study found that denosumab inhibits MALAT1 expression by inhibiting RANKL, thereby upregulating miR-124 expression, which ultimately inhibits the pseudo osteoclastogenesis caused by the MCF-7 cell line (Figure 5). High expression of MALAT1 and low expression of miR-124 in BC tumor tissues may be reasons why denosumab cannot be used to effectively treat and prevent BM in BC patients.
Figure 5.
Molecular mechanism of denosumab inhibiting MCF-7-induced spontaneous osteoclastogenesis. RANKL, receptor activator of NF-κB ligand; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; TRAP, tartrate-resistant acid phosphatase; NFATc1, activated T-cell nuclear factor 1.
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of The Fourth Hospital of Hebei Medical University (No. 2019123).
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.03.17). The authors have no conflicts of interest to declare.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 3.Yates LR, Knappskog S, Wedge D, et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell 2017;32:169-84.e7. 10.1016/j.ccell.2017.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye X, Brabletz T, Kang Y, et al. Upholding a role for EMT in breast cancer metastasis. Nature 2017;547:E1-3. 10.1038/nature22816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosseini H, Obradovic MMS, Hoffmann M, et al. Early dissemination seeds metastasis in breast cancer. Nature 2016;540:552-8. 10.1038/nature20785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pulido C, Vendrell I, Ferreira AR, et al. Bone metastasis risk factors in breast cancer. Ecancermedicalscience 2017;11:715. 10.3332/ecancer.2017.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott LJ, Muir VJ. Denosumab in the prevention of skeletal-related events in patients with bone metastases from solid tumors: profile report. BioDrugs 2011;25:397-400. 10.2165/11207650-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 8.Henry DH, Costa L, Goldwasser F, et al. Randomized, double-blind study of denosumab versus zoledronic acid in the treatment of bone metastases in patients with advanced cancer (excluding breast and prostate cancer) or multiple myeloma. J Clin Oncol 2011;29:1125-32. 10.1200/JCO.2010.31.3304 [DOI] [PubMed] [Google Scholar]
- 9.Barton MK. Denosumab an option for patients with bone metastasis from breast cancer. CA Cancer J Clin 2011;61:135-6. 10.3322/caac.20116 [DOI] [PubMed] [Google Scholar]
- 10.Shizuku M, Kurata N, Mori T, et al. Urine NTx changes predict the survaival in breast cancer patients with bone metastasis receiving denosumab. Ann Oncol 2015;26:vii128. 10.1093/annonc/mdv472.101 [DOI] [Google Scholar]
- 11.Chou J, Wang B, Zheng T, et al. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem Biophys Res Commun 2016;472:262-9. 10.1016/j.bbrc.2016.02.102 [DOI] [PubMed] [Google Scholar]
- 12.Bamodu OA, Huang WC, Lee WH, et al. Aberrant KDM5B expression promotes aggressive breast cancer through MALAT1 overexpression and downregulation of hsa-miR-448. BMC Cancer 2016;16:160. 10.1186/s12885-016-2108-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Che W, Dong Y, Quan HJC, et al. RANKL inhibits cell proliferation by regulating MALAT1 expression in a human osteoblastic cell line hFOB 1.19. Cell Mol Biol (Noisy-le-grand) 2015;61:7-14. [PubMed] [Google Scholar]
- 14.Liang YJ, Wang QY, Zhou CX, et al. MiR-124 targets Slug to regulate epithelial-mesenchymal transition and metastasis of breast cancer. Carcinogenesis 2013;34:713-22. 10.1093/carcin/bgs383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ben Gacem R, Ben Abdelkrim O, Ziadi S, et al. Methylation of miR-124a-1, miR-124a-2, and miR-124a-3 genes correlates with aggressive and advanced breast cancer disease. Tumour Biol 2014;35:4047-56. 10.1007/s13277-013-1530-4 [DOI] [PubMed] [Google Scholar]
- 16.Feng T, Shao F, Wu Q, et al. miR-124 downregulation leads to breast cancer progression via LncRNA-MALAT1 regulation and CDK4/E2F1 signal activation. Oncotarget 2016;7:16205-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao J, Zhang J, Ling Y, et al. Mitochondrial sirtuin 4 resolves immune tolerance in monocytes by rebalancing glycolysis and glucose oxidation homeostasis. Front Immunol 2018;9:419. 10.3389/fimmu.2018.00419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang XH, Jin X, Malladi S, et al. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell 2013;154:1060-73. 10.1016/j.cell.2013.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khosla S. Minireview: the OPG/RANKL/RANK system. Endocrinology 2001;142:5050-5. 10.1210/endo.142.12.8536 [DOI] [PubMed] [Google Scholar]
- 20.Dougall WC. Molecular pathways: osteoclast-dependent and osteoclast-independent roles of the RANKL/RANK/OPG pathway in tumorigenesis and metastasis. Clin Cancer Res 2012;18:326-35. 10.1158/1078-0432.CCR-10-2507 [DOI] [PubMed] [Google Scholar]
- 21.Lacey DL, Boyle WJ, Simonet WS, et al. Bench to bedside: elucidation of the OPG-RANK-RANKL pathway and the development of denosumab. Nat Rev Drug Discov 2012;11:401-19. 10.1038/nrd3705 [DOI] [PubMed] [Google Scholar]
- 22.Canon JR, Roudier M, Bryant R, et al. Inhibition of RANKL blocks skeletal tumor progression and improves survival in a mouse model of breast cancer bone metastasis. Clin Exp Metastasis 2008;25:119-29. 10.1007/s10585-007-9127-1 [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Suarez E, Jacob AP, Jones J, et al. RANK ligand mediates progestin-induced mammary epithelial proliferation and carcinogenesis. Nature 2010;468:103-7. 10.1038/nature09495 [DOI] [PubMed] [Google Scholar]
- 24.Smith MR, Saad F, Coleman R, et al. Denosumab and bone-metastasis-free survival in men with castration-resistant prostate cancer: results of a phase 3, randomised, placebo-controlled trial. Lancet 2012;379:39-46. 10.1016/S0140-6736(11)61226-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee K, Kim MY, Ahn H, et al. Blocking of the ubiquitin-proteasome system prevents inflammation-induced bone loss by accelerating M-CSF receptor c-Fms degradation in osteoclast differentiation. Int J Mol Sci 2017. doi: . 10.3390/ijms18102054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma QL, Fang L, Jiang N, et al. Bone mesenchymal stem cell secretion of sRANKL/OPG/M-CSF in response to macrophage-mediated inflammatory response influences osteogenesis on nanostructured Ti surfaces. Biomaterials 2018;154:234-47. 10.1016/j.biomaterials.2017.11.003 [DOI] [PubMed] [Google Scholar]
- 27.Rao AKDM, Rajkumar T, Mani S. Perspectives of long non-coding RNAs in cancer. Mol Biol Rep 2017;44:203-18. 10.1007/s11033-017-4103-6 [DOI] [PubMed] [Google Scholar]
- 28.Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol 2014;7:3174-81. [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L, Bai H, Deng Y, et al. High MALAT1 expression predicts a poor prognosis of cervical cancer and promotes cancer cell growth and invasion. Eur Rev Med Pharmacol Sci 2015;19:3187-93. [PubMed] [Google Scholar]
- 30.Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010;39:925-38. 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun KK, Hu PP, Xu F. Prognostic significance of long non-coding RNA MALAT1 for predicting the recurrence and metastasis of gallbladder cancer and its effect on cell proliferation, migration, invasion, and apoptosis. J Cell Biochem 2018;119:3099-110. 10.1002/jcb.26451 [DOI] [PubMed] [Google Scholar]
- 32.Bantignies F, Roure V, Comet I, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 2011;144:214-26. 10.1016/j.cell.2010.12.026 [DOI] [PubMed] [Google Scholar]
- 33.Bernstein E, Duncan EM, Masui O, et al. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol 2006;26:2560-9. 10.1128/MCB.26.7.2560-2569.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang F, Yi F, Han X, et al. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett 2013;587:3175-81. 10.1016/j.febslet.2013.07.048 [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Chung MR, Zhou S, et al. STAT3 controls osteoclast differentiation and bone homeostasis by regulating NFATc1 transcription. J Biol Chem 2019;294:15395-407. 10.1074/jbc.RA119.010139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Dai Q, Zeng F, et al. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the Rac1/JNK pathway via targeting MiR-509. Oncol Res 2018. [Epub ahead of print]. 10.3727/096504017X14957939026111 [DOI] [PubMed] [Google Scholar]
- 37.Liu M, Sun W, Liu Y, et al. The role of lncRNA MALAT1 in bone metastasis in patients with non-small cell lung cancer. Oncol Rep 2016;36:1679-85. 10.3892/or.2016.4909 [DOI] [PubMed] [Google Scholar]
- 38.Lee Y, Kim HJ, Park CK, et al. MicroRNA-124 regulates osteoclast differentiation. Bone 2013;56:383-9. 10.1016/j.bone.2013.07.007 [DOI] [PubMed] [Google Scholar]
- 39.Cai WL, Huang WD, Li B, et al. microRNA-124 inhibits bone metastasis of breast cancer by repressing interleukin-11. Mol Cancer 2018;17:9. 10.1186/s12943-017-0746-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang L, Yin Y, Liu J, et al. MiR-124 attenuates osteoclastogenic differentiation of bone marrow monocytes via targeting Rab27a. Cell Physiol Biochem 2017;43:1663-72. 10.1159/000484027 [DOI] [PubMed] [Google Scholar]
- 41.Nakamachi Y, Ohnuma K, Uto K, et al. MicroRNA-124 inhibits the progression of adjuvant-induced arthritis in rats. Ann Rheum Dis 2016;75:601-8. 10.1136/annrheumdis-2014-206417 [DOI] [PubMed] [Google Scholar]