Abstract
Lung cancer is one of the highest incidence cancer types worldwide and one with the lowest 5-year survival rate of all cancer types. Despite recent insights into lung cancer pathobiology, including novel biomarker-targeted therapies and immunotherapies, most of lung patients are diagnosed at late stages with limited and ineffective treatments. Therefore, more approaches are needed to eradicate lung cancer. In the last years, small extracellular vesicles (EVs) secreted by tumor cells have been gaining relevance. These intercellular signal mediators, called exosomes, contain a huge range of biological elements, including lipids, nucleic acids and miRNAs, among others, that carry relevant information. The role of exosomes in cancer progression is dependent on cancer type, molecular characteristics and stage. MicroRNAs molecules are a big part of the content of exosomes cargo and probably the most studied ones. Due to the regulatory role in gene expression, miRNAs may provide information of the molecular characteristics of the tumor and be also able to reprogram distant target cells. Exosomal miRNAs can modulate different biological processes in cancer such as growth, progression, invasion, angiogenesis, metastasis and drug resistance; playing a critical role in modifying the microenvironment of non-small cell lung cancer (NSCLC). Therefore, they can act by regulating tumor resistance and also be useful to monitoring the response/relapse to targeted therapies. In this work, we summarize the relevant advances on the potential role of exosomal miRNAs in NSCLC pathobiogenesis, highlighting the clinical utility of exosomal microRNAs as biomarkers for the NSCLC diagnosis, prognosis, drug resistance and therapeutic strategies.
Keywords: Exosomes, miRNAs, liquid biopsy, biomarker, non-small cell lung cancer (NSCLC)
Introduction
Lung cancer is the most frequent cause of death by cancer worldwide (1). The 5-year survival rate in lung cancer ranges from 73% (in stage IA) to 13% (in stage IV). Nevertheless, approximately 85% of lung cancer diagnosis is in advanced-disease (stage III or IV) with limited therapeutic options. Non-small cell lung cancer (NSCLC) is one of the most frequent diagnoses, representing 85% of lung cancer cases, and it includes adenocarcinoma (ADC), squamous cell carcinoma (SCC), and large-cell carcinoma (LCC) as the main NSCLC subtypes (2). Moreover, small cell lung cancer (SCLC) represents 14–15% of all lung cancers. SCLC is classified as small cell carcinoma, when it presents a pure histology; in other cases, it may happen that small cell carcinoma is combined with large cell carcinoma (3).
New insights into the molecular mechanisms of lung cancer have meant considerable progress in the development of new biomarkers for targeted therapies, such as therapies targeting the epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), the proto-oncogene B-Raf (BRAF), Ret Proto-Oncogene (RET), hepatocyte growth factor receptor (MET) and Neurotrophic Receptor Tyrosine Kinase 1 (NTRK), as well as for the most pioneering immunotherapies, based in checkpoint inhibitors targeting the programmed cell death 1 (PD-1) and programmed death-ligand 1 (PD-L1) pathways (4).
Despite recent advances in cancer treatments and screening methods, most lung cancer patients are still diagnosed in advanced stages, when the disease is already unresectable. Consequently, all treatment-determining diagnoses must be made in small biopsies and/or cytology-type samples. Adding these facts to the lack of a variety in effective treatments, rare 5-year survival percentages are produced (5). Taking the previous into account, the identification of new diagnostic, prognostic and predictive biomarkers is the utmost requirement to achieve effective therapeutic response in NSCLC (6). Recently, liquid biopsy has arisen as a new non-invasive approach that may provide multiple indicators in cancer, including diagnosis, prognosis, resistance to therapy and monitoring of response (7).
Liquid biopsy refers to the molecular analysis, with minimally invasive methods, used to study biomarkers in different body fluids (peripheral blood, urine, saliva, pleural liquid), being a reliable alternative to conventional tissue biopsies. The important biotechnological development in the field of detecting the components of liquid biopsy has allowed relevant clinical applications for cancer patients. The main components analysed in lung cancer liquid biopsies are circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), exosomes, circulating microRNAs, circulating RNA, platelets or plasma/serum metabolites (8).Currently, liquid biopsy is not yet able to replace tissue biopsy in the daily clinical practice, although it offers additional data that cannot be obtained in any other way. For example, in cases in which patients cannot be biopsied or biopsies do not provide enough tissue for mutational analysis, liquid biopsy is the only alternative. This novel tool is already being used in clinical practice through the analysis of cfDNA-based tumor genotyping for the management of targeted therapies in lung cancer (9,10). In patients with advanced disease, it is not feasible to obtain biopsies of every metastatic location; however, body fluids such as blood can include elements from the primary tumor and the metastases. Moreover, liquid biopsy samples can be repeatedly obtained without the risk of comorbidities and used to monitor the disease as well as early detection of response, relapse or resistance to a determined therapy (11).
For all the aforementioned reasons, the development and implementation of more sensitive and minimally invasive techniques that allow the study of specific biomarkers in body fluids at low concentration, are necessary to carry out a new approach in cancer management.
Exosomes are very small vesicles, secreted by tumor cells, capable of transferring their molecular cargo to distant cells. MicroRNAs are small non-coding RNAs often deregulated in cancer, which act as master regulators of the genome. This type of small RNA can represent a large percentage of the exosome content, acting as valuable element in the development of lung cancer. In this review we focus on the role of exosomal microRNAs in the pathobiogenesis of NSCLC.
Exosomes as new source of biomarkers in cancer
Exosomes are small extracellular vesicles (EVs) with a diameter around 40–160 nm which are produced and secreted from most eukaryotic cells and are involved in cell-to-cell communications. They are formed by a lipid bilayer structure and contain inside a wide variety of cellular components such as proteins, mRNAs, small RNAs, DNAs and lipids, among others (12).
Exosomes are produced in a process that consists of a double invagination of the plasma membrane and the formation of intracellular multivesicular bodies (MVBs), which contain intraluminal vesicles (ILVs). ILVs are finally secreted as exosomes (with a maximum size 160 nm) through MVB fusion to the plasma membrane followed by exocytosis. The amount of biogenesis of exosomes between different cells depends on their physiological and pathological states and can be found in the blood, urine, semen and other body fluids (13). In last years, knowledge about the role of exosomes in cancer has come a long way compared to research into their activity in other non-neoplastic diseases.
Different works have elucidated that exosomes are implied in many important biological processes such as intra/extracellular communication, immune system modulation (14), cell growth and maintenance of energy pathways (15). This type of EVs can have a pleiotropic role in cancer, being associated with several hallmarks of this disease (16). Moreover, exosomes derived from cancer cells influence tumor growth and metastasis (17), immune evasion and drug resistance (18).
The role of exosomes in cancer progression is dynamic and specific to cancer type, genetics, and stage. A great number of relevant studies have been performed in exosomes from NSCLC for improving the management and knowledge of the disease. Additionally, collected results show that cancer-derived exosomal miRNAs play a key role in the activation of molecules and reprogramming of the tumor environment (19).
Exosome-derived microRNAs: a valuable tool in NSCLC study
MicroRNAs (miRNAs) are a short and endogenous type of non-coding RNAs. The majority, are transcribed from DNA sequences into primary miRNAs, then transformed into precursors, and finally forming the mature miRNAs. In most cases, miRNAs interact with the 3′ untranslated region (3′ UTR) of target mRNAs to induce mRNA degradation and translational repression. However, the relationship of miRNAs with other regions, including the 5′ UTRs, coding sequences and promoters, have also been revealed (20-22).
MiRNAs may be secreted in extracellular fluids and transported to target cells via protein binding or within microvesicles such as exosomes (23,24). These molecules of RNA are a considerable part of the content of exosomes cargo and probably the most studied ones due to their involvement in regulation of gene expression at post-transcriptional/translational levels and modulation of cell functions in several types of cancer (25).
One of the most important aspects of exosomal miRNAs is that they represent a potential valuable tool for non-invasive early diagnosis, treatment selection and prediction of therapy resistance (26).
Exosomal miRNAs as biomarkers of relevant tumor processes (angiogenesis, EMT and metastasis) in NSCLC
The formation of new vasculature is essential for tumor development, spread and metastasis. Tumors can modulate angiogenesis through exosomes. Concretely, some studies have shown that exosomal miRNAs stimulate angiogenesis. Liu et al. revealed that exosomal miR-21 leads to STAT3 activation, increasing VEGF levels and inducing malignant transformation of human bronchial epithelium (27).
Other authors demonstrated that exosomal miR-9 results in the activation of JAK/STAT pathway, promoting angiogenesis and cell migration. Moreover, Hsu et al. showed that exosomal miR-23a modulates tumor vasculature under normoxic and hypoxic conditions, demonstrating that lung cancer cells transfer genetic information to distant cells (28).
Exosomal miRNAs can also be involved in epithelial-to-mesenchymal transition (EMT) and metastasis processes. EMT is a process through which adherent epithelial cells acquire the ability to migrate, losing cell-to-cell adhesion and cellular polarity. Metastasis is a multifaceted process by which cells separate from the primary tumor and invade adjacent or distant tissues (29).
There seems to be different approaches of exosomal miRNA transfer during cancer metastasis. Firstly, exosomal miRNAs are transferred from invasive cells to less invasive cells promoting the metastasis. For instance, ADC cell-derived exosomal miR-494 and miR-524-3p showed capacity to target non-transformed cells and modulate pre-metastatic organ cells (17). Another example, exosomes derived from ADC cells were able to transfer to osteoclast progenitor cells and target programmed cell death 4 (PDCD4) to facilitate osteoclastogenesis through exosomal miR-21. Secondly, exosomal miRNAs can be secreted by primary tumor cells to the tumor microenvironment, contacting other cells. Fabbri et al. revealed that tumor-secreted miR-21 and miR-29a could activate Toll-like receptors TLR7 and TLR8 in immune cells, triggering tumor growth and metastasis (30). Furthermore, lung cancer cells can influence metastatic bone colonization via exosomal miR-192 (31). Thirdly, tumor cells can modify his behaviour because of unique miRNAs released by normal cells. For instance, exosomal miR-193a-3p, miR-210-3p and miR-5100 can be transferred from bone mesenchymal stem cells (BMSCs) to lung cancers and activate STAT3 signalling, which leads to EMT and metastasis (32).
Potential role of exosomal miRNAs as biomarkers for clinical applications
Effective therapies and early detection of lung cancer ought to be priorities and remain the most efficient approaches to reduce the number of deaths from this disease. Exosomal microRNAs could be proposed as new non-invasive biomarkers in neoplastic disease. In this section, we review the new outcomes on exosomal miRNA involvement as diagnostic, prognostic and predictive biomarkers for NSCLC.
Exosomal miRNA for early detection and diagnosis of NSCLC
As previously mentioned, one of the main problems of lung cancer today is a late diagnosis due to the lack of efficient screening methods. Currently, a common early screening method is low-dose computed tomography. However, this type of screening presents some limitations, which include enhanced costs, high rate of benign nodule detection, over-diagnosis and radiation exposure (33).
In most cases, patients are diagnosed at advanced stages when surgery is not an option. In addition, in many of these patients, there is not even an effective therapeutic option, which leads to a high mortality rate. Therefore, early detection and diagnosis are the most effective strategies for prevention and treatment of lung cancer (34,35). For these reasons, liquid biopsy takes on a key role in order to be a source of biomarkers. Concretely, exosomes have become a valuable source of biomarkers, obtained in a minimally invasive and effective way. The study of the extensive content of molecules present in these microvesicles makes elements such as miRNAs to be postulated as tools for the early detection of this pathology (36).
Interestingly, preceding studies about the diagnostic use of miRNAs associated with lung biopsies revealed the utility of 12 particular miRNAs: miR-17-3p, miR-21, miR-106a, miR-146, miR-155, miR-191, miR-192, miR-203, miR-205, miR-210, miR-212, and miR-214 (37). Afterwards, Rabinowits et al., focused on these 12 miRNAs, demonstrated the resemblance of circulating exosomal miRNA and tumor-derived miRNAs profiling, suggesting that exosomal miRNAs could be used as a screening tool for detection of lung ADC (38).
It has also been demonstrated that miR-378a, miR-379, miR-139-5p, and miR-200b-5p are useful for the discrimination of nodule (lung ADCs and granulomas) and non-nodule (healthy ex-smokers) (97.5% sensitivity, 72.0% specificity). Furthermore, same study revealed that a set of 6 exosomal miRNAs derived from human plasma (miR-629, miR-30a-3p, miR-100, miR-200b-5p, miR-154-3p and miR-151a-5p) differentiated granulomas from lung ADC (96% sensitivity and 60% specificity), and a group of 4 exosomal miRNAs (miR-200b-5p, miR-378a, miR-139-5p and miR-379) distinguished nodules from non-nodules with 96.0% of sensitivity and 60.0% of specificity (39). Zhou et al. also found a group of exosomal miRNAs (miR-19-3p, miR-21-5p and miR-221-3p) for discriminating patients with lung ADC from healthy subjects with sensitivity and specificity rates of 67–73% and 66–80%, respectively (40).
In terms of small cell lung carcinoma (SCLC) and NSCLC histology, Poroyko et al. revealed that miRNAs exosomal cargo is different between patients with SCLC and NSCLC. In particular, 13 miRNAs were suitable to differentiate the two histologies. Three of them (miR-331-5p, miR-451a, miR-363-3p) were capable of distinguishing SCLC and NSCLC cases with highest rates of specificity and sensitivity. What is more, miR-203, suitable for discerning NSCLC and SCLC cases (sensitivity 80%, specificity 100%), was previously reported by Rabinowits et al. in blood-derived exosomes as a biomarker for lung ADC (38).These authors described 7 exosomal miRNAs (miR-451a, miR-486-5p, miR-363-3p, miR-660-5p, miR-15b-5p, miR-25-3p and miR-16-2-3p) specific to differentiate between NSCLC patients and healthy subjects (41).
Another study reported that exosomal miR-126 could be a potential diagnostic biomarker for NSCLC (42). Furthermore, it has been described a group of 3 miRNAs (miR-21, miR-205 and miR-155) for the early detection of NSCLC (43).
Focusing in the histologic heterogeneity of NSCLC, Jin et al. performed a remarkable study which demonstrated that four exosomal miRNAs (miR-let-7b-5p, miR-let-7e-5p, miR-23a-3p and miR-486-5p) were potential diagnostic biomarkers of stage I NSCLC patients (sensitivity 80.5% and specificity 92.31%). Besides, they also revealed that miR-181-5p, miR-30a3p, miR-30e-3p, and miR-361-5p were ADC-specific miRNAs, whereas miR-10b-5p, miR-15b-5p and miR-320b were squamous-specific with areas under the curve (AUC) of 0.936 and 0.911, respectively (44). Previously, Aushev et al. revealed that the levels of a specific panel of exosomal miRNAs (miR-205, miR-19a, miR-19b, miR-30b and miR-20a) decreased in the blood of patients after lung SCC surgery (45).
Exosomes are not only present in blood. Many studies have been focused on others fluids like bronchoalveolar lavage (BAL) and pleural fluid to find new diagnostic biomarkers. A study confirmed that exosomal miRNA levels are increased in plasma and BAL samples from patients with NSCLC in contrast to non-tumor patients (46).
Additionally, some authors identified 9 exosomal miRNAs exosomal miRNAs in pleural fluid (miR-141-3p, miR-200a-3p, miR-200b-3p, miR-200c-3p, miR-205-5p, miR-375, miR-483-5p, miR-429, miR-203a-3p), which were predominantly present in lung cancer over tuberculous and other benign lesions (47,48). Furthermore, exosomal miR-200 was also described in pleural effusions to distinguish ADC from patients with benign effusions (49). More recently, it has been revealed expression of miR-182 and miR-210 in malignant pleural fluid from ADC over benign pleural effusions (50) (Table 1).
Table 1. Main studies focused on exosomal miRNAs used as diagnostic biomarkers in NSCLC.
| Exosomal miRNAs | Source | Clinical value | Reference |
|---|---|---|---|
| miR-378a, miR-379, miR-139-5p, and miR-200b-5p | Plasma | Diagnostic biomarkers to differentiate adenocarcinoma and healthy subjects | (39) |
| miR-19-3p, miR-21-5p and miR-221-3p | Plasma | Diagnostic biomarkers for lung adenocarcinoma | (40) |
| miR-331-5p, miR-451a, miR-363-3p | Plasma | Diagnostic biomarkers for distinguishing SCLC from NSCLC | (41) |
| miR-451a, miR-486-5p, miR-363-3p, miR-660-5p, miR-15b-5p, miR-25-3p and miR-16-2-3p | Plasma | Diagnostic biomarkers to differentiate adenocarcinoma from healthy subjects | (41) |
| miR-126 | Plasma | Diagnostic biomarker for NSCLC | (42) |
| miR-21, miR-205 and miR-155 | Plasma | Diagnostic biomarkers for early-stage NSCLC | (43) |
| miR-let-7b-5p, miR-let-7e-5p, miR-23a-3p and miR-486-5p | Plasma | Diagnostic biomarkers of stage I NSCLC patients | (44) |
| miR-181-5p, miR-30a3p, miR-30e-3p and miR-361-5p | Plasma | Diagnostic biomarkers for lung adenocarcinoma | (44) |
| miR-10b-5p, miR-15b-5p and miR-320b | Plasma | Diagnostic biomarkers for SCC | (44) |
| miR-205, miR-19a, miR-19b, miR-30b and miR-20a | Plasma | Diagnostic biomarkers for SCC after surgery | (45) |
| miR-205-5p, miR-483-5p, miR-375, miR-200c-3p, miR-429, miR-200b-3p, miR-200a-3p, miR-203a-3p and miR-141-3p | Pleural fluid | Diagnostic biomarkers for discriminating lung adenocarcinoma over tuberculosis and other benign lesions | (47,48) |
| miR-200 | Pleural fluid | Diagnostic biomarker for discriminating lung adenocarcinoma from benign effusions | (49) |
| miR-182 and miR-210 | Pleural fluid | Diagnostic biomarkers for distinguishing lung adenocarcinoma from benign effusions | (50) |
Exosomal miRNAs as prognosis biomarkers in NSCLC
TNM stage is the most important determinant for predicting recurrence rates and survival times in NSCLC. Most of NSCLC patients are diagnosed at advanced stages leading to unfavourable prognosis (51).
An accurate prognosis prediction might reduce mortality and improve patient’s survival time. In resected early-stage cases, exosomal miRNAs might be useful for detecting minimal residual disease, allowing patients to take advantage of adjuvant treatments. To accomplish this, it would be necessary the clinical validation of exosomal miRNAs on the prognosis of early-stage disease. Up to date, some exosomal circulating miRNAs have been discovered as reliable biomarkers for NSCLC prognosis. Zhang et al. showed that high levels of serum exosomal miR-378 correlated with positive lymph node metastasis and advanced TNM stage. In consequence, serum exosomal miR-378 might be recognized as a biomarker of poor survival in NSCLC (52).
Another group demonstrated that high serum exosomal miRNA-214 was associated with advanced disease and, therefore, it could be used as a poor survival indicator in NSCLC patients (53).
It has also been revealed that plasma exosomal miR-10b-5p, miR-23b-3p and miR-21-5p were independent prognostic biomarkers in NSCLC (54). Moreover, the prognosis of NSCLC can also be affected by miR-195 (55).
Additionally, it has been demonstrated that the expression of exosomal miR-21 and miR-4257 was significantly increased during recurrence of NSCLC patients. Subsequently, the prognostic biomarker value of these miRNAs was confirmed in an extensive cohort of patients (56).
Furthermore, in advanced NSCLC, patients with high recurrence show lower levels of exosomal miR-146a-5p comparing with those with low recurrence (57).
Moreover, low level of exosomal let-7a-5p was recognized as poor survival biomarker in lung ADC. Interestingly, downregulation of this let-7a-5p miRNA in workers who are exposed to dust plays an important role in lung cancer development (58) (Table 2).
Table 2. Main studies focused on exosomal miRNAs used as prognostic and predictive biomarkers in NSCLC.
| Exosomal miRNAs | Source | Clinical value | Reference |
|---|---|---|---|
| miR-378 | Serum | Biomarker associated with poor survival in NSCLC | (52) |
| miRNA-214 | Serum | Biomarker associated with advance disease and poor survival indicator in NSCLC patients | (53) |
| miR-23b-3p, miR-10b-5p and miR-21-5p | Plasma | Independent prognostic biomarker of NSCLC | (54) |
| miR-21 and miR-4257 | Plasma | Prognostic biomarkers in NSCLC | (56) |
| miR-146a-5p | Serum | Predictive biomarker of the efficacy of Cisplatin in NSCLC and drug resistance | (57) |
| let-7a-5p | Serum | Biomarker associated with poor survival in lung adenocarcinoma | (58) |
| miR-29a-3p and miR-150-5p | Plasma | Biomarkers used to predict response or toxicity in radiotherapy treatment in NSCLC | (59) |
| miR-208a | Serum | Predictive biomarker of radiosensitivity in lung cancer | (60) |
| miR-425-3p | Serum | Predictive biomarker of the clinical response to platinum-based-chemotherapy in patients with NSCLC | (61) |
| miR-32 | Plasma | Predictive biomarker associated with the efficacy of platinum-based-chemotherapy and prognosis in NSCLC | (62) |
| miR-222-3p | Serum | Potential prognostic biomarker of Gemcitabine sensitivity in NSCLC | (63) |
| miR-221-3p and miR- 222-3p | Plasma | Predictive biomarkers associated with the response to Osimertinib in EGFR-mutated NSCLC | (64) |
| miR-320d, miR-320c, miR-320b and miR-125b-5p | Plasma | Predictive biomarker of the efficacy of immunotherapy in advance NSCLC | (65) |
Exosomal miRNAs as predictive biomarkers in NSCLC treatments
Despite substantial advances in chemotherapy, radiotherapy, targeted therapies and immunotherapy, many patients remain refractory to these therapies or acquire resistances during treatment. Therefore, the identification of patients who will derive benefit is a priority for treatment optimization. Looking for new biomarkers to guide patient selection and provide indications of efficacy and/or toxicity is essential. Exosomal miRNAs derived from peripheral blood or plasma samples might represent reliable predictive biomarkers for optimal treatment selection of patients with NSCLC.
Regarding radiotherapy treatment, exosomal miR-29a-3p and miR-150-5p were recognized as biomarkers of dose exposure, that could be used as predictive biomarkers of response and/or toxicity (59). Moreover, it has been shown that the presence in serum of exosomal miR-208a can affect the radiosensitivity of lung tumor cells by targeting p21 (60).
With respect to chemotherapy treatments, Yuwen et al. revealed that serum exosomal miR-146a-5p could be used as a biomarker for improving predictions of the efficacy of Cisplatin in NSCLC and monitoring of resistance to therapy (57). Some authors have also demonstrated exosomal miR-425-3p as a potential biomarker for predicting the clinical response to platinum-based-chemotherapy in patients with NSCLC (61). Furthermore, plasma levels of miR-32 are correlated with the efficacy of platinum-based-chemotherapy and prognosis in NSCLC (62).Recently, serum levels of exosomal miR-222-3p have been suggested to have a prognostic value for gemcitabine sensitivity in NSCLC (63).
In reference to targeted-therapies, exosomal plasma levels of miR-221-3p and miR-222-3p were linked to the response to Osimertinib in EGFR-mutated NSCLC patients (64).
More studies are needed to clarify the role of exosomal miRNAs in the regulation of tumors treated with TKIs-targeted therapies.
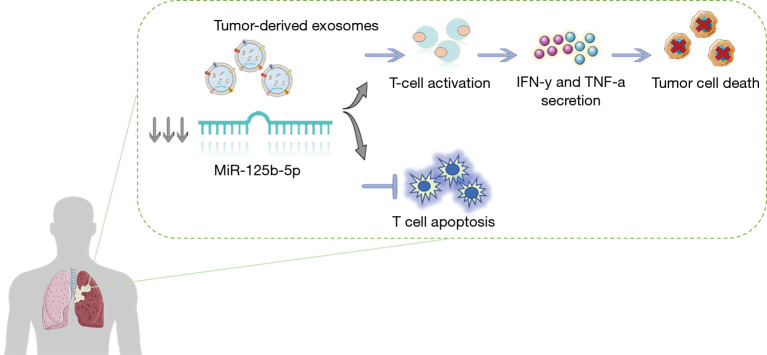
As for the most recent treatment strategies, plasma exosomal miR-320d, miR-320c and miR-320b have been identified as potential biomarkers for predicting the efficacy of immunotherapy in advanced NSCLC. Furthermore, in cases when the T-cell suppressor miR-125b-5p is downregulated during the treatment, patients achieve increased T-cell function and show an excellent response to immunotherapy (65) (Figure 1, Table 2).
Figure 1.
Diagram of the potential regulation process of miR-125-5p on T-cells.
Exosomal miRNAs as drug resistance biomarkers in NSCLC
The comprehension of drug resistance mechanisms is one of the main objectives to reduce the poor prognosis in lung cancer. Development of drug resistance to standardized chemotherapy as well as to targeted therapies, like tyrosine kinase inhibitors, or immunotherapy is one of the main problems in cancer (66-68). Accordingly, the study of the mechanisms of resistance potentially regulated by exosomal microRNAs will help to improve cancer treatments.
Wu et al. reported a significant higher expression of exosomal miR-96 in lung cancer compared to control lung tissues. This microRNA modifies the chemotherapeutic sensitivity by downregulating the drug resistance-related gene lime domain only protein 7 (LMO7). In consequence, by targeting LMO7, circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance (69).
Yuwen et al. demonstrated that higher serum exosomal levels of miR-146a-5p predicts increased therapeutic effect of Cisplatin in NSCLC patients, possibly by inhibiting autophagy-related protein 2 (ATG2) to suppress autophagy (57). Additionally, exosomal miR-512 and miR-373 have also been associated with high sensitivity to Cisplatin and suppression of tumor progression (70).
On the other hand, exosomal miRNAs can also be involved in resistance mechanisms to TKIs. In 2018, Jing et al. reported that exosomal transfer of miR-21 from Gefitinib-resistant H827R cells to Gefitinib-sensitive HCC827 cells activated AKT signalling, leading to Gefitinib resistance (71).
Same year, another group also revealed a significant higher expression of miR-214 in Gefitinib-resistant PC9GR cells and their derived exosomes than in Gefitinib-sensitive PC9 cells and exosomes. The authors reported that sensitive cells acquired resistance to Gefitinib when exosomal miR-214 was transferred from PC9GR cells to PC9 cells. Nevertheless, when PC9GR cells were transfected with a miR-214 antagomir, cells became sensitive to Gefitinib (72).
Accordingly, we can conclude that exosomal miRNAs can provide information on the molecular characteristics of the cells by which they were secreted and be endowed with the capacity to reprogram distant target cells. Moreover, exosomal miRNAs can regulate tumor resistance and represent a potential tool for monitoring the response/relapse of anti-neoplastic therapies.
Exosomal miRNAs as immune-oncology biomarkers in NSCLC microenvironment
The delivering of exosomal miRNAs by tumor and stromal cells in the microenvironment has been confirmed to be associated with tumor initiation, relapse, progression and metastasis. Exosomes can promote tumor progression by transferring immunosuppressive factors such as exosomal miRNAs, which can modulate the function of immune cells such as T-lymphocytes, dendritic cells (DCs) and natural killer (NK) cells (73).
Some exosomal miRNAs, such as miR-183, are TGF-β-inducible and can silence tumor-associated NK cells by targeting and suppressing other activating proteins (74).
Furthermore, the susceptibility to the lysis by cytotoxic T cells is regulated by hypoxia inducible miR-210. Hypoxic tumor-derived exosomes can negatively regulate NK cell function by a mechanism which involves TGF-β and miR-23a (75).
Another example of this immunomodulation in lung cancer cells is exosomal mir-21/29a, that is able to initiate tumor growth and metastasis through activation of TLR7 and TLR8 on immune cells (30). Yin et al. also confirmed communication between lung cancer cells and CD4+ lymphocytes through exosomal mir-214, which efficiently decreased PTEN expression, promoting regulatory T cell (Treg) expansion and tumor growth (76).
Additionally, lung cancer cell invasion has been described to be stimulated by exosomal transfer of miR-223 from platelets, through suppression of erythrocyte membrane protein band 4.1 like 3 (EPB41L3) (77). Finally, exosomal miR-193a-3p, miR-210-3p and miR-5100 from bone marrow-derived mesenchymal stem cells (BMSCs) are also reported to promote lung tumor cell invasion via activation of STAT3 signalling-induced EMT (32).
Conclusions
The discovery of relevant functions of exosomes has been a milestone in the searching of biomarkers in liquid biopsy. MicroRNAs, abundant elements in the exosome cargo, participate in the proliferation, metastasis, EMT, angiogenesis and immunomodulation processes in lung cancer. Furthermore, current researches have demonstrated the relevant use of exosomal miRNAs in the study of lung cancer, providing a significant advance in the clinical management. Due to their great potential, exosomal miRNAs can be used as an excellent non-invasive tool for early diagnosis, prognosis and prediction of treatment success or drug resistances in this pathology.
However, there are still challenges to be achieved. Firstly, because of the heterogeneity in size (different subtypes of vesicles), standardized methodologies must be established for isolation, characterization and study of exosomes cargo. Secondly, a complete comprehension of exosomal miRNA-mediated immunomodulation or drug resistance mechanisms is needed. Therefore, further studies are required to determine specific components and mechanisms of the exosomal miRNA to understand their role in cancer pathogenesis.
In conclusion, exosomal miRNAs have a considerable potential for the optimal management of NSCLC patients. Regardless multiple and recent technical and scientific developments in the field of liquid biopsy, the ultimate goal is to implement a useful and standardized methodology with high sensitivity/specificity for the isolation and analysis of exosomes and their content from minimally invasive samples, which might allow improvements in the prognosis, prediction and monitoring of the disease in the clinical practice.
Acknowledgments
Funding: This work is supported by Instituto de Salud Carlos III (ISCIII), Fondo de Investigación Sanitaria: CB16/12/00350 and PI18/00226. E Duréndez-Sáez has a predoctoral fellowship by Asociación Española Contra el Cáncer, Valencia (AECC Valencia). S. Torres-Martínez is supported by the Generalitat Valenciana and Fondo Social Europeo, fellowship ACIF/2018/275.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editors (Alfons Navarro, Joan Josep Castellano and Marina Díaz-Beyá) for the series “Clinic and Therapeutic Potential of Non-coding RNAs in Cancer” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure forms (available at https://dx.doi.org/10.21037/tcr-20-2815). The series “Clinic and Therapeutic Potential of Non-coding RNAs in Cancer” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. 10.1038/nature25183 [DOI] [PubMed] [Google Scholar]
- 3.Travis WD. Update on small cell carcinoma and its differentiation from squamous cell carcinoma and other non-small cell carcinomas. Mod Pathol 2012;25:S18-30. 10.1038/modpathol.2011.150 [DOI] [PubMed] [Google Scholar]
- 4.Torres S, González Á, Cunquero Tomas AJ, et al. A profile on cobas® EGFR Mutation Test v2 as companion diagnostic for first-line treatment of patients with non-small cell lung cancer. Expert Rev Mol Diagn 2020; 20:575-82. 10.1080/14737159.2020.1724094 [DOI] [PubMed] [Google Scholar]
- 5.Howlader N, Forjaz G, Mooradian MJ, et al. The Effect of Advances in Lung-Cancer Treatment on Population Mortality. N Engl J Med 2020;383:640-9. 10.1056/NEJMoa1916623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgensztern D, Ng SH, Gao F, et al. Trends in stage distribution for patients with non-small cell lung cancer: A national cancer database survey. J Thorac Oncol 2010;5:29-33. 10.1097/JTO.0b013e3181c5920c [DOI] [PubMed] [Google Scholar]
- 7.Mathai RA, Vidya R, Reddy B, et al. Potential Utility of Liquid Biopsy as a Diagnostic and Prognostic Tool for the Assessment of Solid Tumors: Implications in the Precision Oncology. J Clin Med 2019;8:373. 10.3390/jcm8030373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calabuig-Fariñas S, Jantus-Lewintre E, Herreros-Pomares A, et al. Circulating tumor cells versus circulating tumor DNA in lung cancer-which one will win? Transl Lung Cancer Res 2016;5:466-82. 10.21037/tlcr.2016.10.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rolfo C, Russo A. Liquid biopsy for early stage lung cancer moves ever closer. Nat Rev Clin Oncol 2020;17:523-4. 10.1038/s41571-020-0393-z [DOI] [PubMed] [Google Scholar]
- 10.Rolfo C, Cardona AF, Cristofanilli M, et al. Corrigendum to "Challenges and opportunities of cfDNA analysis implementation in clinical practice: Perspective of the International Society of Liquid Biopsy (ISLB)" [Crit. Rev. Oncol. Hematol. 151 (July) (2020) 102978]. Crit Rev Oncol Hematol 2020;154:103058. 10.1016/j.critrevonc.2020.103058 [DOI] [PubMed] [Google Scholar]
- 11.Molina-Vila MA, Mayo-de-Las-Casas C, Giménez-Capitán A, et al. Liquid Biopsy in Non-Small Cell Lung Cancer. Front Med (Lausanne) 2016;3:69. 10.3389/fmed.2016.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science 2020;367:eaau6977. 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pucci M, Taverna S, Reclusa P, et al. Exosomes in semen: opportunities as a new tool in prostate cancer diagnosis. Transl Cancer Res 2017;6:S1331-8. 10.21037/tcr.2017.10.25 [DOI] [Google Scholar]
- 14.Muller L, Simms P, Hong CS, et al. Human tumor-derived exosomes (TEX) regulate Treg functions via cell surface signaling rather than uptake mechanisms. Oncoimmunology 2017;6:e1261243. 10.1080/2162402X.2016.1261243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jella KK, Nasti TH, Li Z, et al. Exosomes, Their Biogenesis and Role in Inter-Cellular Communication, Tumor Microenvironment and Cancer Immunotherapy. Vaccines (Basel) 2018;6:69. 10.3390/vaccines6040069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meehan K, Vella LJ. The contribution of tumour-derived exosomes to the hallmarks of cancer. Crit Rev Clin Lab Sci 2016;53:121-31. 10.3109/10408363.2015.1092496 [DOI] [PubMed] [Google Scholar]
- 17.Rana S, Malinowska K, Zöller M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia 2013;15:281-95. 10.1593/neo.122010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giallombardo M, Taverna S, Alessandro R, et al. Exosome-mediated drug resistance in cancer: the near future is here. Ther Adv Med Oncol 2016;8:320-2. 10.1177/1758834016648276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan Z, Tian Y, Niu G, et al. Role of microRNAs in remodeling the tumor microenvironment (Review). Int J Oncol 2020;56:407-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014;15:509-24. 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- 21.Broughton JP, Lovci MT, Huang JL, et al. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol Cell 2016;64:320-33. 10.1016/j.molcel.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallach S, Calabuig-Fariñas S, Jantus-Lewintre E, et al. MicroRNAs: promising new antiangiogenic targets in cancer. Biomed Res Int 2014;2014:878450. 10.1155/2014/878450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997-1006. 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- 24.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108:5003-8. 10.1073/pnas.1019055108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles 2016;5:31292. 10.3402/jev.v5.31292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pucci M, Reclusa Asiáin P, Duréndez Sáez E, et al. Extracellular Vesicles As miRNA Nano-Shuttles: Dual Role in Tumor Progression. Target Oncol 2018;13:175-87. 10.1007/s11523-018-0551-8 [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Luo F, Wang B, et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett 2016;370:125-35. 10.1016/j.canlet.2015.10.011 [DOI] [PubMed] [Google Scholar]
- 28.Hsu YL, Hung JY, Chang WA, et al. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene 2017;36:4929-42. 10.1038/onc.2017.105 [DOI] [PubMed] [Google Scholar]
- 29.Aiello NM, Kang Y. Context-dependent EMT programs in cancer metastasis. J Exp Med 2019;216:1016-26. 10.1084/jem.20181827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabbri M, Paone A, Calore F, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A 2012;109:E2110-6. 10.1073/pnas.1209414109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valencia K, Luis-Ravelo D, Bovy N, et al. miRNA cargo within exosome-like vesicle transfer influences metastatic bone colonization. Mol Oncol 2014;8:689-703. 10.1016/j.molonc.2014.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Sai B, Wang F, et al. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol Cancer 2019;18:40. 10.1186/s12943-019-0959-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono K, Hiraoka T, Ono A, et al. Low-dose CT scan screening for lung cancer: comparison of images and radiation doses between low-dose CT and follow-up standard diagnostic CT. Springerplus 2013;2:393. 10.1186/2193-1801-2-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varela G, Thomas PA. Surgical management of advanced non-small cell lung cancer. J Thorac Dis 2014;6 Suppl 2:S217-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balata H, Fong KM, Hendriks LE, et al. Prevention and Early Detection for NSCLC: Advances in Thoracic Oncology 2018. J Thorac Oncol 2019;14:1513-27. 10.1016/j.jtho.2019.06.011 [DOI] [PubMed] [Google Scholar]
- 36.Tamkovich SN, Tutanov OS, Laktionov PP. Exosomes: Generation, structure, transport, biological activity, and diagnostic application. Biochem (Mosc) Suppl Ser A Membr Cell Biol 2016;10:163-73. [Google Scholar]
- 37.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006;9:189-98. 10.1016/j.ccr.2006.01.025 [DOI] [PubMed] [Google Scholar]
- 38.Rabinowits G, Gerçel-Taylor C, Day JM, et al. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 2009;10:42-6. 10.3816/CLC.2009.n.006 [DOI] [PubMed] [Google Scholar]
- 39.Cazzoli R, Buttitta F, Di Nicola M, et al. MicroRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol 2013;8:1156-62. 10.1097/JTO.0b013e318299ac32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou X, Wen W, Shan X, et al. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget 2017;8:6513-25. 10.18632/oncotarget.14311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poroyko V, Mirzapoiazova T, Nam A, et al. Exosomal miRNAs species in the blood of small cell and nonsmall cell lung cancer patients. Oncotarget 2018;9:19793-806. 10.18632/oncotarget.24857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grimolizzi F, Monaco F, Leoni F, et al. Exosomal miR-126 as a circulating biomarker in non-small-cell lung cancer regulating cancer progression. Sci Rep 2017;7:15277. 10.1038/s41598-017-15475-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai X, Friedman A. Exosomal miRs in lung cancer: A mathematical model. PLoS One 2016;11:e0167706. 10.1371/journal.pone.0167706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin X, Chen Y, Chen H, et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res 2017;23:5311-9. 10.1158/1078-0432.CCR-17-0577 [DOI] [PubMed] [Google Scholar]
- 45.Aushev VN, Zborovskaya IB, Laktionov KK, et al. Comparisons of microRNA Patterns in Plasma before and after Tumor Removal Reveal New Biomarkers of Lung Squamous Cell Carcinoma. PLoS One 2013;8:e78649. 10.1371/journal.pone.0078649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodríguez M, Silva J, López-Alfonso A, et al. Different exosome cargo from plasma/bronchoalveolar lavage in non-small-cell lung cancer. Genes Chromosomes Cancer 2014;53:713-24. 10.1002/gcc.22181 [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Xu YM, Zou YQ, et al. Identification of differential expressed PE exosomal miRNA in lung adenocarcinoma, tuberculosis, and other benign lesions. Medicine (Baltimore) 2017;96:e8361. 10.1097/MD.0000000000008361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin J, Wang Y, Zou YQ, et al. Differential miRNA expression in pleural effusions derived from extracellular vesicles of patients with lung cancer, pulmonary tuberculosis, or pneumonia. Tumour Biol 2016; 37:15835-45. 10.1007/s13277-016-5410-6 [DOI] [PubMed] [Google Scholar]
- 49.Hydbring P, De Petris L, Zhang Y, et al. Exosomal RNA-profiling of pleural effusions identifies adenocarcinoma patients through elevated miR-200 and LCN2 expression. Lung Cancer 2018;124:45-52. 10.1016/j.lungcan.2018.07.018 [DOI] [PubMed] [Google Scholar]
- 50.Tamiya H, Mitani A, Saito A, et al. Exosomal MicroRNA expression profiling in patients with lung adenocarcinoma-associated malignant pleural effusion. Anticancer Res 2018;38:6707-14. 10.21873/anticanres.13039 [DOI] [PubMed] [Google Scholar]
- 51.Woodard GA, Jones KD, Jablons DM. Lung Cancer Staging and Prognosis. Cancer Treat Res 2016;170:47-75. 10.1007/978-3-319-40389-2_3 [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Xu H. Serum exosomal miR-378 upregulation is associated with poor prognosis in non–small-cell lung cancer patients. J Clin Lab Anal 2020;34:e23237. 10.1002/jcla.23237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong H, Lu M, Wu F, et al. Exosomal microRNA-214 expression and its prognostic significance in non-small cell lung cancer patients. J King Saud Univ Sci 2020; 32:1060-4. 10.1016/j.jksus.2019.10.001 [DOI] [Google Scholar]
- 54.Liu Q, Yu Z, Yuan S, et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 2017;8:13048-58. 10.18632/oncotarget.14369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L, Feng T, Zhang W, et al. MicroRNA Biomarker hsa-miR-195-5p for Detecting the Risk of Lung Cancer. Int J Genomics 2020;2020:7415909. 10.1155/2020/7415909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dejima H, Iinuma H, Kanaoka R, et al. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol Lett 2017;13:1256-63. 10.3892/ol.2017.5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuwen DL, Sheng BB, Liu J, et al. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur Rev Med Pharmacol Sci 2017;21:2650-8. [PubMed] [Google Scholar]
- 58.Zhang L, Hao C, Zhai R, et al. Downregulation of exosomal let-7a-5p in dust exposed- workers contributes to lung cancer development. Respir Res 2018;19:235. 10.1186/s12931-018-0949-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dinh TKT, Fendler W, Chałubińska-Fendler J, et al. Circulating miR-29a and miR-150 correlate with delivered dose during thoracic radiation therapy for non-small cell lung cancer. Radiat Oncol 2016;11:61. 10.1186/s13014-016-0636-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang Y, Cui Y, Li Z, et al. Radiation-induced miR-208a increases the proliferation and radioresistance by targeting p21 in human lung cancer cells. J Exp Clin Cancer Res 2016;35:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuwen D, Ma Y, Wang D, et al. Prognostic role of circulating exosomal miR-425-3p for the response of NSCLC to platinum-based chemotherapy. Cancer Epidemiol Biomarkers Prev 2019;28:163-73. 10.1158/1055-9965.EPI-18-0569 [DOI] [PubMed] [Google Scholar]
- 62.Xu S, Li J, Chen L, et al. Plasma miR-32 levels in non-small cell lung cancer patients receiving platinum-based chemotherapy can predict the effectiveness and prognosis of chemotherapy. Medicine (Baltimore) 2019;98:e17335. 10.1097/MD.0000000000017335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei F, Ma C, Zhou T, et al. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol Cancer 2017;16:132. 10.1186/s12943-017-0694-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giallombardo M, Chacartegui JJ, Reclusa P, et al. Follow up analysis by exosomal miRNAs in EGFR mutated non-small cell lung cancer (NSCLC) patients during osimertinib (AZD9291) treatment: A potential prognostic biomarker tool. J Clin Oncol 2016;34:suppl.e23035. 10.1200/JCO.2016.34.15_suppl.e23035 [DOI] [Google Scholar]
- 65.Peng XX, Yu R, Wu X, et al. Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR/ALK wild-type advanced non-small cell lung cancer. J Immunother Cancer 2020;8:e000376. 10.1136/jitc-2019-000376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin JJ, Shaw AT. Recent Advances in Targeting ROS1 in Lung Cancer. J Thorac Oncol 2017;12:1611-25. 10.1016/j.jtho.2017.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pottier C, Fresnais M, Gilon M, et al. Tyrosine Kinase Inhibitors in Cancer: Breakthrough and Challenges of Targeted Therapy. Cancers (Basel) 2020;12:731. 10.3390/cancers12030731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bai R, Chen N, Li L, et al. Mechanisms of Cancer Resistance to Immunotherapy. Front Oncol 2020;10:1290. 10.3389/fonc.2020.01290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu H, Zhou J, Mei S, et al. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J Cell Mol Med 2017;21:1228-36. 10.1111/jcmm.13056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adi Harel S, Bossel Ben-Moshe N, Aylon Y, et al. Reactivation of epigenetically silenced miR-512 and miR-373 sensitizes lung cancer cells to cisplatin and restricts tumor growth. Cell Death Differ 2015;22:1328-40. 10.1038/cdd.2014.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jing C, Cao H, Qin X, et al. Exosome-mediated gefitinib resistance in lung cancer HCC827 cells via delivery of miR-21. Oncol Lett 2018;15:9811-7. 10.3892/ol.2018.8604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, Li M, Hu C. Exosomal transfer of miR-214 mediates gefitinib resistance in non-small cell lung cancer. Biochem Biophys Res Commun 2018;507:457-64. 10.1016/j.bbrc.2018.11.061 [DOI] [PubMed] [Google Scholar]
- 73.Greening DW, Gopal SK, Xu R, et al. Exosomes and their roles in immune regulation and cancer. Semin Cell Dev Biol 2015;40:72-81. 10.1016/j.semcdb.2015.02.009 [DOI] [PubMed] [Google Scholar]
- 74.Donatelli SS, Zhou JM, Gilvary DL, et al. TGF-β-inducible microRNA-183 silences tumor-associated natural killer cells. Proc Natl Acad Sci U S A 2014;111:4203-8. 10.1073/pnas.1319269111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Noman MZ, Buart S, Romero P, et al. Hypoxia-inducible miR-210 regulates the susceptibility of tumor cells to lysis by cytotoxic T cells. Cancer Res 2012;72:4629-41. 10.1158/0008-5472.CAN-12-1383 [DOI] [PubMed] [Google Scholar]
- 76.Yin Y, Cai X, Chen X, et al. Tumor-secreted miR-214 induces regulatory T cells: A major link between immune evasion and tumor growth. Cell Res 2014;24:1164-80. 10.1038/cr.2014.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liang H, Yan X, Pan Y, et al. MicroRNA-223 delivered by platelet-derived microvesicles promotes lung cancer cell invasion via targeting tumor suppressor EPB41L3. Mol Cancer 2015;14:58. 10.1186/s12943-015-0327-z [DOI] [PMC free article] [PubMed] [Google Scholar]