Abstract
Although Francisella tularensis is a well-known, highly virulent bacterium that causes tularemia in humans, other Francisella species have been associated with sporadic human infections. We describe a human cutaneous infection with bacteremia caused by F. salimarina, a Francisella species recently identified from seawater and fishes, in an immunocompromised patient in France.
Keywords: Francisella salimarina, Francisella species, opportunistic infections, France, bacteria, zoonoses
Although the taxonomy of the genus Francisella includes a wide diversity of species, only F. tularensis subspecies tularensis and F. tularensis subsp. holarctica cause the potentially life-threatening disease tularemia (1). Several Francisella spp., including F. philomiragia, F. novicida, F. opportunistica, and F. hispaniensis, are occasional opportunistic human pathogens; the other Francisella spp. are not associated with human infections (1). We describe a human infection caused by F. salimarina, recently identified from aquatic environments and fishes.
In June 2017, a 76-year-old man received a diagnosis of acute myelomonocytic leukemia and was admitted to Poitiers University Hospital (Poitiers, France). The patient lived in a small town 30 km from the Atlantic Ocean, had not travelled abroad recently, and had no pets. The day after admission, first-line chemotherapy of subcutaneous azacitidine was started for 7 days. After 3 days of chemotherapy, piperacillin/tazobactam was introduced for 5 days because of febrile aplasia. The patient was then discharged with an antibiotic prophylaxis (sulfamethoxazole/trimethoprim at 800 mg/160 mg 3×/wk). On July 26, the second azacitidine treatment was not administered because the patient again experienced febrile aplasia. Physical examination revealed skin lesions on 2 left-hand fingers that had appeared 3 weeks earlier. These lesions were erythematous and crusty but not purulent (Figure, panel A). They were associated with a left axillary lymphadenopathy. Antibiotic treatment with piperacillin/tazobactam and teicoplanin was started but was changed to imipenem/cilastatin and daptomycin after 5 days because of poor clinical response. Aerobic blood cultures performed at admission tested positive on July 31 and Gram stain showed a small gram-negative coccobacillus (Figure, panel B). Antibiotic treatment was changed to cefepime, administered for 3 days. No identification could be obtained by MALDI-TOF (matrix-assisted laser desorption/ionization time-of-flight) mass spectrometry (Vitek MS; bioMérieux, https://www.biomerieux.com). The strain was identified as a Francisella spp. by 16s rDNA amplification and sequencing. A cutaneous biopsy was performed because of persistent fever and worsening skin lesions in the patient; the same Francisella spp. strain was isolated. Doxycycline (100 mg 2×/d) was administered for 8 days, followed by sulfamethoxazole/trimethoprim, which led to apyrexia.
Figure.
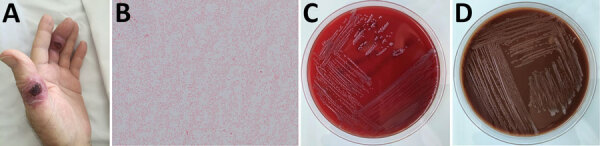
Skin ulcers and bacteremia caused by Francisella salimarina in an immunocompromised patient and isolated bacteria morphology, France. A) Skin lesion on 2 left-hand fingers. B) Small gram-negative coccobacillus isolated from blood and skin lesions (original magnification ×1,000). C) Growth on blood agar after 2 days of incubation at 35°C in 5% CO2. D) Growth on chocolate agar after 2 days of incubation at 35°C in 5% CO2.
The Francisella spp. strain (referred to as CHUGA-F75) was sent to the French National Reference Centre for Francisella for further characterization. The strain was strictly aerobic and grew well on chocolate agar supplemented with IsoVitaleX (bioMérieux), blood agar, and tryptic soy agar, yielding gray mucoid colonies after 24 h of incubation at 35°C in 5% CO2, but not on Drigalski agar (Figure, panels C, D). Biochemical testing revealed a positive oxidase, a weakly positive catalase, and a negative urease test. The strain was also halotolerant; it could grow in modified Mueller-Hinton broth with up to 8% NaCl. ISFtu2, Tul4, and type B real-time PCR tests, which detected most Francisella spp., F. tularensis, and F. tularensis subsp. holarctica, were all negative for DNA extracted from this strain (2,3). Species identification could not be obtained by using MALDI-TOF mass spectrometry, either with the routine database (MBT IVD Library DB-7171), the Biotox database (MBT SR Library; both from Bruker, https://www.bruker.com), or the French National Reference Centre for Francisella database containing F. tularensis, F. novicida, and F. philomiragia (4). Therefore, we performed whole-genome sequencing by using second and third next-generation sequencing platforms MiSeq (Illumina, https://www.illumina.com) and MinION (Oxford Nanopore Technologies, https://nanoporetech.com). Hybrid assembly of the sequencing data using Unicycler software on the Galaxy web platform (https://usegalaxy.org) enabled circularization of a 1,940,863 bp bacterial chromosome (Genbank accession no. CP076680). Whole-genome–based identification of the strain was assessed by using the Type Strain Genome Server (https://tygs.dsmz.de) (5). The CHUGA-F75 strain clustered in the same branch as the F. salimarina SYSU SYW-1, the F. marina E95-16, and the F. salina TX07-7308 strains (Appendix Figure), probably representing the same species because of high genetic homology, although different species names have been published (6–8). Digital DNA-DNA hybridization >70%, average nucleotide identity >95%, and difference in percent guanine-cytosine content <1 percent between the CHUGA-F75 strain and the 3 F. salimarina, F. marina, and F. salina strains confirmed the CHUGA-F75 isolate belonged to the same species. Because the only validly published species name according to the International Code of Nomenclature of prokaryotes is F. salimarina, we identified CHUGA-F75 as F. salimarina. Using the broth microdilution method in cation-adjusted Mueller-Hinton broth as recommended by the Clinical and Laboratory Standards Institute, we found that the CHUGA-F75 strain was sensitive to gentamicin (MIC = 0.125 mg/L), doxycycline (MIC = 1 mg/L), and ciprofloxacin (MIC = 0.016 mg/L) and resistant to sulfamethoxazole/trimethoprim (MIC = 32 mg/L).
F. marina was described as responsible for systemic disease in fishes (Lutjanus guttatus, the cultured spotted rose snapper) in Central America, whereas 4 F. salimarina strains have been isolated from costal seawater in Guangdong Province, China, and 1 strain of F. salina has been grown from brackish seawater and seaweed off the coast of Galveston, Texas, USA (6–8). To our knowledge, these Francisella spp. were not responsible for human infection so far. This report, like previous descriptions of human infections caused by emergent Francisella spp., highlights that environmental or fish-related Francisella spp. could be responsible for opportunistic human infections resembling tularemia.
Additional information about ulceroglandular infection and bacteremia caused by Francisella salimarina in immunocompromised patient, France
Acknowledgments
The Direction Générale de l’Armement of France funded this research (ANR-17-ASTR-0024).
Biography
Dr. Hennebique is a clinical microbiologist in the bacteriology laboratory of Grenoble University Hospital, Grenoble, France, which also hosts the French National Reference Center for Francisella tularensis. Her primary research interests are the tularemia agent, with an emphasis on its mechanisms of environmental survival in water, susceptibility to antibiotics, and virulence.
Footnotes
Suggested citation for this article: Hennebique A, Caspar Y, Maurin M, Boisset S, Pelloux I, Gallego-Hernanz MP, et al. Ulceroglandular infection and bacteremia caused by Francisella salimarina in immunocompromised patient, France. Emerg Infect Dis. 2022 Feb [date cited]. https://doi.org/10.3201/eid2802.211380
These authors contributed equally to this article.
References
- 1.Hennebique A, Boisset S, Maurin M. Tularemia as a waterborne disease: a review. Emerg Microbes Infect. 2019;8:1027–42. 10.1080/22221751.2019.1638734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Versage JL, Severin DDM, Chu MC, Petersen JM. Development of a multitarget real-time TaqMan PCR assay for enhanced detection of Francisella tularensis in complex specimens. J Clin Microbiol. 2003;41:5492–9. 10.1128/JCM.41.12.5492-5499.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kugeler KJ, Pappert R, Zhou Y, Petersen JM. Real-time PCR for Francisella tularensis types A and B. Emerg Infect Dis. 2006;12:1799–801. 10.3201/eid1211.060629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Regoui S, Hennebique A, Girard T, Boisset S, Caspar Y, Maurin M. Optimized MALDI TOF mass spectrometry identification of Francisella tularensis subsp. holarctica. Microorganisms. 2020;8:E1143. 10.3390/microorganisms8081143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. 10.1038/s41467-019-10210-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Challacombe JF, Petersen JM, Gallegos-Graves V, Hodge D, Pillai S, Kuske CR. Whole-genome relationships among Francisella bacteria of diverse origins define new species and provide specific regions for detection. Appl Environ Microbiol. 2017;83:e02589–16. 10.1128/AEM.02589-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L-H, Luo H-M, Feng J-H, Ming Y-Z, Zheng M-L, Deng G-Y, et al. Francisella salimarina sp. nov., isolated from coastal seawater. Int J Syst Evol Microbiol. 2020;70:3264–72. 10.1099/ijsem.0.004164 [DOI] [PubMed] [Google Scholar]
- 8.Soto E, Griffin MJ, Morales JA, Calvo EB, de Alexandre Sebastião F, Porras AL, et al. Francisella marina sp. nov., etiologic agent of systemic disease in cultured Spotted Rose Snapper (Lutjanus guttatus) in Central America. Appl Environ Microbiol. 2018;84:e00144–18. 10.1128/AEM.00144-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information about ulceroglandular infection and bacteremia caused by Francisella salimarina in immunocompromised patient, France


