Abstract
Linear B-cell epitopes of the Rhodococcus equi virulence-associated protein (VapA) were mapped using a synthetic peptide bank in this study. The peptides were screened in an enzyme-linked immunosorbent assay (ELISA) with a total of 70 sera from foals with current R. equi disease (51 sera), as well as from foals that had either recovered from R. equi infection 10 months previously (3 sera) or that had no known history of R. equi disease (16 sera). An epitope with the sequence NLQKDEPNGRA was identified and was universally recognized by all 51 sera from foals with R. equi disease and was not recognized by any of the other sera. There was poor reactivity between all sera and peptides relating to other areas of the VapA protein. It is proposed that an ELISA based upon a defined peptide epitope may be used in an improved serological diagnostic test for R. equi infection in foals.
Rhodococcus equi is a facultatively aerobic, gram-positive bacterium considered to be a soil saprophyte (3). R. equi is a significant pathogen in foals, causing a pyogranulomatous pneumonia which is sometimes accompanied by extrapulmonary manifestations such as bacteremia, lymphadenitis, and enteritis (2, 16). R. equi is also known to sometimes cause severe pulmonary and disseminated disease in immunocompromised humans, particularly in AIDS patients (4).
Currently, an important virulence-associated factor identified in R. equi disease in foals is considered to be the virulence-associated protein, VapA (10, 22). This protein is 15 to 17 kDa and is encoded by a gene present on a large 85- to 90-kb plasmid. Previous studies have shown that the majority of clinical R. equi isolates from foals, unlike environmental isolates, typically contain this virulence plasmid (8, 20). Giguere et al. have shown that this plasmid plays a role in the intracellular survival and replication of R. equi in host macrophages and consequently is an important factor for the development of R. equi disease in foals (5). Importantly, studies have shown that the detection of significant levels of antibody to semipurified VapA is a reasonable marker of virulent R. equi infection in foals (14; S. A. Hines and S. K. Hietala, Editorial, Equine Vet. J. 28:339–340, 1996).
The aim of this study was to identify linear B-cell epitopes of VapA by screening a synthetic peptide library based upon this protein with sera from R. equi infected foals in an enzyme-linked immunosorbent assay (ELISA). Similar studies have been carried out on other antigens associated with bacterial pathogens with success (12, 13).
Peptide assay.
Biotinylated peptides synthesized by Mimotopes, Victoria, Australia, were used in all of the assays. The peptide bank used in the initial screening of sera was designed based upon the published sequence of VapA (GenBank accession no. D21236) (18). A total of 50 overlapping peptides, each 11 amino acid residues in length (offset by 3 residues at a time), were synthesized beginning from the predicted signal peptide cleavage site between amino acids 31 and 32 up to and including the C terminus.
A second set of peptides was used to further define the region between peptides 11 and 14 (LQKDEPNGRASD) of the VapA protein. A total of 19 peptides were designed based upon this region; 12 were truncated peptides and contained single stepwise amino acid deletions starting from either the N or the C terminus. Six peptides were overlapping 6-mers and covered the sequence offset by one residue at a time beginning at the N terminus. A final peptide KDEPNGR was designed based upon the core sequence of the B-cell epitope identified in assays using the previous 18 peptides.
A total of 70 foal sera, most of these from animals aged between 4 to 12 weeks, were used to screen the peptides. Fifty-one sera were from foals with current R. equi disease (positive sera). Sixteen sera were from foals with no known history of R. equi infection, and three sera were from foals that had recovered from R. equi infection 10 months previously (negative sera). Thirty-nine of the positive sera and all negative sera were obtained from studs in South Australia and New South Wales, Australia. The remaining sera were obtained elsewhere (Table 1).
TABLE 1.
Sera used in this study and their reactivity in peptide and whole-cell assays
| Serum (n = 70) | Locationa | Clinical backgroundb | Positive peptide(s) in region of B-cell epitope | Results with whole-cell ELISA |
|---|---|---|---|---|
| LP1 | SA | Clinical R. equi diseasec | 11, 12, 13 | Positive |
| LP2 | SA | Clinical R. equi disease | 12, 16 | Positive |
| LP3 | SA | Clinical R. equi disease | 12, 13 | Positive |
| LP4 | SA | Clinical R. equi disease | 11, 12, 13 | Negative |
| LP5 | SA | Clinical R. equi disease | 12, 13 | Negative |
| LP6 | SA | Clinical R. equi disease | 11, 12, 13 | Positive |
| LP7 | SA | Clinical R. equi disease | 12 | Positive |
| LP8 | SA | Clinical R. equi disease | 11, 12, 13 | Positive |
| LP9 | SA | Clinical R. equi disease | 12, 13 | Positive |
| LP10 | SA | Clinical R. equi disease | 11, 12, 13 | Negative |
| LP11 | SA | Clinical R. equi disease | 11, 12, 13 | Positive |
| LP12 | SA | Clinical R. equi disease | 11, 12, 13 | Positive |
| LP13 | SA | Clinical R. equi disease | 11, 12, 13 | Positive |
| LP14 | SA | Clinical R. equi disease | 11, 12, 13, 14 | Positive |
| LP15 | SA | Clinical R. equi disease | 11, 12, 13 | Negative |
| LP16 | SA | Clinical R. equi disease | 12, 13 | Positive |
| LP17 | SA | Clinical R. equi disease | 12, 13 | Positive |
| LP18 | SA | No history of R. equi disease | None | Positive |
| LP19 | SA | No history of R. equi disease | None | Negative |
| LP20 | SA | No history of R. equi disease | None | Negative |
| LP21 | SA | No history of R. equi disease | None | Negative |
| LP22 | SA | No history of R. equi disease | None | Positive |
| LP23 | SA | No history of R. equi disease | None | Positive |
| LP24 | SA | No history of R. equi disease | None | Negative |
| LP25 | SA | No history of R. equi disease | None | Positive |
| LP26 | SA | No history of R. equi disease | None | Negative |
| LP27 | SA | No history of R. equi disease | None | Positive |
| LP28 | SA | No history of R. equi disease | None | Positive |
| LP29 | SA | No history of R. equi disease | None | Negative |
| LP30 | SA | R. equi infection 10 mo previously; animal recovered completely | None | Positive |
| LP31 | SA | R. equi infection 10 mo previously; animal recovered completely | None | Positive |
| LP32 | SA | R. equi infection 10 mo previously; animal recovered completely | None | Negative |
| LP33 | SA | No history of R. equi disease | None | Positive |
| LP34 | SA | No history of R. equi disease | None | Negative |
| LP35 | SA | No history of R. equi disease | None | Negative |
| LP36 | SA | Clinical R. equi disease | 11, 12, 13 | Negative |
| GL1 | SA | Clinical R. equi disease | 12, 13 | Positive |
| GL2 | SA | No history of R. equi disease | None | Positive |
| SC11420 | NSW | Clinical R. equi disease | 12, 13 | Positive |
| SC11968 | NSW | Clinical R. equi disease | 11, 12, 13 | Positive |
| SC12164 | NSW | Clinical R. equi disease | 11, 12, 13 | Positive |
| SC12171 | NSW | Clinical R. equi disease | 11, 12, 13 | Positive |
| SC9275 | NSW | Clinical R. equi disease | 11, 12, 13 | Positive |
| SC9278 | NSW | Clinical R. equi disease | 11, 12, 13 | Positive |
| SC12149 | NSW | Clinical R. equi disease | 11, 12, 13, 14 | Positive |
| SC9276 | NSW | Clinical R. equi disease | 12, 14 | Negative |
| SC9277 | NSW | Clinical R. equi disease | 12, 13 | Positive |
| SC1 | NSW | Clinical R. equi disease | 12, 13, 14 | Negative |
| SC6626 | NSW | Clinical R. equi disease | 11, 12, 13, 14 | Negative |
| SC2 | NSW | Clinical R. equi disease | 12, 13 | Positive |
| SC7104 | NSW | Clinical R. equi disease | 11, 12, 13, 14 | Positive |
| SC7053 | NSW | Clinical R. equi disease | 11, 12, 13, 14 | Positive |
| SC3 | NSW | Clinical R. equi disease | 11, 12, 13 | Negative |
| SC6586 | NSW | Clinical R. equi disease | 11, 12, 13, 14 | Positive |
| SC4 | NSW | Clinical R. equi disease | 11, 12, 13 | Positive |
| SC5 | NSW | Clinical R. equi disease | 12, 13 | Positive |
| SC6278 | NSW | Clinical R. equi disease | 11, 12, 13 | Positive |
| SC6794 | NSW | Clinical R. equi disease | 11, 12, 13, 14 | Positive |
| JA10 | Japan | Clinical R. equi disease | 11, 12, 13, 14 | Positive |
| JA11 | Japan | Clinical R. equi disease | 11, 12, 13 | Negative |
| JA2 | Japan | Clinical R. equi disease | 11, 12, 13 | Positive |
| JA5 | Japan | Clinical R. equi disease | 11, 12, 13, 14 | Positive |
| JA3 | Japan | Clinical R. equi disease | 11, 12, 13 | Positive |
| JA1 | Japan | Clinical R. equi disease | 11, 12, 13, 14 | Positive |
| JA9 | Japan | Clinical R. equi disease | 11, 12, 13, 14 | Negative |
| JA6 | Japan | Clinical R. equi disease | 11, 12 | Positive |
| JA4 | Japan | Clinical R. equi disease | 12 | Negative |
| JA7 | Japan | Clinical R. equi disease | 11, 12, 13, 14 | Negative |
| JA8 | Japan | Clinical R. equi disease | 11, 12, 13, 14 | Positive |
| US1 | United States | Clinical R. equi disease | 11, 12, 13, 14 | Positive |
SA, South Australia; NSW, New South Wales (Australia).
Diagnosis of R. equi disease was based upon symptoms, culture, and/or radiography.
The ELISA using the biotinylated peptides was performed by the method recommended by the manufacturer with the following modifications: Neutravidin (Pierce Chemical Company) at a concentration of 0.3 μg/well was used to coat the plates. Plates were blocked for 2 h at 4°C. The conjugate diluent used contained 1% casein to decrease the nonspecific binding. The secondary antibody used was caprine anti-horse immunoglobulin G (Bethyl Laboratories). The whole-cell antigen was prepared with R. equi ATCC 33701 (known to contain the VapA-encoding virulence plasmid) using a previously described method (1). Briefly, bacteria cultured on plates for 48 h were harvested, washed with physiological saline, and fixed overnight in 1% formalin in saline. The following day the bacteria were washed three times in saline, resuspended in bicarbonate coating buffer, and used in the ELISA. The whole-cell preparation was not tested for the presence of VapA or any other specific antigens.
Tetramethyl benzidine was used as the chromogenic substrate in all assays, and optical density (OD) values were read in an ELISA plate reader at a wavelength of 450 nm (reference wavelength, 630 nm).
Interpretation of data.
In the initial assay using the 50 peptides in the overlapping bank of the entire VapA protein, a positive result was assigned by using a cutoff value of twice the background OD. The background OD was the mean of the lowest 50% of all OD values obtained with that particular serum, and all OD readings that were twice this value were considered positive. The background ranged from 0.04 to 0.3, indicating a high degree of variability in the reactivity of sera with the peptide bank.
In the assay to identify the most reactive peptides containing elements of region LQKDEPNGRASD, the cutoff OD value for a positive result was determined using the mean value of the lowest 25% of all OD values obtained with that serum (range, 0.13 to 0.78) plus three times their standard deviation. All OD readings above the cutoff point were considered positive.
A positive result in the R. equi whole-cell ELISA was based upon twice the background OD. The background OD (0.07 to 0.37) was the OD value of the well containing all reagents and sera used in the corresponding test assay without the whole-cell antigen preparation. All OD values above the background OD were considered positive.
Screening of sera with peptide bank of VapA sequence.
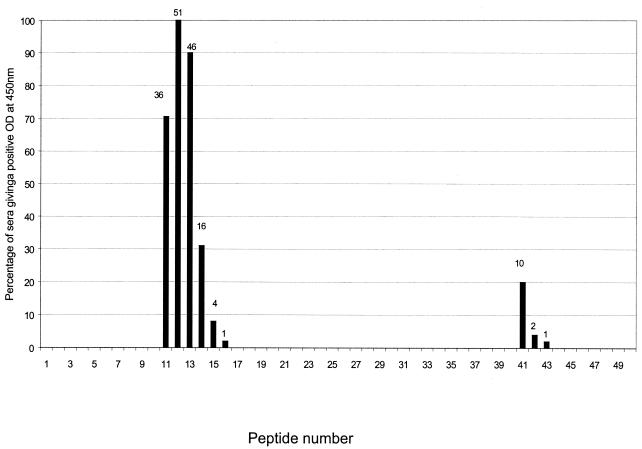
The 51 positive sera screened against 50 peptides recognized an epitope between amino acids 62 to 81 of the VapA sequence corresponding to peptides 11 to 14 (OD values between 0.25 and 1.5). The amino acid sequences of these peptides are TSLNLQKDEPN (peptide 11), NLQKDEPNGRA (peptide 12), KDEPNGRASDT (peptide 13), and PNGRASDTAGQ (peptide 14). Peptide 12 was universally recognized by all 51 sera associated with current R. equi infection. Forty-nine of these sera recognized at least two peptides in this region, and two sera were positive with only peptide 12 (Table 1). Thirteen sera were positive with all four peptides, i.e., peptides 11 to 14. Four sera were positive with peptides 15 or 16 in addition to reacting with at least one peptide in the region from peptides 11 to 14 (Fig. 1).
FIG. 1.
Fifty-one positive sera used to screen the fifty overlapping peptides derived from the mature form of VapA in an ELISA. A positive result is considered to be at least twice the background OD at 450 nm (background OD range, 0.04 to 0.3). Numbers above columns indicate the actual number of sera reacting with a given peptide.
In addition, 11 of the positive sera also reacted positively with one or two of the peptides from 41 to 43 (region between amino acids 152 and 168 of VapA). The sequence of this secondary epitope did not have any similarity to the sequence encompassed by peptides 11 to 14. Ten of these sera gave a positive result with peptide 41, which corresponded to sequence YLNINFFDSSG (Fig. 1).
Apart from these peptides, six sera reacted positively with peptides from other regions of VapA (data not shown) and generally had OD readings ranging from 0.13 to 0.6; however, these readings were much less than those obtained with peptides 11 to 14 (0.25 to 1.5).
All sera from animals with no known history of R. equi infection gave a negative result with peptides 11 to 14.
These assays show that a major linear epitope of VapA lies in the region between peptides 11 and 14 corresponding to amino acids 62 to 81 of the VapA precursor protein sequence. Based on the universal reactivity of peptide NLQKDEPNGRA, it is likely that a B-cell epitope is in this region of VapA.
The region between peptides 11 and 14 of VapA contains predominantly hydrophilic residues, and analysis of the precursor VapA sequence using the Hopp and Woods hydrophobicity algorithm indicated that the region corresponding to peptides 11 to 14 was the most hydrophilic region of the entire protein (11). Studies have shown VapA to be a lipid-modified, hydrophobic, surface-expressed protein (19). Therefore, it would be expected that the hydrophilic region of this protein would lie on the cell surface and consequently be more likely to interact with the host immune system. Interestingly, the minor epitope identified between peptides 41 and 43 was within the hydrophobic region of the VapA protein. This may mean that occasionally non-cell-surface-exposed regions of VapA do interact with the host immune system, although to a much lesser extent than the major cell surface domain of the protein.
Further definition of the B-cell epitope within NLQKDEPNGRASD.
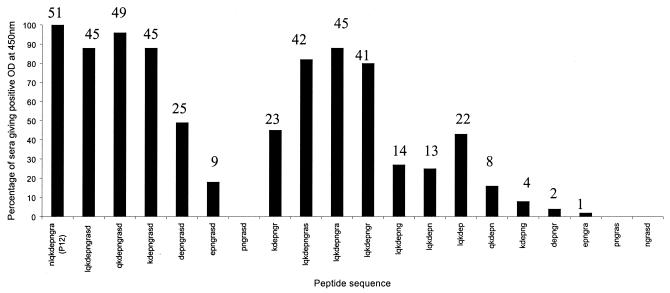
The 51 positive sera were tested against the peptides LQKDEP, QKDEPN, KDEPNG, DEPNGR, EPNGRA, PNGRAS, NGRASD, LQKDEPNGRASD, LQKDEPNGRAS, LQKDEPNGRA, LQKDEPNGR, LQKDEPNG, LQKDEPN, QKDEPNGRASD, KDEPNGRASD, DEPNGRASD, EPNGRASD, and PNGRASD. All of these sera gave relatively high OD values (between 0.5 and 1.17) with peptides containing KDEPNGR as part of their sequence. However, none of the peptides was universally recognized by all of the positive sera tested, unlike peptide 12 from the initial VapA peptide bank (Fig. 2).
FIG. 2.
Further definition of the B-cell epitope within LQKDEPNGRASD using peptides derived from that sequence. Positive peptides were defined as having an OD of at least three times the standard deviation above the lowest 25% of all OD values at a 450-nm wavelength (OD range, 0.13 to 0.78). Numbers above columns indicate actual numbers of sera reacting with a given peptide. The universally reactive peptide 12 (NLQKDEPNGRA) has been included for comparison.
These results indicated that it was not possible to further define or shorten the epitope NLQKDEPNGRA since shortening of the peptide in this region resulted in decreased OD readings with some sera. Similar results relating to studies on various non-R. equi antigens have been reported by some researchers (S. Rodda, personal communication) and may occur because some antibodies in a particular serum react more avidly with certain amino acid residues than with others within the epitope. This would mean that a shortened peptide may only react with a proportion of VapA antibodies present in a particular serum, resulting in a weak-positive or false-negative reaction. Previous studies relating to the epitope mapping of Mycoplasma bovis variable surface lipoproteins have shown B-cell epitopes to be around three to seven amino acid residues long (17). However, other studies have shown universally reactive B-cell epitopes relating to bacterial antigens to be as long as 13 or even 15 residues (6, 25), similar to the size of the 11-mer epitope NLQKDEPNGRA described here.
Comparison of peptide-based ELISA with a whole-R. equi-cell-based ELISA.
Thirteen positive sera in the peptide assay were negative in an ELISA based upon whole R. equi cells. These sera gave lower OD readings (0.25 to 0.7) compared with the remainder of the positive sera (0.7 to 1.7) in the peptide assay. This indicates that with weakly positive sera, the whole-cell assay was not as sensitive as the peptide assay.
Importantly, 10 negative sera in the peptide-based assay were positive with the whole-cell ELISA. These may be false-positive results due to the whole-cell assay detecting the presence of antibodies to non-VapA antigens, expressed by nonpathogenic environmental R. equi, that the foals had been exposed to.
The results clearly indicate that the peptide-based ELISA is superior to that based upon whole cells because of its greater sensitivity. In addition, the whole-cell assay was more time-consuming and labor-intensive to perform compared to the peptide-based test. The peptide assay also has the advantage in that the target peptide antigen can be more readily quality controlled and easily produced in contrast to a whole-cell extract.
Significance of the linear B-cell epitope of VapA.
There have been other ELISA techniques developed for R. equi serology in the past; these have mainly been assays using antigens of unknown identity from R. equi, including Tween 20 extracts of a VapA-negative R. equi ATCC 6939 (7, 21) and lyophilized supernatant antigens of various strains of R. equi (9). Consequently, these assays did not specifically detect antibodies to VapA. Furthermore, earlier studies have demonstrated the presence of antibody against presumably nonpathogenic R. equi in healthy horses and foals. This highlights the importance of detection of VapA-specific antibodies in order to differentiate between a clinical R. equi infection and environmental exposure to nonvirulent R. equi (9). A VapA-based ELISA developed previously was found to be more useful with respect to specificity; however, it was not suitable for use as a routine diagnostic test, since VapA protein extraction was laborious and possibly contained other antigens in addition to VapA (14). Therefore, an ELISA based on universally reactive synthetic peptide epitopes of the VapA protein has the potential to be used as an easy-to-perform and specific routine diagnostic test.
Recently, Takai et al. described six other virulence-associated proteins—VapC, VapD, VapE, VapF, VapG, and VapH—encoded by genes on the 80- to 90-kb R. equi virulence plasmid (24). In addition, VapB associated with intermediately virulent R. equi on a second plasmid has also been described (23). None of these proteins contained regions of significant homology to the B-cell epitope identified in this study (23, 24).
In conclusion, the B-cell epitope of VapA identified in this study may be used in a convenient and reliable diagnostic test for R. equi disease. Previous studies in the murine model indicate that VapA may be important in the immune response to R. equi (15), it is therefore possible that the epitope identified in this study may be used as a component of a vaccine for the prevention of R. equi disease in foals, particularly as a component in a multivalent combination vaccine.
Acknowledgments
We thank the Rural Industries Research and Development Corporation and the University of South Australia for funding this project.
We thank Institute of Medical and Veterinary Science for the use of equipment and facilities. We are grateful to Campbell Baker, Angela Begg, Shinji Takai, Barbara Byrne, Glenn Browning, Phil Houston, and Anna Morton for kindly providing the sera used in this study and Stuart Rodda of Mimotopes for helpful discussions.
REFERENCES
- 1.Barka N, Tomasi J P, Stadtsbaeder S. ELISA using whole Legionella pneumophila cell as antigen. Comparison between monovalent and polyvalent antigens for the serodiagnosis of human legionellosis. J Immunol Methods. 1986;93:77–81. doi: 10.1016/0022-1759(86)90435-7. [DOI] [PubMed] [Google Scholar]
- 2.Barton M D, Embury D H. Studies of the pathogenesis of Rhodococcus equi infection in foals. Aust Vet J. 1987;64:332–339. doi: 10.1111/j.1751-0813.1987.tb06061.x. [DOI] [PubMed] [Google Scholar]
- 3.Barton M D, Hughes K L. Ecology of Rhodococcus equi. Vet Microbiol. 1984;9:65–76. doi: 10.1016/0378-1135(84)90079-8. [DOI] [PubMed] [Google Scholar]
- 4.Capdevila J A, Bujan S, Gavalda J, Ferrer A, Pahissa A. Rhodococcus equi pneumonia in patients infected with the human immunodeficiency virus. Report of 2 cases and review of the literature. Scand J Infect Dis. 1997;29:535–541. doi: 10.3109/00365549709035890. [DOI] [PubMed] [Google Scholar]
- 5.Giguere S, Hondalus M K, Yager J A, Darrah P, Mosser D M, Prescott J F. Role of the 85-kilobase plasmid and plasmid-encoded virulence-associated protein A in intracellular survival and virulence of Rhodococcus equi. Infect Immun. 1999;67:3548–3557. doi: 10.1128/iai.67.7.3548-3557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harboe M, Malin A S, Dockrell H S, Wiker H G, Ulvund G, Holm A, Jorgensen M C, Andersen P. B-cell epitopes and quantification of the ESAT-6 protein of Mycobacterium tuberculosis. Infect Immun. 1998;66:717–723. doi: 10.1128/iai.66.2.717-723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higuchi T, Hashikura S, Gojo C, Inui T, Satoh S, Yoshida M, Ishiyama T, Yamada H, Takai S. Clinical evaluation of the serodiagnostic value of enzyme-linked immunosorbent assay for Rhodococcus equi infection in foals. Equine Vet J. 1997;29:274–278. doi: 10.1111/j.2042-3306.1997.tb03123.x. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi T, Hashikura S, Gojo S, Hagiwara S, Inui T, Satoh S, Yoshida M, Fujii M, Hidaka D, Tsubaki S, Takai S. Isolation of virulent Rhodococcus equi from transtracheal aspirates of foals serodiagnosed by enzyme-linked immunosorbent assay. J Vet Med Sci. 1997;59:1097–1101. doi: 10.1292/jvms.59.1097. [DOI] [PubMed] [Google Scholar]
- 9.Hietala S K, Ardens A A, Sansome A. Detection of Corynebacterium equi-specific antibody in horses by enzyme-linked immunosorbent assay. Am J Vet Res. 1985;46:13–15. [PubMed] [Google Scholar]
- 10.Hondalus M K, Mosser D M. Survival and replication of Rhodococcus equi in macrophages. Infect Immun. 1994;62:4167–4175. doi: 10.1128/iai.62.10.4167-4175.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopp T P, Woods K R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norton R E, Heuzenroeder M, Manning P A. Antigenic epitope mapping of the M24 protein of Streptococcus pyogenes: implications for serodiagnosis of rheumatic fever. FEMS Immunol Med Microbiol. 1996;16:267–271. doi: 10.1111/j.1574-695X.1996.tb00145.x. [DOI] [PubMed] [Google Scholar]
- 13.Panchanathan V, Naidu B R, Devi S, Di Pasquale A, Mason T, Pang T. Immunogenic epitopes of Salmonella typhi GroEL heat shock protein reactive with both monoclonal antibody and patients sera. Immunol Lett. 1998;62:105–109. doi: 10.1016/s0165-2478(98)00028-5. [DOI] [PubMed] [Google Scholar]
- 14.Prescott J F, Fernandez A S, Nicholson V M, Patterson M C, Yager J, Viel A L, Perkins G. Use of a virulence-associated protein based enzyme-linked immunosorbent assay for Rhodococcus equi serology in horses. Equine Vet J. 1996;28:344–349. doi: 10.1111/j.2042-3306.1996.tb03103.x. [DOI] [PubMed] [Google Scholar]
- 15.Prescott J F, Patterson M C, Nicholson V M, Morein B, Yager J A. Assessment of the immunogenic potential of Rhodococcus equi virulence associated protein (VapA) in mice. Vet Microbiol. 1997;56:213–225. doi: 10.1016/s0378-1135(97)00090-4. [DOI] [PubMed] [Google Scholar]
- 16.Prescott J F, Hoffman A M. Rhodococcus equi. Vet Clin N Am Equine Pract. 1993;9:375–384. doi: 10.1016/s0749-0739(17)30404-2. [DOI] [PubMed] [Google Scholar]
- 17.Sachse K, Helbig J H, Lysnyansky I, Grajetzki C, Muller W, Jacobs E, Yogev D. Epitope mapping of immunogenic and adhesive structures in repetitive domains of Mycoplasma bovis variable surface lipoproteins. Infect Immun. 2000;68:680–687. doi: 10.1128/iai.68.2.680-687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekizaki T, Takai S, Egawa Y, Ikeda T, Ito H, Tsubaki S. Sequence of the Rhodococcus equi gene encoding the virulence-associated 15–17-kDa antigens. Gene. 1995;155:135–136. doi: 10.1016/0378-1119(95)00009-u. [DOI] [PubMed] [Google Scholar]
- 19.Tan C, Prescott J F, Patterson M C, Nicholson V M. Molecular characterization of a lipid-modified virulence-associated protein of Rhodococcus equi and its potential in protective immunity. Can J Vet Res. 1995;59:51–59. [PMC free article] [PubMed] [Google Scholar]
- 20.Takai S. Epidemiology of Rhodococcus equi infections: a review. Vet Microbiol. 1997;56:167–176. doi: 10.1016/s0378-1135(97)00085-0. [DOI] [PubMed] [Google Scholar]
- 21.Takai S, Kawazu S, Tsubaki S. Immunoglobulin and specific antibody responses to Rhodococcus (Corynebacterium) equi infection in foals as measured by enzyme-linked immunosorbent assay. J Clin Microbiol. 1986;23:943–947. doi: 10.1128/jcm.23.5.943-947.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takai S, Koike K, Ohbushi S, Izumi C, Tsubaki S. Identification of 15- to 17-kilodalton antigens associated with virulent Rhodococcus equi. J Clin Microbiol. 1991;29:439–443. doi: 10.1128/jcm.29.3.439-443.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takai S, Anzai T, Fujita Y, Akita O, Shoda M, Tsubaki S, Wada R. Pathogenicity of Rhodococcus equi expressing a virulence-associated 20-kDa protein (VapB) in foals. Vet Microbiol. 2000;76:71–80. doi: 10.1016/s0378-1135(00)00226-1. [DOI] [PubMed] [Google Scholar]
- 24.Takai S, Hines S A, Sekizaki T, Nicholson V M, Alperin D A, Osaki M, Takamatsu D, Nakamura M, Suzuki K, Ogino N, Kakuda T, Dan H, Prescott J F. DNA sequence and comparison of virulence plasmids from Rhodococcus equi ATCC 33701 and 103. Infect Immun. 2000;68:6840–6847. doi: 10.1128/iai.68.12.6840-6847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaguchi H, Osaki T, Kai M, Taguchi H, Kamiya S. Immune response against a cross-reactive epitope on the heat shock protein 60 homologue of Helicobacter pylori. Infect Immun. 2000;68:3448–3454. doi: 10.1128/iai.68.6.3448-3454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]