Abstract
Background
Inhibitor growth protein 1 (ING1) is a tumor suppressor, and its down-regulation is involved in the progression and aggressive phenotypes of human malignancies through its interactions with the H3K4me3 and p53.
Methods
We collected datasets to analyze the relationship between ING1b mRNA expression and accumulative survival rate, and carried out immunohistochemistry analyses to determine the expression profiles of the p33ING1 protein on the mouse, normal human, and human cancer tissue microarrays.
Results
Compared with normal tissues, the ING1b mRNA was highly expressed in various types of cancer tissues, including, colorectal, lung, and breast cancers, and was positively correlated with the overall survival rate of gastric cancer patients. In mouse tissues, the subcellular location of p33ING1 was frequently nuclear; however, it was occasionally cytoplasmic or nucleocytoplasmic. There was a positive detection in the neuron body, a part of glial cells, the glandular epithelium of the stomach, intestines, breast, hepatocytes, heart, skeletal muscle cells, the bronchial and alveolar epithelium, and nephric tubules. In human tissues, the p33ING1 protein, apart from its cytoplasmic distribution, was distributed in the nuclei of the tongue, esophagus, stomach, intestine, lung, trachea, skin, appendix, cervix, endometrium, ovary, and breast. p33ING1 immunoreactivity was strongly detected in the stomach, trachea, skin, cervix, and breast, while it was weak in the other tissues. The positive rate of p33ING1 was 41.0% in the tested cancer entities (489/1,194). In general, p33ING1 expression was restricted to only the cytoplasm for all cancers, whereas it was found in the nucleus of renal clear cells, ovarian and colorectal cancers. Among them, p33ING1 was expressed in more than half of squamous cell carcinomas derived from the esophagus and cervix, while it was rarely expressed in hepatocellular (21.0%) and renal clear cell carcinoma (19.4%).
Conclusions
The findings suggest that p33ING1 might be participated in the repair and regeneration of organs or tissues the repair and regeneration of organs or tissue, and the carcinogenesis of the highly proliferative epithelium.
Keywords: Inhibitor growth protein 1 (ING1), expression profile, immunohistochemistry, bioinformatics, mouse, human, cancer
Introduction
The inhibitor of growth (ING) family, class II tumor suppressors, consists of 5 members, and function as the reader and writer of the histones epigenetic code, regulating DNA damage repair, chromatin remodeling, cellular senescence, the cell cycle, apoptosis, and cell differentiation. The human ING1 gene has 4 exons (1a, 1b, 1c, and 2) and 2 introns. In the literature, it has been reported that there are multiple alternatively spliced transcript variants encoding distinct isoforms from 3 different promoter regions, which include the p24ING1c (Isoform B: 210aa and E:262aa), p33ING1b (Isoform A:279aa), p47ING1a (Isoform D:422aa), and ING1d (Isoform C: 235aa), which share an identical C terminus with the conserved plant homeodomain (PHD) finger motif (1,2). In recent years, many researchers have become interested in p33ING1b because of its function as a tumor suppressor.
The p33ING1 protein has nuclear localization sequences (NLS, aa142-192) and physically interacts with p53 in the nucleus, whose complex is speculated to bind to Bax and p21 promoter and up-regulate their expression (3). p33ING1 interacts with proliferating cell nuclear antigens (PCNA) via the PCNA-binding protein domain to maximally induce apoptosis in a stress-induced manner (4). The cellular senescence-inhibited gene (CSIG) protein was identified as a binding partner for p33ING1 in the nucleolus (5). The p33ING1 protein binds to and regulates the activities of histone acetyltransferase (HAT) and histone deacetylase (HDAC) chromatin-remodeling complexes, which are responsible for the modulation of gene expressions in response to a variety of stresses, like angiopoietin (6,7). In contrast, p33ING1interacts with members of the14-3-3 family through Ser199 phosphorylation, resulting in its nucleus to plasmic translocalization, such as in mitochondria where the ING1 protein colocalizes and interacts with Bax to induce apoptosis (8). The tyrosine kinase Src can physically associate with phosphorylating ING1 (tyrosine 55 and 212), causes nuclear-to-cytoplasmic relocation of ING1 and reduces stability of ING1 (9).
ING1 mRNA was found to be universally expressed in various human tissues as 2 major bands at 2.2 and 2.5 kb by Northern blot (10). Most tissues were found to show various degrees of expression of ING1band p33ING1c, but not ING1a (11). Coles et al. (12) found that ING1 deletion in mice reveals a p53-independent role of ING1 for the suppression of cell proliferation and tumorigenesis, and apoptotic induction by up-regulating Bax expression. Kichina et al. (13) reported that mouse ING1 deletion results in decreased body size, hypersensitivity to radiation, and increased lymphomas incidence rates. A significant decrease in ING1 expression was also observed in gastric (14) and hepatocellular carcinoma (15), esophageal squamous cell carcinoma (16) tissues, when it was compared with the corresponding normal tissues. Rare mutations of ING1 in these tumors suggest that other mechanisms may be contributed to down-regulating ING1 expression in colorectal (17) and breast (18) cancers, head neck squamous cell carcinoma (19), lung cancer (20), basal cell carcinoma (21), and brain tumors (22).
Identification of the normal tissue or cell types that express p33ING1 would contribute to the clarification of its physiological functions, while the observation of its expression patterns and heterogeneity between tumor cases will benefit targeted gene therapies and the establishment of an animal model of conditional p33ING1 knockout. In the present study, the intermittent microwave irradiation was employed for immunohistochemistry of p33ING1, during which microwaving causes minute vibrations of more than 2.4 billion times/s, and enhances specificity (23). The protein expression of p33ING1 has been detected in normal mouse and human tissues, and human cancer tissues. Additionally, ING1b mRNA was investigated using the TCGA and Kaplan-Meier databases.
Methods
Specimen and tissue microarray
Three male and female C57BL/6 mice (8 weeks old) were sacrificed under sodium pentobarbital anesthesia, and the brain, lung, heart, breast, stomach, liver, spleen, kidney, and intestine were collected. Ten percent neutral formalin soaked all tissues for 48 hours. Then all tissues were embedded in paraffin, and cut into 4 µm sections. The human normal and cancer tissue arrays were purchased from Shanghai Outdo Biotech (Co., Ltd., Shanghai, China). The human normal tissues included cerebellum, brain stem, tongue, heart, lung, aorta, thyroid, esophagus, stomach, intestine, pancreas, liver, trachea, appendix, smooth muscle, skeletal muscle, testis, bladder, and prostate; and the cancer tissues included 62 renal clear cell carcinomas, 62 hepatocellular carcinomas, 45 esophageal squamous cell carcinomas, 62 pancreatic carcinomas, 31 cervical squamous cell carcinomas. The human breast, cervix, endometrium, and ovary tissues were sampled from surgical samples in our hospital. One hundred and ninety-six gastric cancer, 96 colorectal cancer, 208 ovarian cancer, 96 endometrial cancer, 144 breast cancer and 192 lung cancer were collected from our hospital. Tissues were subjected to the performance of tissue microarray using Tissue Microarrayer (AZUMAYA KIN-1, Tokyo, Japan). The cancer patients did not receive chemotherapy, radiation therapy, or adjuvant therapy before the operation. The patients or their relatives provided written consent for the use of tumor tissue for clinical research, and the research protocol was approved by the Ethical and Animal Experimentation Committees of Shengjing Hospital of China Medical University.
Immunohistochemistry
Consecutive sections were dewaxed with xylene and rehydrated with alcohol. Sections were then soaked in target retrieval solution (TRS, Dako, USA) with a microwave oven for 20 minutes (Oriental Rotor Ltd. Co., Tokyo, Japan). Sections were blocked in 5% bovine serum albumin for 30 minutes and incubated with mouse anti-p33ING1 antibody (Cab3, sc-21728, 1:50, Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 15 minutes, followed by incubation with anti-mouse secondary antibody conjugated to horseradish peroxidase (ready for use, Dako, USA) for 15 minutes. All incubations were put into a microwave oven to allow intermittent irradiation (23). The slides were washed with TBST (3×1 minute) after each treatment. The p33ING1 protein was visualized using 3, 3'-diaminobenzidine. After being counterstained with Mayer’s hematoxylin, the sections were dehydrated, cleared, and mounted. Instead of the primary antibody, normal mouse IgG as a negative control.
Immunostaining evaluation
As indicated in Figures 1-3, p33ING1 protein was localized to the cytoplasm and/or nuclei. Firstly, we (Zhao S and Zheng HC) selected the strong expression field under the low magnification, and randomly counted 100 cells from 5 different representative fields of each section. Secondly, the inconsistent data were confirmed by both persons. The percentages of counted cells were scored as follows: 0–10%, negative (−); 11–100%, positive (+).
Figure 1.
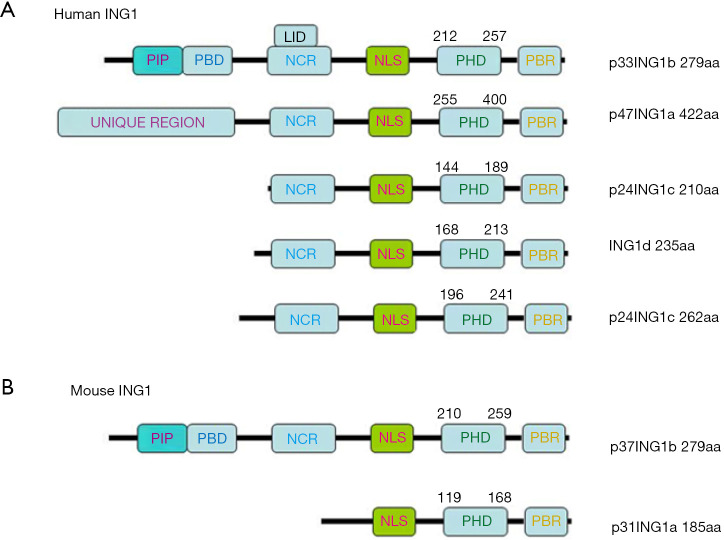
The schematic diagram of human and mouse ING1 protein structure. (A) Human ING1 protein; (B) mouse ING1 protein. ING1, inhibitor growth protein 1.
Figure 2.
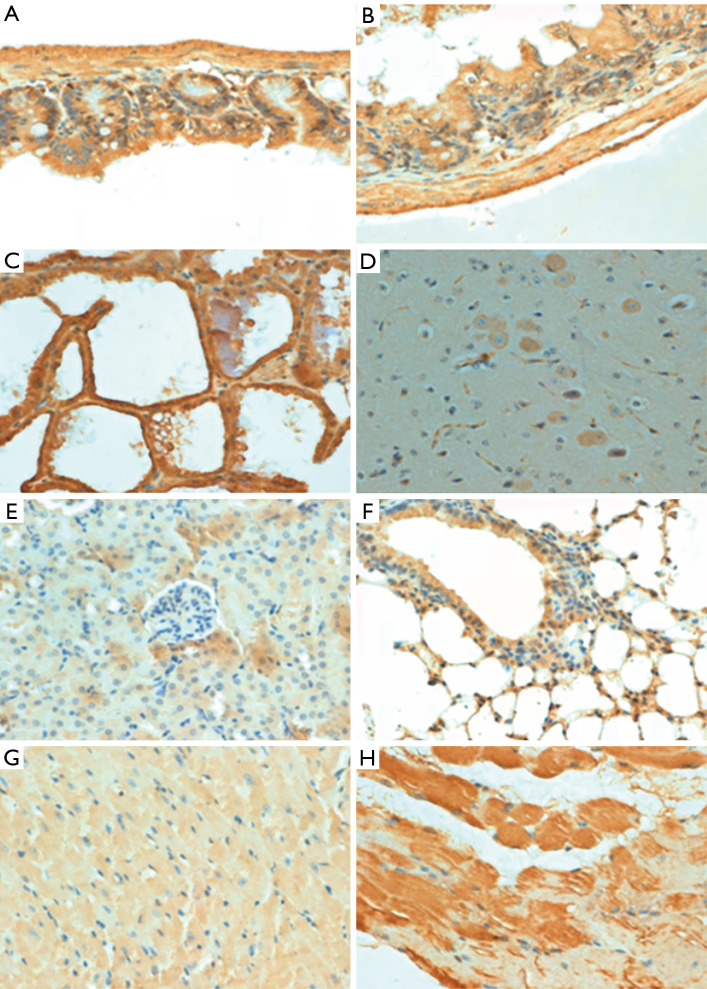
Immunohistochemical reactivity of normal mouse tissues with anti-p33ING1. (A) stomach; (B) intestine; (C) breast; (D) brain; (E) liver; (F) lung and bronchi; (G) heart muscle; (H) striated muscle. All the pictures were captured by a 200× microscope.
Figure 3.
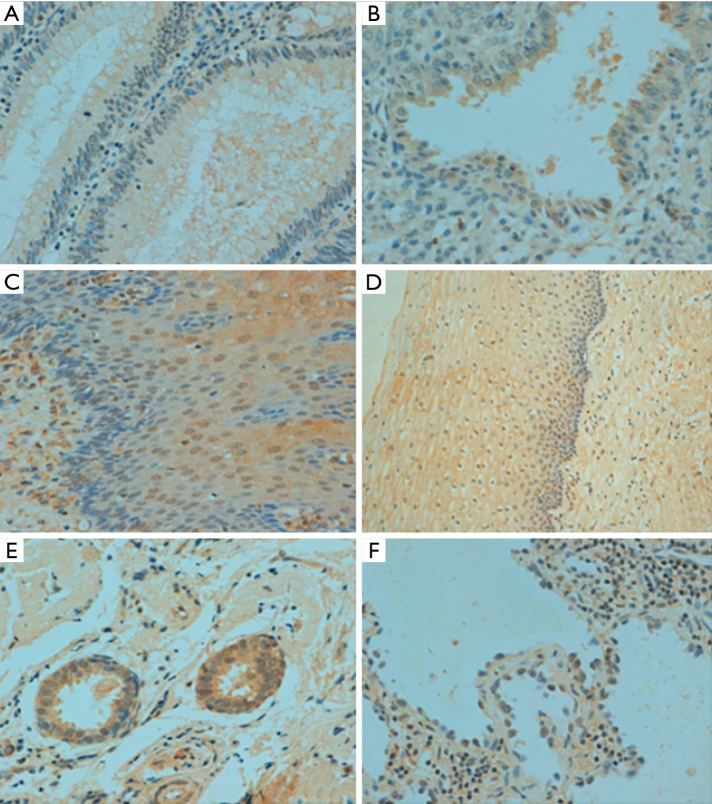
Immunohistochemical reactivity of normal human tissues with anti-p33ING1. (A) Intestine; (B) endometrium; (C) esophagus; (D) cervix; (E) breast; (F) lung and bronchi; (G) heart. All the pictures were captured by a 200× microscope.
Database analysis
p33ING1 mRNA expression and prognostic significance were analyzed in the Oncomine (www.oncomine.org) and Kaplan-Meier plotter (http://kmplot.com) databases.
Results
The schematic diagram of human and mouse ING1 protein structure
As Figure 1A shows the humanING1 protein includes 5 isoforms: p33ING1b (279aa), p47ING1a (422aa), p24ING1c (210aa and 262aa), and ING1d (235aa), which have an identical C terminus with a conserved PHD finger motif. The mouse ING1 protein includes 2 isoforms: p37ING1b (279aa) and p31ING1a (185aa) (Figure 1B).
p33ING1b protein expression in normal mouse tissues
As Figure 2 shows, the subcellular location of p33ING1 was often nuclear but occasionally cytoplasmic or nucleus to plasmic in the mouse tissues with either a sporadic or local pattern, although expression levels differed among tissues and cell populations. The distribution of p33ING1expression is summarized in Table 1. p33ING1 protein was sporadically localized in the skin and spleen. In other organs, it was positively detected in the neuron body, part of the glial cells, the glandular epithelium of the stomach, the intestine and breast, hepatocytes, the heart and striated muscle cells, the bronchial and alveolar epithelium, and nephric tubules.
Table 1. p33ING1 expression in normal mouse tissues.
| Cell types | Tissue |
|---|---|
| Glandular epithelium | Stomach, intestine, breast, lung |
| Neurons cells | Brain |
| Glial cells | Brain |
| Cardiocytes | Heart |
| Nephric tubules | Kidney (weak) |
| Striated muscle cells | Muscle |
| Hepatocytes | Liver (weak) |
| Sporadic | Spleen, skin |
p33ING1b protein expression in normal human tissues
In human tissues, p33ING1 protein was detected in the nucleus of the lung, tongue, esophagus, stomach, intestine, trachea, skin, appendix, breast, cervix, endometrium, and ovary (Figure 3) although its cytoplasmic localization was observed in all tissues. p33ING1 immunoreactivity was strongly detected in the stomach, trachea, skin, cervix, and breast, while it was weakly expressed in the cerebrum, cerebellum, brain stem, thymus, thyroid, pancreas, skeletal muscle, testis, and bladder (Table 2).
Table 2. p33ING1 expression in normal human tissues.
| p33ING1 expression | Tissue |
|---|---|
| Nucleus | – |
| Cytoplasm | Prostate, bladder, testis, heart, skeletal muscle, smooth muscle, pancreas, liver, thyroid, aorta, brain stem, cerebellum, cerebrum |
| Nucleus and cytoplasm | Breast, ovary, endometrium, cervix, appendix, trachea, lung, intestine, stomach, esophagus, tongue |
p33ING1b protein expression in human cancer tissues
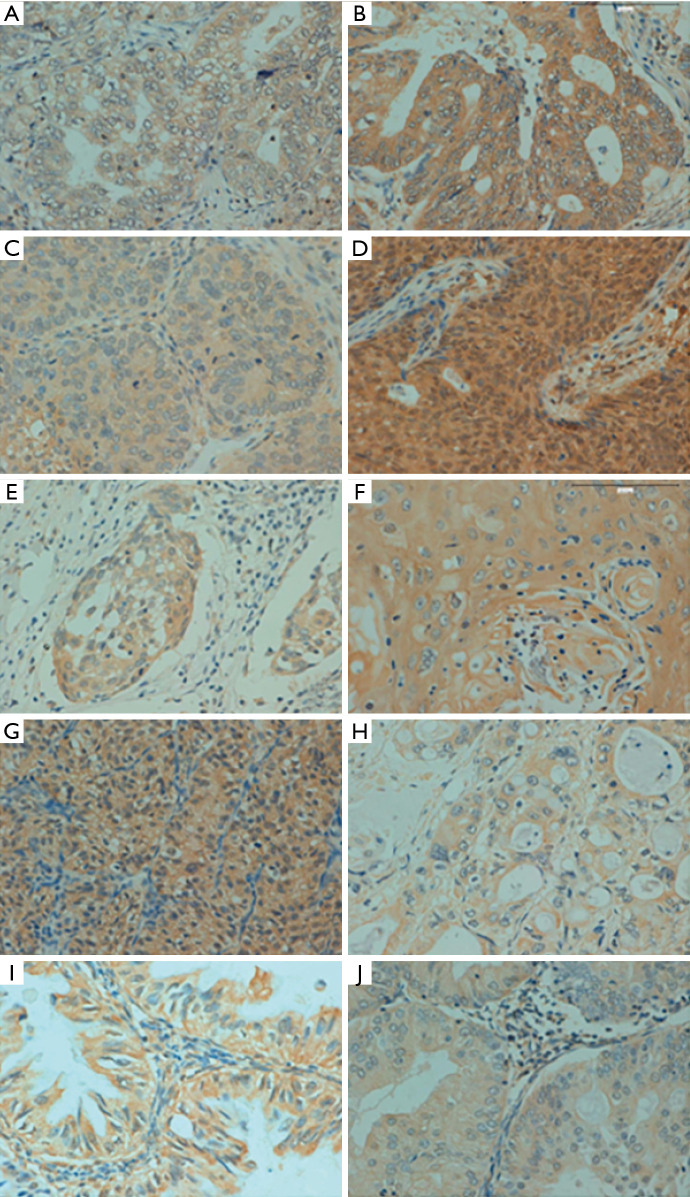
p33ING1-positive specimens were found in 489 of 1,194 evaluated cancer entities (41.0%), with homogeneity in the expression pattern (Figure 4 and Table 3). In general, p33ING1 expression was found to be restricted to the cytoplasm of all cancers, and occasionally in the nucleus of some cancer tissues, like in renal clear cell carcinoma, ovarian carcinoma, and colorectal carcinoma. p33ING1 was expressed in more than half of squamous cell carcinomas derived from the esophagus and cervix, although the highest rate was detected in breast cancer (57.6%, 83/144). p33ING1-positive cases were rare in hepatocellular carcinoma (21.0%, 13/62) and renal clear cell carcinoma (19.4%, 12/62).
Figure 4.
p33ING1 protein expression in human cancer by immunohistochemistry. (A) Gastric carcinoma; (B) colorectal carcinoma; (C) hepatocellular carcinoma; (D) breast carcinoma; (E) cervical carcinoma; (F) esophageal carcinoma; (G) renal carcinoma; (H) pancreatic carcinoma; (I) ovarian carcinoma; (J) endometrial carcinoma. All the pictures were captured by a 200× microscope.
Table 3. p33ING1 expression in various human carcinomas.
| Cancer type | Total cases | Positive cases | Positive rate (%) | p33ING1 expression | |
|---|---|---|---|---|---|
| Nucleus | Cytoplasm | ||||
| Hepatocellular carcinoma | 62 | 13 | 21.0 | − | + |
| Renal clear cell carcinoma | 62 | 12 | 19.4 | + | + |
| Pancreatic carcinoma | 62 | 24 | 38.7 | − | + |
| Esophageal carcinoma | 45 | 23 | 51.1 | − | + |
| Cervical carcinoma | 31 | 17 | 54.8 | − | + |
| Breast carcinoma | 144 | 83 | 57.6 | + | + |
| Gastric carcinoma | 196 | 57 | 29.1 | − | + |
| Colorectal carcinoma | 96 | 51 | 53.1 | + | + |
| Ovarian carcinoma | 208 | 96 | 46.2 | + | + |
| Endometrial carcinoma | 96 | 44 | 45.8 | − | + |
| Lung carcinoma | 192 | 69 | 35.9 | − | + |
ING1b mRNA expression in human cancers tissues
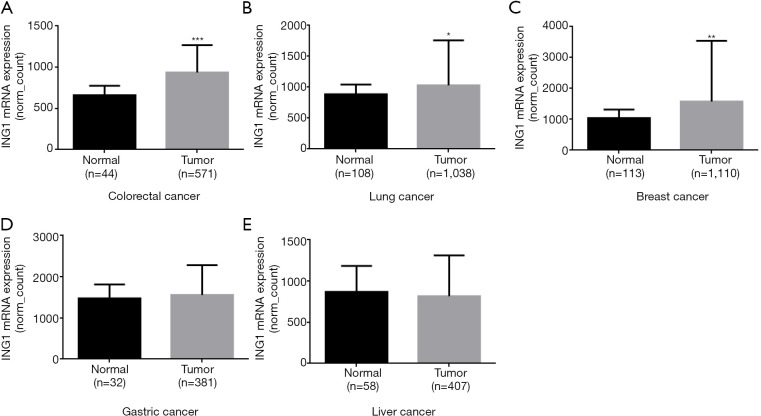
In the TCGA data, ING1b mRNA expression was higher in colorectal cancer, lung cancer, breast cancer than in the normal tissues (Figure 5A,B,C, P<0.05). But ING1b mRNA expression was no significant difference between gastric cancer and gastric normal tissues (Figure 5D, P>0.05). ING1b mRNA expression was also no significant difference between liver cancer and liver normal tissues (Figure 5E, P>0.05).
Figure 5.
ING1 mRNA expression in human cancer. (A) Colorectal carcinoma; (B) lung carcinoma; (C) breast carcinoma; (D) gastric carcinoma; (E) liver carcinoma. ING1, inhibitor growth protein 1.
The relationship between ING1b mRNA expression and accumulated survival rate in human tumors
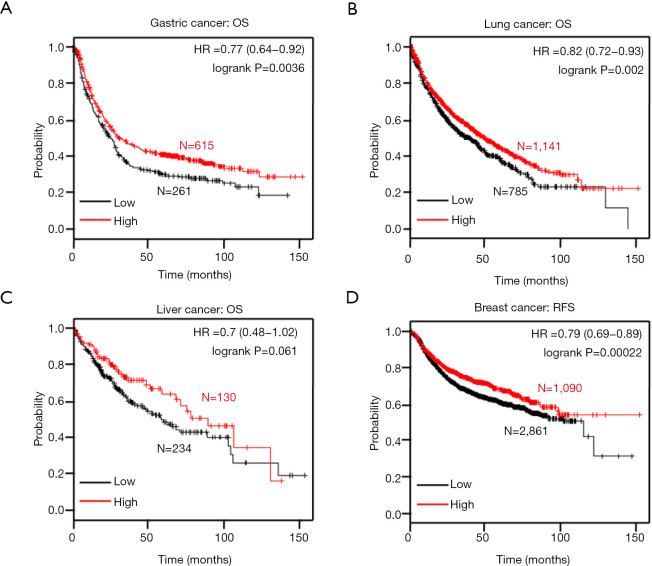
In the Kaplan-Meier plotter data, ING1b mRNA was positively correlated with the overall survival rates in gastric and lung cancer patients (Figure 6A,B, P=0.0036). There was no correlation between ING1b mRNA expression in liver cancer (Figure 6C, P=0.061). However, there was a lack of survival data for colorectal cancer. In addition, ING1b mRNA was positively related to progression-free survival rates in breast cancer patients (Figure 6D, P=0.061).
Figure 6.
Relationship between ING1 mRNA expression and accumulate survival rate in human tumors. (A) gastric carcinoma; (B) lung carcinoma; (C) hepatocellular carcinoma; (D) breast carcinoma. HR, hazard ratio.
Conclusions
In a study by Nouman et al. (24), two monoclonal antibodies were produced by a standard murine hybridoma technique, including MAb GN1 recognizing p33ING1bN-terminal (aa 1–32) and GN2 recognizing its C-terminal (aa 33–279). The immunostaining results showed that the immunoreactivity of GN1was principally distributed to the nucleus and cytoplasm, although MAb GN2 only demonstrated cytoplasmic positivity. The nuclear localization signal (NLS) of ING1 and phosphorylation-dependent sites for nucleus to plasmic translocation were localized in the C-terminal. In combination with a rare mutation of ING1, we believe that both antibodies recognize different epitopes, which are distinctly exposed to different antigen retrieval solutions or approaches. In the present study, we examined a wide range of tissue samples and found that the positive signal of p33ING1b was detected in the cytoplasm of normal mouse and human tissues and human cancer tissues, but haphazard in both the cytoplasm and nucleus, which is in consistent with the study mentioned above (24). The discrepancy might be attributed to the different antibody and immunostaining methods. Also, we speculated that Santa Cruz p33ING1b might react with the C-terminal of p33ING1b. Finally, we think that the non-specific reaction of antigen-antibody might give another explanation for the differences between these findings.
Due to the amino acid sequence alignment being revealed to have a high degree of similarity between mouse ING1b and human ING1b since they are 89% identity (2), we also did not find differences in patterns of p33ING1 expression between mouse and human tissues, except for the subcellular location in several cell types. In human tissues, p33ING1b protein was strongly detected in the stomach, trachea, skin, cervix, and breast, but weakly in the cerebrum, cerebellum, brain stem, thymus, thyroid, pancreas, skeletal muscle, testis, and bladder, indicating the functional involvement of p33ING1b in specific cell types and a specific functional status. Subsequently, we conditionally deleted p33ING1b using the tissue-specific promoter to construct the animal model of cancer. In the articles, the antiproliferative and apoptotic function of p33ING1b has been emphasized (4,8). The p33ING1b overexpression in the stomach, trachea, skin, cervix, and breast might be closely linked to the higher regenerative ability regardless of either the glandular or squamous epithelium, which is confirmed by the lower expression in the organs with weak repair and renewal, like the brain, testis, and spleen.
Here, we investigated the more frequent epithelial tumors and demonstrated that breast and cervical cancers had a high positive rate of p33ING1b expression, which is in agreement with the data in normal human tissues. In contrast, hepatocellular and renal clear cell carcinomas were shown to have less p33ING1b expression at a positive rate of 20% or so. This would significantly advance the understanding of cancer patients who could potentially benefit from p33ING1b-targeting therapy. Previously, deletion of nuclear p33ING1b was observed in melanoma, seminoma, papillary thyroid carcinoma, ductal breast carcinoma, and acute lymphoblastic leukemia, while cytoplasmic p33ING1b was more restricted, being detected in around 30% of neoplastic tissues. These findings suggest that p33ING1b function may be changed by its translocation to the cytoplasm in human cancer cells (11). Vieyra et al. (25) found that ING1 protein aberrantly localized to the cytoplasm, and slightly lower than to the nucleus of glioma cells, which is in line with our findings. A high frequency of loss of heterozygosity (LOH) in the ING1 chromosomal region, 13q34, has been found in non-small cell lung carcinoma, head and neck carcinoma, and esophageal squamous cell carcinoma (16,20,26), but its mutation is very rare, suggesting that LOH might result in down-regulated ING1b expression in human malignancies.
Interestingly, the results of expression analysis showed that human ING1 was regulated at a transcriptional level. Thus, ING1b mRNA is ubiquitously expressed in all tissues and here was analyzed in gastric, colorectal, liver, breast, and lung cancer. There is a clear link between the survival rate of cancer patients and ING1b mRNA expression. This observation may indicate that the survival rate is dependent on the human ING1 gene, while the biological significance of this transcriptional regulation is not yet clear. Previous studies found decreased ING1 expression in several human cancers, including in cervical cancer, renal carcinoma, acute lymphocytic leukemia (8,27,28), and other cancers. We noticed that most of the current studies referred to tumors of the embryo, gynecological urinary tract, and nervous system, and our results were related to gastrointestinal carcinoma. Therefore, we supposed that the different types of carcinoma might serve as a reason for this controversial result.
In summary, our research clarifies the variable expression of p33ING1b in normal mouse and human tissues, and human cancer tissues, and demonstrates the differential expression and/or subcellular location of p33ING1b among tissues, suggesting its differential functional involvement. According to our data, we hypothesize that ING1 might be participated in the repair and regeneration of tissues and may contribute to the carcinogenesis of the highly proliferative epithelium.
Acknowledgments
Funding: This study was financed by the National Natural Scientific Foundation of China (81672700), the Natural Scientific Foundation of Liaoning Province (20180530105), and Shenyang Science and Technology Grant (18-013-0-59).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The human tissue collection was approved by the Ethics Committee of Shengjing Hospital of China Medical University (Shenyang, Liaoning province). All patients consented to participate in this research.
Footnotes
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.04.28). The authors have no conflicts of interest to declare.
References
- 1.Cheung KJ, Jr, Li G. The tumor suppressor ING1: structure and function. Exp Cell Res 2001;268:1-6. 10.1006/excr.2001.5258 [DOI] [PubMed] [Google Scholar]
- 2.Guérillon C, Larrieu D, Pedeux R. ING1 and ING2: multifaceted tumor suppressor genes. Cellular and Molecular Life Sciences 2013;70:3753-72. 10.1007/s00018-013-1270-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng X, Hara Y, Riabowol K. Different HATS of the ING1 gene family. Trends Cell Biol 2002;12:532-8. 10.1016/S0962-8924(02)02391-7 [DOI] [PubMed] [Google Scholar]
- 4.Scott M, Bonnefin P, Vieyra D, et al. UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J Cell Sci 2001;114:3455-62. [DOI] [PubMed] [Google Scholar]
- 5.Abad M, Moreno A, Palacios A, et al. The tumor suppressor ING1 contributes to epigenetic control of cellular senescence. Aging Cell 2011;10:158-71. 10.1111/j.1474-9726.2010.00651.x [DOI] [PubMed] [Google Scholar]
- 6.Thakur S, Feng X, Qiao Shi Z, et al. ING1 and 5-azacytidine act synergistically to block breast cancer cell growth. PLoS One 2012;7:e43671. 10.1371/journal.pone.0043671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tallen G, Farhangi S, Tamannai M, et al. The inhibitor of growth 1 (ING1) proteins suppress angiogenesis and differentially regulate angiopoietin expression in glioblastoma cells. Oncol Res 2009;18:95-105. 10.3727/096504009789954645 [DOI] [PubMed] [Google Scholar]
- 8.Gong W, Russell M, Suzuki K, et al. Subcellular targeting of p33ING1b by phosphorylation-dependent 14-3-3 binding regulates p21WAF1 expression. Mol Cell Biol 2006;26:2947-54. 10.1128/MCB.26.8.2947-2954.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu L, Thakur S, Leong-Quong RY, et al. Src regulates the activity of the ING1 tumor suppressor. PLoS One 2013;8:e60943. 10.1371/journal.pone.0060943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimada Y, Saito A, Suzuki M, et al. Cloning of a novel gene (ING1L) homologous to ING1, a candidate tumor suppressor. Cytogenet Cell Genet 1998;83:232-5. 10.1159/000015188 [DOI] [PubMed] [Google Scholar]
- 11.Nouman GS, Anderson JJ, Mathers ME, et al. Nuclear to cytoplasmic compartment shift of the p33ING1b tumour suppressor protein is associated with malignancy in melanocytic lesions. Histopathology 2002;40:360-6. 10.1046/j.1365-2559.2002.01369.x [DOI] [PubMed] [Google Scholar]
- 12.Coles AH, Liang H, Zhu Z, et al. Deletion of p37Ing1 in mice reveals a p53-independent role for Ing1 in the suppression of cell proliferation, apoptosis, and tumorigenesis. Cancer Res 2007;67:2054-61. 10.1158/0008-5472.CAN-06-3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kichina JV, Zeremski M, Aris L, et al. Targeted disruption of the mouse ing1 locus results in reduced body size, hypersensitivity to radiation and elevated incidence of lymphomas. Oncogene 2006;25:857-66. 10.1038/sj.onc.1209118 [DOI] [PubMed] [Google Scholar]
- 14.Oki E, Maehara Y, Tokunaga E, et al. Reduced expression of p33(ING1) and the relationship with p53 expression in human gastric cancer. Cancer Lett 1999;147:157-62. 10.1016/S0304-3835(99)00288-8 [DOI] [PubMed] [Google Scholar]
- 15.Ohgi T, Masaki T, Nakai S, et al. Expression of p33(ING1) in hepatocellular carcinoma: relationships to tumour differentiation and cyclin E kinase activity. Scand J Gastroenterol 2002;37:1440-8. 10.1080/003655202762671332 [DOI] [PubMed] [Google Scholar]
- 16.Chen L, Matsubara N, Yoshino T, et al. Genetic alterations of candidate tumor suppressor ING1 in human esophageal squamous cell cancer. Cancer Res 2001;61:4345-9. [PubMed] [Google Scholar]
- 17.Chen LS, Wei JB, Zhou YC, et al. Genetic alterations and expression of inhibitor of growth 1 in human sporadic colorectal cancer. World J Gastroenterol 2005;11:6120-4. 10.3748/wjg.v11.i39.6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toyama T, Iwase H, Watson P, et al. Suppression of ING1 expression in sporadic breast cancer. Oncogene 1999;18:5187-93. 10.1038/sj.onc.1202905 [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Cespedes M, Okami K, Cairns P, et al. Molecular analysis of the candidate tumor suppressor gene ING1 in human head and neck tumors with 13q deletions. Genes Chromosomes Cancer 2000;27:319-22. [DOI] [PubMed] [Google Scholar]
- 20.Luo ZG, Tang H, Li B, et al. Genetic alterations of tumor suppressor ING1 in human non-small cell lung cancer. Oncol Rep 2011;25:1073-81. [DOI] [PubMed] [Google Scholar]
- 21.Chen B, Campos EI, Crawford R, et al. Analyses of the tumour suppressor ING1 expression and gene mutation in human basal cell carcinoma. Int J Oncol 2003;22:927-31. [PubMed] [Google Scholar]
- 22.Tallen G, Kaiser I, Krabbe S, et al. No ING1 mutations in human brain tumours but reduced expression in high malignancy grades of astrocytoma. Int J Cancer 2004;109:476-9. 10.1002/ijc.11715 [DOI] [PubMed] [Google Scholar]
- 23.Kumada T, Tsuneyama K, Hatta H, et al. Improved 1-h rapid immunostaining method using intermittent microwave irradiation: practicability based on 5 years’ application in Toyama Medical and Pharmaceutical University Hospital. Mod Pathol 2004;17:1141-9. 10.1038/modpathol.3800165 [DOI] [PubMed] [Google Scholar]
- 24.Nouman GS, Angus B, Lunec J, et al. Comparative assessment expression of the inhibitor of growth 1 gene (ING1) in normal and neoplastic tissues. Hybrid Hybridomics 2002;21:1-10. 10.1089/15368590252917584 [DOI] [PubMed] [Google Scholar]
- 25.Vieyra D, Senger DL, Toyama T, et al. Altered subcellular localization and low frequency of mutations of ING1 in human brain tumors. Clin Cancer Res 2003;9:5952-61. [PubMed] [Google Scholar]
- 26.Gunduz M, Gunduz E, Rivera RS, et al. The inhibitor of growth (ING) gene family: potential role in cancer therapy. Curr Cancer Drug Targets 2008;8:275-84. 10.2174/156800908784533454 [DOI] [PubMed] [Google Scholar]
- 27.Russell MW, Soliman MA, Schriemer D, et al. ING1 protein targeting to the nucleus by karyopherins is necessary for activation of p21. Biochem Biophys Res Commun 2008;374:490-5. [DOI] [PubMed] [Google Scholar]
- 28.Ohmori M, Nagai M, Tasaka T, et al. Decreased expression of p33ING1 mRNA in lymphoid malignancies. Am J Hematol 1999;62:118-9. [DOI] [PubMed] [Google Scholar]