Abstract
Background
The mesoderm induction early response 1, family member 3 (MIER3) gene has been recognized as potentially being associated with cancer. However, in relation to the development of non-small cell lung cancer (NSCLC), the expression pattern and the role of MIER3 are yet to be reported. The aim of this research was to investigate the rate of expression of MIER3 in NSCLC cells and tissues and to investigate the role of MIER3 in NSCLC.
Methods
Seventeen patients received NSCLC tissues and corresponding healthy tissues. MTT assay was used to determine cell proliferation. For detecting mRNA and protein expression, we used both quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot method. To measure cell apoptosis and cell cycle distribution, we applied the flow cytometry technique. We used a wound-healing assay and a Transwell invasion assay to study cell migration and invasion.
Results
In comparison with adjacent normal tissues, the expression of MIER3 was down-regulated in NSCLC tissues. In addition, the level of MIER3 in NSCLC cell lines was also lower than in pulmonary epithelial cell BEAS-2B. Moreover, when MIER3 was overexpressed, cell proliferation, migration, and invasion were significantly inhibited, apoptosis increased, and cell cycle arrest was induced in A549 and H460 cells. MIER3 overexpression also suppressed tumor growth in NSCLC xenograft mouse models. Furthermore, our study demonstrated that MIER3 down-regulated the Wnt/β-catenin signaling pathway in NSCLC cells. More importantly, MIER3 decreased the activity of histone acetyltransferase (HAT) p300, which may have contributed to its regulation on β-catenin and tumorigenesis.
Conclusions
The data suggests MIER3 takes on the tumor-suppressor role in the progression of NSCLC and, therefore, could prove to be a valuable clinical marker in the prognosis of the disease.
Keywords: Mesoderm induction early response 1, family member 3 (MIER3); non-small cell lung cancer (NSCLC); β-catenin; acetylation; histone acetyltransferase (HAT); p300
Introduction
As one of the most common forms of malignant tumors, lung cancer is recognized worldwide as the leading cause of deaths attributed to cancer (1). Non-small cell lung cancer (NSCLC) affects around eighty percent of lung cancer patients (2). Although there are advancements in diagnostic and therapeutic techniques, NSCLC patients with distant metastases can expect a poor prognosis and a five-year survival (3). The molecular mechanism behind the NSCLC occurrence remains unclear. Therefore, there is an urgent call for newer and more effective targets against NSCLC progression to be identified in the hope that new diagnostic strategies that target these markers can be developed.
In the early response 1 (MIER) family of mesoderms, there are three genes encoding similar proteins with preserved primary sequence, particularly in the domains of ELM2 and SANT (4,5). The best known member of the family isMIER1, which is recognised as an early response gene for fibroblast development and is activated by the mesoderm differentiation process in Xenopus (6). In the recruitment of histone deacetylase (HDAC) 1/2 activity MIER1α and MIER1β were shown to function as transcriptional repressors (7). Less is known about MIER2 and MIER3, and there remains much to be discovered about their biological characteristics and functions. MIER3 has been reported to have a high rate of mutation in hypermutated colorectal tumors (8) and is a candidate gene for susceptibility of breast cancer, which could play roles in tumorigenesis (9). A recent study found that the development of colorectal cancer is suppressed through the down-regulation of Sp1 and by inhibiting epithelial mesenchymal transition (10). However, in NSCLC, neither the expression pattern nor the mechanism of MIER3 is understood.
Two classes of enzymes with opposing roles catalyze the changes in protein acetylation: HAT and HDAC (11,12). Histone acetylation has been shown to play a critical role in stemness and tumor originicity (13-15) in a number of studies. Also reported was a correlation between changes in post-translational histone modifications, a lack of different histone acetylation/methylation markers, and breast cancer tumorigenesis (16). In colon cancer therapy (17), inhibition of the Wnt/β-Catenin pathway and HAT activity is seen as a target.
In this study, we explored the relationship between MIER3 and the progression of NSCLC in vitro and in vivo and studied its underlying mechanism. We specifically examined the effects of MIER3 on the Wnt/β-catenin signaling pathway and the activity of histone acetyl-transferase.
Methods
Reagents
In this analysis, the following reactives (suppliers) were used: antibody to MIER3 (Abcam, Cambridge, MA, USA); antibodies to bax, BCL-2, cleaved caspase3, β-catenin, cyclin D1, c-Myc, MMP7 and p300 (Santa Cruz Biotechnology, CA, USA); antibodies to acetylated Histone3, acetylated Histone4, Histone3 and Histone4 (Cell Signaling Technology, Danvers, MA, USA); antibody to β-actin and HRP-conjugated secondary antibodies (SunShine Biotechnology, Nanjing, China). An ELISA kit for p300 (R&D Systems, Minneapolis, MN, USA) was also used.
Patients
NSCLC tissues and adjacent normal tissues were obtained from 17 patients diagnosed with advanced NSCLC (IV stage) at Linyi Central Hospital (Linyi, China). Informed consent was obtained from all patients, and the study was approved by the Research Ethics Committee of Linyi Central Hospital.
Cell culture
BES-2B, A549, H460, H1299, and H1975 cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were grown in DMEM complemented with 10% FBS (Life Technologies, Grand Island, USA), 100 IU/ml penicillin (Sigma, USA), and 100 µg/mL streptomycin (Sigma, USA) at 37 °C in a 5% CO2 incubator.
Quantitative real-time polymerase chain reaction (qRT-PCR)
TRIzol reagent (Invitrogen, Carlsbad, USA), and according to the manufacturer’s protocol, was used to extract total RNA from cells or tissues. Equal amounts of RNA were reversely-transcribed to cDNA with Reverse Transcriptase (Takara, Dalian, China). The level of mRNA in MIER3 was analyzed using the SYBR Prime-Script RT-PCR kit (Takara, Dalian, China) in the QuantStudio 3 Real-Time PCR System (Applied Biosystems, USA). β-actin was utilized as an endogenous control. Fold changes were calculated through relative quantification (2−ΔΔCt) (18). The primers for MIER3 were F:5'-CTTTGGGTGGGACGGTAAATGCT-3' and R:5'-CAGACGGTTGCTACACTGTT GGT-3'. The primers for β-actin were F:5'-ACTCGTCATACTCCTGCT-3' and R:5'-GAAACTACCTTCAA CTCC-3'. In each case, the assay was performed three times to account for technical variability.
Western blot analysis
A RIPA lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China) was used to extract proteins from cells and tissues, according to the manufacturer’s protocol. Equal proportions of protein samples were separated using 10% SDS-PAGE gel and then transferred onto polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA). After blocking with 5% non-fat milk, the membranes, along with respective primary antibodies and secondary antibodies, underwent a period of incubation. The antibody-bound proteins were visualized using enhanced chemical luminescence (ECL, Bio-Rad Laboratories) and quantified by ImageJ software.
MIER3 overexpression
For ectopic expression of MIER3, the full-length human MIER3 cDNA was PCR-amplified and subcloned into pcDNA3.1 (Invitrogen, Shanghai, China) vector. The primers for cloning MIER3 were: F:5'GGTACCATGGCGGAGGCTTCTTTTGG3' and R:5'GAATTCTCACTCAGAGTGTAGGGCCG3'. An empty pcDNA3.1 vector was used as a control. The constructions were transfected into cells by Lipofectamine 3000 reagent (Invitrogen, Shanghai, China) according to the manufacturer’s instructions. Stable cell lines that expressed MIER3 were treated with 0.5 mg/mL puromycin for 10 days (10).
Cell viability analysis
A 96-well plate was used to culture A549 and H460 cells. After transfection of pcDNA3.1-MIER3, cell incubation with MTT solution took place for 4 hours at 37 °C. The medium was then taken away, and the formazan crystals were dissolved using dimethyl sulfoxide (DMSO). A microplate reader was used to calculate absorbance at 490 nm.
Cell apoptosis assay
A549 and H460 cells were transfected with pcDNA3.1-MIER3 for 24 hours. The cells were washed with ice-cold PBS before being resuspended in Annexin V to facilitate binding and being incubated at room temperature for 15 minutes using FITC-conjugated Annexin V/PI (ThermoFisher, USA). An Attune NxT Flow Cytometer (ThermoFisher Scientific, USA) was then used to analyze the cells.
Cell cycle analysis
Incubation of the A549 and H460 cells at the logarithmic growth phase was carried out with a serum-free medium for 24 hours after 50% confluence was reached. Following MIER3 overexpression, cells were harvested and resuspended with cold 70% ethanol. Propidium iodide (PI) was then used to incubate the suspension, and flow cytometry (ThermoFisher Scientific, USA) was used for analysis. The cell number in the G0, G0/G1, S, and G2/M phases was recorded.
Wound healing assay
A 24-well chamber was used to culture a confluent cell monolayer, which was then transfected with pcDNA3.1-MIER3. Detached or damaged cells were removed through a process of washing and stroking with a plastic pipette tip. After incubation for 24 hours, a microscope was used to monitor cell migrations, and the migration distance was measured using ImageJ software.
Transwell invasion assay
The Transwell invasion assay was used to study the invasion capacity of the A549 and H460 cells. Cells transfected with pcDNA3.1-MIER3 or pcDNA3.1-vector were seeded in the upper chamber for a short time with a serum-free medium. Twenty percent FBS was then added to the medium in the lower chamber. Following incubation for 24 hours, a cotton swab was used to remove non-invading cells from the top well, while the bottom cells were fixed in 95% ethanol and stained with hematoxylin. Under a microscope, the number of invaded cells was counted on 10 random fields.
Tumor xenograft mouse models
Athymic C57BL/6 mice (20–25 g body weight) were obtained from the Animal Center of Linyi Central Hospital (Linyi, China). The Animal Care and Research Committee of Linyi Central Hospital approved all of the animal experiments in this study. Mice were housed under controlled conditions (25±2 °C, 70% humidity and 12-hlight-dark periods), fed on a regular sterile chow diet, and had free access to water. A549 cells transfected with pcDNA3.1-MIER3 or pcDNA3.1-vector were administered to the mice via a subcutaneous injection. The weight and volume of the tumors were measured every 5 days post-injection. Tumor volume (V) was calculated as follows: V (mm3) = length × width2/2. The volumes of the tumors were measured every 5 days. The mice were sacrificed at day 35, and the tumor weight was calculated.
Immunohistochemistry
The mice were euthanized by intraperitoneal injection of pentobarbital sodium (200 mg/kg body weight) at 35 days post-injection. Tumor sections were prepared according to standard protocol. They were incubated with the MIER3 antibody (1:500, #ab127688, Abcam) overnight after rehydration and antigen retrieval had been carried out. Incubation with a secondary antibody then took place for 30 minutes. Diaminobenzidine was used to perform color reactions. Sections were visualized using a fluorescent microscope.
Enzyme-linked immunosorbent assay (ELISA)
The activity of p300 was measured by ELISA according to the manufacturer’s instructions.
Co-immunoprecipitation
For co-immunoprecipitation, cell lysates were prepared using the processes described above. The cell lysates were incubated with pre-blocked Protein A-Sepharose beads (Abcam, Cambridge, MA, USA). The membrane was blocked with a 5% milk solution in tris-buffered saline with Tween (TBST) for 1 hour, before probing with the following antibodies: anti-MIER3 rabbit polyclonal antibody (Abcam, Cambridge, Inc., ab127688 Recombinant fragment, corresponding to a region within amino acids 234–476 of human MIER3) (10); anti-p300 (Abcam, ab54984) overnight at 4 °C. The harvesting of complexes was then carried out using protein A-Sepharose (GE Healthcare, USA). Bound proteins were separated with SDS-PAGE before being visualized using the Western blotting technique.
Statistical analysis
SPSS 19.0 software was used to analyze data, and the results were expressed as mean ± standard deviation (SD). The statistical significance among groups was analyzed using one-way ANOVA, followed by Bonferroni’s post hoc test. Student’s t-test was used to compare the difference between 2 groups. A difference was considered significant at P<0.05.
Results
MIER3 is down-regulated in human NSCLC
To gain insight into the biological significance of MIER3 in NSCLC, we first investigated the MIER3 expression in NSCLC tissues and cells. NSCLC tissues and adjacent normal tissues from 17 patients were analyzed using qPCR and Western blotting. As shown in Figure 1A, the mRNA level of MIER3 was significantly decreased in NSCLC tumor tissues. Western blot of four random samples verified this result (Figure 1B,C). Besides, the mRNA and protein levels of MIER3 in NSCLC cell lines, including A549, H460, H1299, and H1975, were also lower than in the pulmonary epithelial cell BEAS-2B (Figure 1D,E,F). These results indicated that MIER3 is down-regulated in human NSCLC. A540 and H460 cells were studied further because they displayed the most significant decrease in MIER3.
Figure 1.
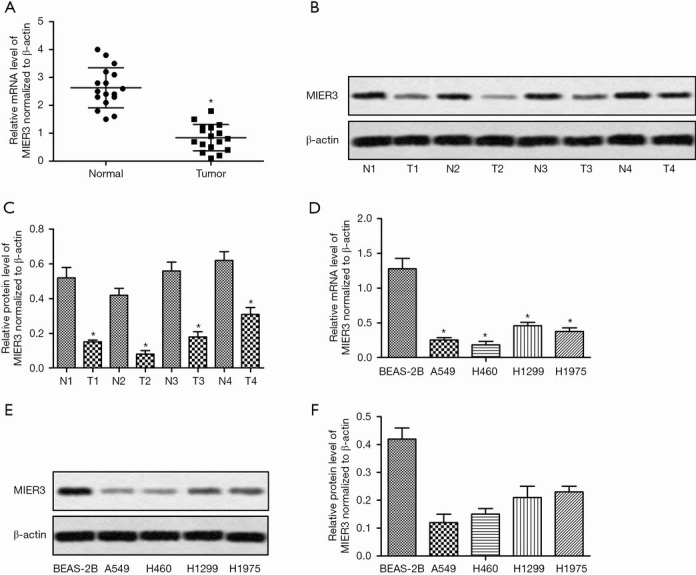
MIER3 is down-regulated in human NSCLC. (A) The mRNA levels of MIER3 in NSCLC tissues and adjacent normal tissues from 17 patients were analyzed by qRT-PCR. (B) The protein levels of MIER3 in four random samples were detected by Western blot. (C) Quantification of Figure 1B. (D,E) The expressions of MIER3 in BEAS-2B cell and four NSCLC cell lines including A549, H460, H1299, and H1975, were detected by qPCR (D) and Western blot (E). (F) Quantification of Figure 1E. *, P<0.05 versus normal tissues or BEAS-2B cells. NSCLC, non-small cell lung cancer; MIER3, mesoderm induction early response 1, family member 3; qRT-PCR, quantitative real-time polymerase chain reaction.
MIER3 overexpression inhibits proliferation and promotes apoptosis in human NSCLC cells
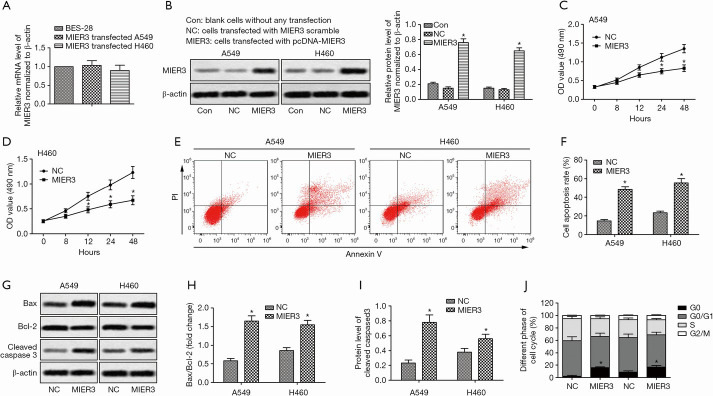
Based on the observations above, we performed gain-of-function studies. The pcDNA3.1-MIER3 plasmid was transfected to A549 and H460 cells to induce overexpression of MIER3. Relative expression of MIER3 was measured in A549 and H460 cells compared with BES-2B cells. The results demonstrated that there was no significant difference detected in the three cell lines (Figure 2A). The results suggested that MIER3 was successfully overexpressed in A549 and H460 cells. As shown in Figure 2B, pcDNA3.1-MIER3 transfection significantly increased the expression of MIER3. The MTT assay showed that MIER3 overexpression significantly inhibited the proliferation of A549 and H460 cells (Figure 2C,D). Besides, MIER3 overexpression notably promoted the apoptosis of A549 and H460 cells (Figure 2E,F), and also altered the expression of proteins associated with apoptosis, including Bax, Bcl-2 and cleaved-caspase3 (Figure 2G,H,I). Furthermore, MIER3 overexpression induced a cell cycle arrest, which was shown by the accumulation of cells in the G0 phase (Figure 2J). These results taken together suggest that MIER3 is a tumor suppressor of NSCLC.
Figure 2.
MIER3 overexpression acts to inhibit proliferation and increase apoptosis in human NSCLC cells. A549 and H460 cells were transfected with pcDNA3.1-vector or pcDNA3.1-MIER3 for 24 hours. (A) Relative expression of MIER3 was detected by qRT-PCR in BES-2B, A549 and H460 cells. (B) The protein level of MIER3 was detected by western blot. Quantification of Figure 2A. (C,D) MTT assay was used to analyze the viability of cells. (E) Cell apoptosis was measured by flow cytometry. (F) Quantification of apoptotic cells. (G) The expressions of Bax, Bcl-2, and cleaved caspase 3 were detected by Western blot. (H,I) Quantification of Figure 2G. (J) Cells were stained with PI (20 µg/mL) to determine the percentages of cells in the G0, G0/G1, S and G2/M phases. *, P<0.05 versus vector control. MIER3, mesoderm induction early response 1, family member 3; NSCLC, non-small cell lung cancer; qRT-PCR, quantitative real-time polymerase chain reaction.
MIER3 overexpression suppresses migration and invasion of NSCLC cells
The wound-healing assay and Matrigel Transwell assay were carried out to allow the investigation of the effect of MIER3 on cell migration and invasion. The results demonstrated that MIER3 significantly decreased the capabilities of migration (Figure 3A,B) and invasion (Figure 3C,D) in A549 and H460 cells.
Figure 3.
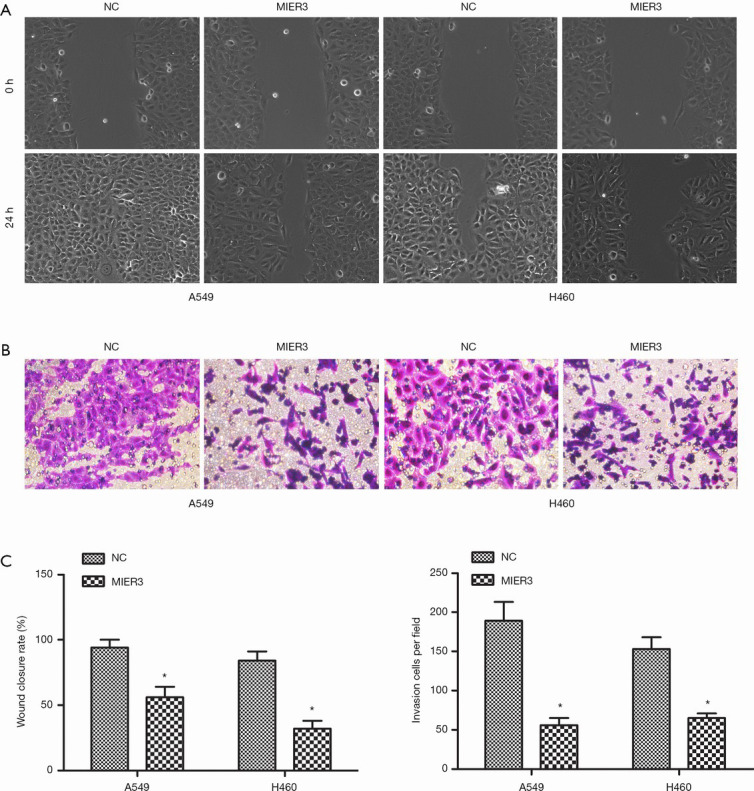
MIER3 overexpression suppresses migration and invasion of NSCLC cells. A549 and H460 cells were transfected with pcDNA3.1-vector or pcDNA3.1-MIER3 for 24 h. (A) Cell migration was detected by wound healing assay. (B) The migration distance was quantified by Image J software, and the wound closure rate was presented. (C) Cell invasion was measured by Matrigel Transwell assay, images were magnified at ×200. (D) Invasion cells per field was presented. *, P<0.05 versus vector control. MIER3, mesoderm induction early response 1, family member 3; NSCLC, non-small cell lung cancer.
MIER3 overexpression inhibits tumor growth in vivo
Mice were inoculated with A549 cells, which were transfected with pcDNA3.1-MIER3 or pcDNA3.1-vector to build tumor xenograft models to explore the function of MIER3 in vivo. Relative expression of MIER3 was measured by qRT-PCR, and the results displayed that the mRNA level of MIER3 was sharply elevated in the MIER3 group in comparison with the NC group (Figure 4A). The immunohistochemistry result showed that MIER3 was successfully overexpressed in tumor tissues (Figure 4B). More importantly, MIER3 overexpression dramatically inhibited tumor growth (Figure 4C,D,E).
Figure 4.
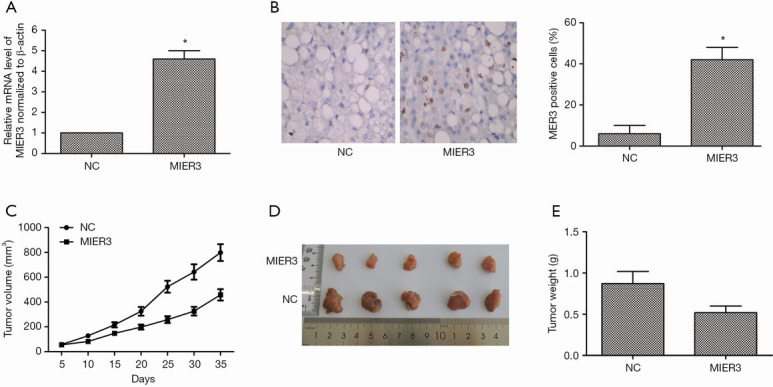
MIER3 overexpression inhibits tumor growth in vivo. Mice were inoculated with A549 cells, which have transfected with pcDNA3.1-MIER3 or pcDNA3.1-vector. (A) Relative expression of MIER3 was detected by qRT-PCR. (B) Immunohistochemistry detected MIER3 expression in tumor tissues. (C) Tumor volumes were calculated per 5 days. (D) Image of tumors from two groups. (E) Tumor weights were measured after 35 days of injection. n=5. *, P<0.05 versus vector control. MIER3, mesoderm induction early response 1, family member 3; qRT-PCR, quantitative real-time polymerase chain reaction.
MIER3 down-regulates Wnt/β-catenin signaling
As Wnt/β-catenin signaling is a crucial factor in the progression of NSCLC, we next examined the effect of MIER3 on β-catenin expression. As shown in (Figure 5A,B,C) MIER3 overexpression significantly decreased the expression of β-catenin in A549 and H460 cells. Besides, MIER3 also reduced the protein levels of some well-known Wnt/β-Catenin target genes, such as Cyclin D1, c-Myc, and metalloprotease (MMP)7. These results suggest that the Wnt/β-Catenin signaling pathway has an involvement in the effects MIER3 has on tumor suppression.
Figure 5.
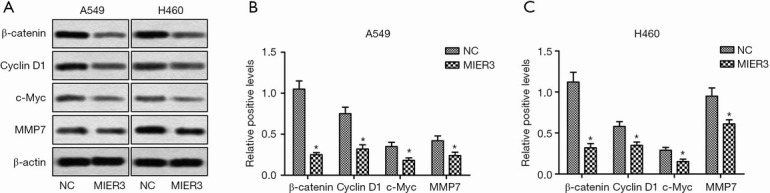
MIER3 down-regulates Wnt/β-catenin signaling. A549 and H460 cells were transfected with pcDNA3.1-vector or pcDNA3.1-MIER3 for 24 h. (A) The protein levels of β-catenin, Cyclin D1, c-Myc, and MMP7 were detected by Western blot. (B) Quantification of relative protein levels in A549. (C) Quantification of relative protein levels in H460. *, P<0.05 versus vector control. MIER3, mesoderm induction early response 1, family member 3.
MIER3 inhibits HAT p300 activity and decreases the level of acetyl-Histone
Previous studies have demonstrated that the activity of HAT p300 and histone acetylation is critical for β-Catenin activation (19). Based on this, we investigated the activity of p300 using ELISA. The results showed that MIER3 overexpression significantly decreased the activity of HAT p300 (Figure 6A). Besides, MIER3 overexpression also decreased the acetylation level of Histone3 and Histone4 (Figure 6B,C,D). Furthermore, the co-immunoprecipitation assay demonstrated that MIER3 and p300 could directly bond together, which may contribute to the inhibition of p300 (Figure 6E).
Figure 6.
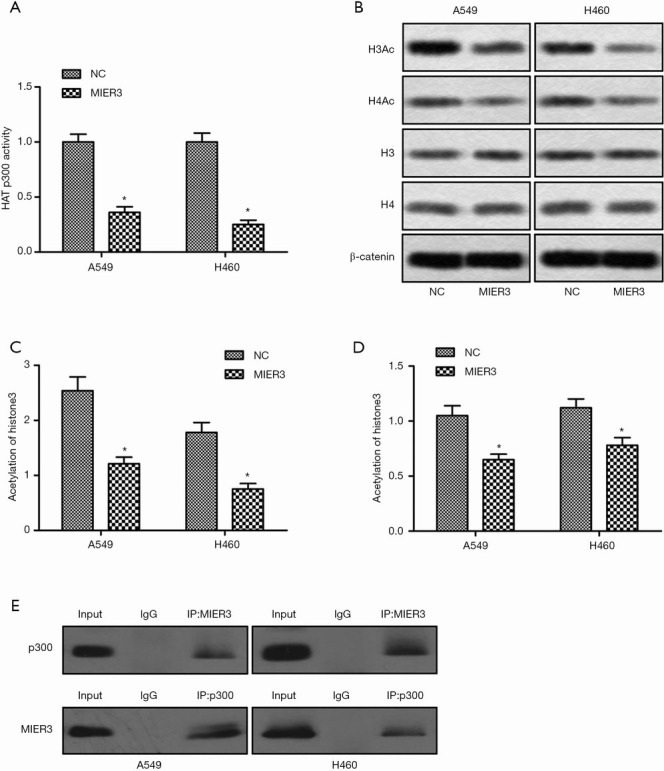
MIER3 inhibits HAT p300 activity and decreases the level of acetyl-Histone. A549 and H460 cells were transfected with pcDNA3.1-vector or pcDNA3.1-MIER3 for 24 h. (A) The activity of HAT p300 was measured by ELISA. (B) The acetylation levels of Histon3 and Histone4 were detected by Western blot. (C,D) Quantification of Figure 6B. (E) MIER3 co-immunoprecipitates with p300. *, P<0.05 versus vector control. MIER3, mesoderm induction early response 1, family member 3; HAT, histone acetyltransferase.
Discussion
MIER3, a mostly uncharacterized gene, is one of the three members of the MIER family, which also includes MIER1 and MIER2 (5). In recent years, MIER3 was considered as an oncogene due to its increased levels of expression in breast carcinomas in comparison to pathologically normal breast tissues (9). However, further studies have since found that the expression of MIER3 mRNA and protein is down-regulated in colorectal cancer tissues (20).There is a link between low levels of MIER3 expression and advanced stage of clinical disease, classification of tumor T, and poor rates of survival in patients with colorectal cancer (10). In this study, we discovered that in human NSCLC cells and tissues, the mRNA and protein levels of MIER3 are down-regulated.
Further studies have demonstrated that overexpression of MIER3 inhibits proliferation, increases apoptosis, and suppresses migration and invasion in NSCLC cells. In vivo, our study also showed MIER3 overexpression to inhibit tumor growth. Thus, our study indicates that MIER3 is a tumor suppressor gene that takes up an inhibitory role in NSCLC development and progression. The results differ from previous studies on breast cancer (9). Such contrasting findings suggest that the specificity and sophistication of the function and associated pathways of MIER3 vary depending on the nature of the tumor.
The Wnt/β-catenin signaling pathway regulates various biological events in tumorigenesis, such as gene expression, cell proliferation, metabolism, apoptosis and metastasis (21,22). When Wnt/β-catenin pathway is activated, the free cytosolic β-catenin collects in the nucleus and binds with Lymphoid enhancer-binding Factor and T Cell Factor proteins, and subsequently facilitates the transcription of c-Myc, Cyclin D1, MMPs, and other target genes (23,24). c-Myc, a multifunctional, nuclear phosphoprotein, acts as a player in cell cycle progression, proliferation, apoptosis and cellular transformation (25). Cyclin D1 is a protein needed for progression through the G1 phase of the cell cycle (26). MMPs are associated with various physiological or pathological processes such as morphogenesis, angiogenesis, and metastasis (27). In this study, MIER3 overexpression was discovered to decrease the expression of β-catenin and its downstream proteins, including c-Myc, Cyclin D1, and MMP7. These results suggest that Wnt/β-catenin may be a downstream signaling pathway through which MIER3 exerts its tumor-suppressive function.
Acetylation is an essential protein modification in the physiology of cells. Histone acetylation and deacetylation occur on lysine residues in the N-terminal tail as part of gene regulation (28). Typically, enzymes with HAT or HDAC activity catalyze these reactions, although HATs and HDACs can also act as modifiers on the acetylation status of non-histone (29). p300 is a multifunctional transcriptional coactivator that interacts with numerous transcription factors and exhibits protein/HAT activity (30). The interaction between β-catenin and p300 is a critical part of proximal-distal axis determination in β-catenin activation (19,31,32). Specific inhibition of p300/β-catenin interaction results in decreased transcription of Wnt target genes (19,32,33). It was reported that Rimonabant, a Cannabinoid Receptor 1 inverse agonist inhibited Wnt/β-Catenin pathway and HAT activity, which could be a target for colon cancer therapy (17). In our study, we determined that when MIER3 is overexpressed, the activity of p300 decreases, and, subsequently, the acetylation levels of Histone3 and Histone4 are down-regulated. These results propose the more direct target of MIER3 regulation in the Wnt/β-Catenin pathway.
It is well known that MIER1, owing to its ability to recruit HDAC1 and HDAC2, contributes to the repression of transcription (7). A recent study demonstrated that MIER2, but not MIER3, can recruit HDACs (4). However, the significance of MIER3 in histone acetylation is not well explored. Our study indicated that MIER3 interacts with HAT p300 and decreases its activity, which suggests that the mechanism by which MIER3 regulates histone acetylation differs from that of MIER1 and MIER2.
In conclusion, our study demonstrated that MIER3 acts as a tumor suppressor in NSCLC. The results suggest that MIER3 overexpression attenuated NSCLC progression in vitro and in vivo. Mechanically, MIER3 deactivated HAT p300 and down-regulated the Wnt/β-catenin signaling pathway, which may contribute to its anticarcinogenic function. To our knowledge, the results from our study represent the first evidence to suggest the significance of MIER3 in NSCLC progression and histone acetylation. Our ability to understand the exact molecular mechanism behind NSCLC is crucial to unlocking strategies for early diagnosis and therapy for patients of the disease. The findings of this study also suggest that MIER3 may be a promising therapeutic target for NSCLC.
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Research Ethics Committee of Linyi Central Hospital (SYXK-Lu-20190017). Informed consent was obtained from all patients.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.01.07). The authors have no conflicts of interest to declare.
References
- 1.Economopoulou P, Mountzios G. The emerging treatment landscape of advanced non-small cell lung cancer. Ann Transl Med 2018;6:138. 10.21037/atm.2017.11.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 3.Köhler J. Second-Line Treatment of NSCLC-The Pan-ErbB Inhibitor Afatinib in Times of Shifting Paradigms. Front Med (Lausanne) 2017;4:9. 10.3389/fmed.2017.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X, Wei SH, Shi XN, et al. Effects of Msp on the Proliferation, Migration and Invasion of Human Non-small Cell Lung Cancer Cells. Sichuan Da Xue Xue Bao Yi Xue Ban 2017;48:41-5. [PubMed] [Google Scholar]
- 5.Strausberg RL, Feingold EA, Grouse LH, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A 2002;99:16899-903. 10.1073/pnas.242603899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paterno GD, Li Y, Luchman HA, et al. cDNA cloning of a novel, developmentally regulated immediate early gene activated by fibroblast growth factor and encoding a nuclear protein. J Biol Chem 1997;272:25591-5. 10.1074/jbc.272.41.25591 [DOI] [PubMed] [Google Scholar]
- 7.Ding Z, Gillespie LL, Paterno GD. Human MI-ER1 alpha and beta function as transcriptional repressors by recruitment of histone deacetylase 1 to their conserved ELM2 domain. Mol Cell Biol 2003;23:250-8. 10.1128/MCB.23.1.250-258.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Network . Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.denDekker AD, Xu X, Vaughn MD, et al. Rat Mcs1b is concordant to the genome-wide association-identified breast cancer risk locus at human 5q11.2 and MIER3 is a candidate cancer susceptibility gene. Cancer Res 2012;72:6002-12. 10.1158/0008-5472.CAN-12-0748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng M, Hu Y, Song W, et al. MIER3 suppresses colorectal cancer progression by down-regulating Sp1, inhibiting epithelial-mesenchymal transition. Sci Rep 2017;7:11000. 10.1038/s41598-017-11374-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin W, Li Y, Chen F, et al. Klotho preservation via histone deacetylase inhibition attenuates chronic kidney disease-associated bone injury in mice. Sci Rep. 2017;7:46195. 10.1038/srep46195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huynh NC, Everts V, Ampornaramveth RS. Histone deacetylases and their roles in mineralized tissue regeneration. Bone Rep. 2017;7:33-40. 10.1016/j.bonr.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez GJ, Richmond PA, Bunker EN, et al. Genome-wide dose-dependent inhibition of histone deacetylases studies reveal their roles in enhancer remodeling and suppression of oncogenic super-enhancers. Nucleic Acids Res 2018;46:1756-76. 10.1093/nar/gkx1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Liu XR, Jin Z. Effects of Interference with UCA1 and Inhibition of miR-185-5p on Activation, Autophagy and Survival of β-Catenin Pathway in Non-small Cell Lung Cancer. Sichuan Da Xue Xue Bao Yi Xue Ban 2019;50:157-63. [PubMed] [Google Scholar]
- 15.Aztopal N, Erkisa M, Erturk E, et al. Valproic acid, a histone deacetylase inhibitor, induces apoptosis in breast cancer stem cells. Chem Biol Interact 2018;280:51-8. 10.1016/j.cbi.2017.12.003 [DOI] [PubMed] [Google Scholar]
- 16.Elsheikh SE, Green AR, Rakha EA, et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res 2009;69:3802-9. 10.1158/0008-5472.CAN-08-3907 [DOI] [PubMed] [Google Scholar]
- 17.Proto MC, Fiore D, Piscopo C, et al. Inhibition of Wnt/beta-Catenin pathway and Histone acetyltransferase activity by Rimonabant: a therapeutic target for colon cancer. 2017;7:11678. [DOI] [PMC free article] [PubMed]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 19.Rieger ME, Zhou B, Solomon N, et al. p300/beta-Catenin Interactions Regulate Adult Progenitor Cell Differentiation Downstream of WNT5a/Protein Kinase C (PKC). J Biol Chem 2016;291:6569-82. 10.1074/jbc.M115.706416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitule P, Vycital O, Bruha J, et al. Differential expression and prognostic role of selected genes in colorectal cancer patients. Anticancer Res 2013;33:4855-65. [PubMed] [Google Scholar]
- 21.Liu P, Ma S, Liu H, et al. HCFU inhibits cervical cancer cells growth and metastasis by inactivating Wnt/beta-catenin pathway. J Cell Biochem 2017. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 22.Pei Z, Du X, Song Y, et al. Down-regulation of lncRNA CASC2 promotes cell proliferation and metastasis of bladder cancer by activation of the Wnt/beta-catenin signaling pathway. Oncotarget 2017;8:18145-53. 10.18632/oncotarget.15210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lecarpentier Y, Claes V, Vallee A. Interactions between PPAR Gamma and the Canonical Wnt/Beta-Catenin Pathway in Type 2 Diabetes and Colon Cancer. 2017;2017:5879090. [DOI] [PMC free article] [PubMed]
- 24.Zheng Y, Jiang L, Hu Y, et al. Metallothionein 1H (MT1H) functions as a tumor suppressor in hepatocellular carcinoma through regulating Wnt/beta-catenin signaling pathway. BMC Cancer 2017;17:161. 10.1186/s12885-017-3139-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bretones G, Delgado MD, Leon J. Myc and cell cycle control. Biochim Biophys Acta 2015;1849:506-16. 10.1016/j.bbagrm.2014.03.013 [DOI] [PubMed] [Google Scholar]
- 26.Datta D, Anbarasu K, Rajabather S, et al. Nucleolar GTP-binding Protein-1 (NGP-1) Promotes G1 to S Phase Transition by Activating Cyclin-dependent Kinase Inhibitor p21 Cip1/Waf1. J Biol Chem 2015;290:21536-52. 10.1074/jbc.M115.637280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alaseem A, Alhazzani K, Dondapati P, et al. Matrix Metalloproteinases: A challenging paradigm of cancer management. Semin Cancer Biol 2019;56:100-15. 10.1016/j.semcancer.2017.11.008 [DOI] [PubMed] [Google Scholar]
- 28.Chen HP, Zhao YT, Zhao TC. Histone deacetylases and mechanisms of regulation of gene expression. Crit Rev Oncog 2015;20:35-47. 10.1615/CritRevOncog.2015012997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadoul K, Boyault C, Pabion M, et al. Regulation of protein turnover by acetyltransferases and deacetylases. Biochimie 2008;90:306-12. 10.1016/j.biochi.2007.06.009 [DOI] [PubMed] [Google Scholar]
- 30.Choi JR, Lee SY, Shin KS, et al. p300-mediated acetylation increased the protein stability of HIPK2 and enhanced its tumor suppressor function. Sci Rep 2017;7:16136. 10.1038/s41598-017-16489-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bordonaro M, Lazarova DL. Determination of the Role of CBP- and p300-Mediated Wnt Signaling on Colonic Cells. JMIR Res Protoc 2016;5:e66. 10.2196/resprot.5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warner DR, Smith SC, Smolenkova IA, et al. Inhibition of p300 histone acetyltransferase activity in palate mesenchyme cells attenuates Wnt signaling via aberrant E-cadherin expression. Exp Cell Res 2016;342:32-8. 10.1016/j.yexcr.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang SH, Li N, Wei Y, et al. beta-catenin deacetylation is essential for WNT-induced proliferation of breast cancer cells. Mol Med Rep 2014;9:973-8. 10.3892/mmr.2014.1889 [DOI] [PubMed] [Google Scholar]