Abstract
COVID-19 is a fast-spreading pandemic, and early detection is crucial for stopping the spread of infection. Lung images are used in the detection of coronavirus infection. Chest X-ray (CXR) and computed tomography (CT) images are available for the detection of COVID-19. Deep learning methods have been proven efficient and better performing in many computer vision and medical imaging applications. In the rise of the COVID pandemic, researchers are using deep learning methods to detect coronavirus infection in lung images. In this paper, the currently available deep learning methods that are used to detect coronavirus infection in lung images are surveyed. The available methodologies, public datasets, datasets that are used by each method and evaluation metrics are summarized in this paper to help future researchers. The evaluation metrics that are used by the methods are comprehensively compared.
Keywords: COVID-19 detection, DL-Based COVID-19 detection, Lung image classification, Coronavirus pandemic, Medical image processing
Graphical abstract
1. Introduction
The World Health Organization (WHO) declared the spread of the coronavirus infection a pandemic in March 2020, which is called the coronavirus pandemic or COVID-19 pandemic. The coronavirus pandemic is caused by severe acute respiratory syndrome coronavirus 2 (SARS CoV 2). The outbreak originally started in Wuhan, China, and later spread to every country in the world [1]. The coronavirus spreads through respiratory droplets of the infected person that are produced through cough or sneeze. These droplets can further contaminate the surfaces increasing the spread. Coronavirus-infected persons may suffer from mild to severe respiratory illness and may require ventilation support [2]. Older people and people with chronological disorders are easily prone to coronavirus infection. Thus, many governments have closed their borders and locked down people to break the cycle and prevent the spread of the pandemic [3].
With the sequencing of ribonucleic acid (RNA) from the coronavirus, many vaccines are being developed worldwide. The developed vaccines use both traditional and next-generation technology with six vaccine platforms, namely, live attenuated virus, inactivated virus, protein or subunit, viral vector-based, messenger RNA (mRNA), and deoxyribonucleic acid (DNA). Although vaccines can reduce the rapid spread and facilitate the development of immunity via the production of suitable antibodies, the efficacy of the vaccines is still 95%. Many issues are encountered in administering the vaccine, such as supply chain logistical challenges, vaccine hesitancy, and vaccine complacency. A vaccine is a prevention measure rather than a cure [4]. Even with the availability of the vaccine, early detection of the coronavirus is important, as it can facilitate tracing of the people who were in contact directly and indirectly. By tracing these people, further spread of the pandemic can be avoided. COVID-19 infection manifests as lung infection, and computed tomography (CT) and chest X-ray (CXR) images are primarily used in the detection of lung infection of any type [5].
Along with doctors and clinical personnel, researchers and technologists are focusing their efforts on early detection of coronavirus infections. According to PubMed [6], 755 academic articles were published with the search term “coronavirus” in 2019, and this number rose to 1245 in the first 80 days of 2020. Artificial intelligence and deep learning methods are the most commonly used methods by researchers for the detection of coronavirus infection from CT and CXR images. Deep learning methods have shown significant performance in many research applications, such as computer vision [7], object tracking [8], gesture recognition [9], face recognition [10], and steganography [[11], [12], [13]]. Deep learning methods are widely used because of their improved performance compared to traditional methods. In contrast to traditional methods and machine learning methods, the features need not be hand-picked. By changing the parameters and configurations of the deep learning convolutional neural network (CNN) architecture, a model can be trained to learn the best possible features for the dataset in use. Researchers have used deep learning methods to explore the field of medical imaging even before the coronavirus pandemic. With the recent pandemic, the use of deep learning methods for the detection of coronavirus infection from images has increased tremendously.
A detailed survey of the available deep learning approaches for the detection of coronavirus infection from images such as CT scans or CXR images is conducted in this paper. Although other surveys are available in the literature, most of them cover a wider scope. For example, Ulhaq et al. [14] surveyed all methods that address coronaviruses, such as medical image processing, data science methods for pandemic modeling, AI and the Internet of things (IoT), AI for text mining and natural language processing (NLP), and AI in computational biology and medicine. This provides an overall view of what is happening in the research world. A survey on the application of computer vision methods for COVID-19 [15] described the segmentation of lung images. This paper aims to exclusively describe coronavirus detection methods using deep learning methods. In the hope of helping researchers develop better coronavirus detection methods, this paper summarizes all the methods that have been reported in the literature. Along with the methods, the used datasets, commonly used metrics for evaluation and comparison are discussed and future direction are elaborated in this paper.
2. Background
Before discussing the details of the available methods for coronavirus infection detection, it is essential to have a working knowledge of deep convolutional neural networks and popular CNN architectures. In this section, a brief overview of CNN architectures and main points on available CNN architectures are presented.
2.1. Convolutional neural networks
Convolutional neural networks, specifically artificial neural networks, are a branch of deep learning methods that are inspired by the natural visual perception mechanism of living organisms [16]. CNNs are nothing but stacked multilayered neural networks. There are three major categories of layers, namely, convolutional layers, pooling layers and fully connected layers. The first layer of any CNN model is an input layer, where the width, height and depth of the input image are specified as the input parameters. Immediately after the input layer, convolutional layers are defined with the number of filters, filter window size, stride, padding and activation as the parameters. Convolutional layers are used to extract meaningful feature maps for the input location by calculating the weighted sum [17,18]. Then, each feature map is passed through an activation function, and bias is added to form the output. Usually, rectilinear unit (ReLU) activation is used as the activation function [19].
Pooling layers are used to reduce the size of the output from the convolutional layers. As the model increases in size with an increasing number of filters in the convolutional layer, the output dimensionality also increases exponentially, which makes it hard for computers to handle. Pooling layers are added to reduce the dimensions for easy computation and sometimes to suppress noise. The pooling layer can be a max pooling, average pooling, global average pooling, or spatial pooling layer. The most commonly used pooling layer is a max pooling layer [20]. The output is flattened to form a single-array feature vector, which is fed to a fully connected layer. Finally, a classification layer is defined with activation functions such as sigmoid, softmax and tanh functions [21]. The number of classes is specified in this layer, and the extracted features are aggregated into class scores.
Batch normalization layers are applied after the input layer or after the activation layers to standardize the learning process and reduce the training time [22]. Another important parameter is the loss function, which summarizes the error in the predictions during training and validation. The loss is backpropagated to the CNN model after each epoch to enhance the learning process [23].
2.2. Transfer learning and fine-tuning
After designing, creating and building a deep learning model, the number of epochs is set to start training. During training, random weights are initialized, which will be refined during each epoch to make the result closer to the classification score. However, in transfer learning, instead of using random weight values, the model can be initialized with weight values from pretrained models. Transfer learning performs best when there is a limited availability of training data. When performing transfer learning, the last layer of the pretrained model architecture is replaced with a fully connected layer with the same number of classes as the new dataset. The architecture is retrained to use the model for the new dataset [24].
Another method, namely, fine-tuning, is also used when the dataset is small. Similar to transfer learning, the last layer of the architecture is replaced and redefined. The only difference is that in transfer learning, all the layers are retrained, while in fine-tuning, some layers can be redefined and retrained according to the application [25]. One major disadvantage of these methods is that the size of the input image cannot be changed. Therefore, if the pretrained model uses a smaller image dimension and transfer learning has to be conducted on a dataset with a larger image dimension, resizing the image is compulsory. Resizing a large image to a smaller image can affect the performance of the model in some cases. Careful consideration must be taken when transfer learning and fine-tuning are implemented.
2.3. Available architectural families
Several available architectures generalize well irrespective of the dataset or application. Various popular architectures, such as AlexNet, VGG, Inception, ResNet, DenseNet, MobileNet, and Xception, are summarized in this section.
AlexNet is a simple five-layer convolutional neural network. There are two variants of the VGG network – VGG16 and VGG19 [26]. The VGG architecture was originally proposed for image recognition applications. In VGG16 and VGG19, 16 and 19 wt layers are used with a smaller convolutional filter size of 3 × 3. The network won first and second places in the ILSVR (ImageNet) competition [27] in 2014. The size of the input image is fixed to 224 × 224. The model is trained on the ImageNet dataset, which contains millions of images [28].
In contrast to CNN architectures, in which the layers are stacked, a new architecture with an inception block is introduced in InceptionNet [29]. Several variants are available in the inception family. The inception network is also used for image classification and localization and participated in the ILSVR (ImageNet) competition [27] in 2014. Instead of increasing the depth of the model by adding additional layers, the authors apply various filter sizes to the input image simultaneously in the inception block. This leads to the growth of the model width. All the outputs of the inception block are concatenated and fed to the next inception block. Available versions include InceptionV1 (GoogLeNet) [29], InceptionV2 and InceptionV3 [18], InceptionV4 and InceptionResNet [30]. The input image size that is accepted by the model is 224 × 224.
ResNet [31] is also used in image classification methods and was the winner of the ILSVRC 2015 [27]. The ResNet family uses the residual block, which is a network-in-network in their architecture. Five steps with convolutional and identity blocks are used to define the network. Similar to the VGG family, the input image size is 224 × 224. Many variations are available. Inception-ResNet [30] is a hybrid architecture that combines the inception and residual blocks. The input image size for InceptionResNet is 229 × 229.
The DenseNet architecture [32] is a variation of the ResNet architecture. Similar to the ResNet family, a residual identity block is used to build the architecture, except concatenation is conducted in place of summation. Traditional CNN models have L connections for L layers, whereas the DenseNet model has direct connections. Each layer is connected into every other layer in a feed-forward fashion. The feature maps of all the previous layers are used as input to the current layer, and the feature map of the current layer is fed to all the other layers. The size of the accepted input image is 224 × 224.
MobileNets are compact architectures with depthwise separable convolutional layers that can be used in mobile phones and embedded systems [33]. Usually, 2D convolutional layers are used, but in depthwise separable convnets, two 1D convolutional layers are used. Doing so has helped reduce the number of parameters and, hence, decrease the computation and training times and memory usage. There are 54 layers, and the input image size is 224 × 224.
Xception [34] architectures are similar to the Inception family, where inception blocks with depthwise separable convolutional layers are used. The input image size is 229 × 229, and the number of layers is 71.
3. Summary of the research methods
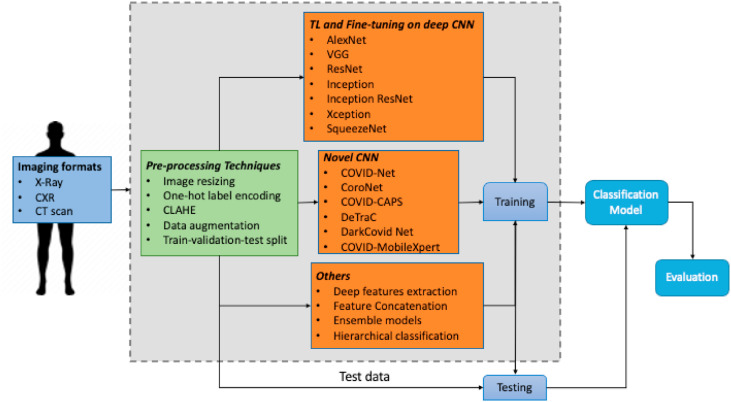
Since COVID-19 is a novel pandemic, only a few datasets with a limited number of samples are publicly available. The best strategy that can be followed with the limited availability of data is either transfer learning or fine-tuning (Section 2.2). Although new CNN architectures can be constructed, to improve the performance, a wider range of images under each class is required. According to this study, the majority of the papers use transfer learning methods, a few rely on fine-tuning, and only a handful propose a novel CNN architecture with comparable performance to transfer learning-based methods. The majority of the works use transfer learning from models that are pretrained on the ImageNet dataset. Additionally, the input image size to the architecture is either 224 × 224 or 229 × 229, but the dataset that is used to train and test the model contains images of various sizes. A simple preprocessing step is used to resize the images in the dataset to fit into the shape of the input layer of the network. In this section, first, transfer learning and fine-tuning-based methods and the CNN architectures that are used will be specified. Then, methods with novel CNN architectures will be described. Finally, methods that do not belong to these categories will be described in detail. Fig. 1 presents an overall summary of all the methods that are reviewed in this paper.
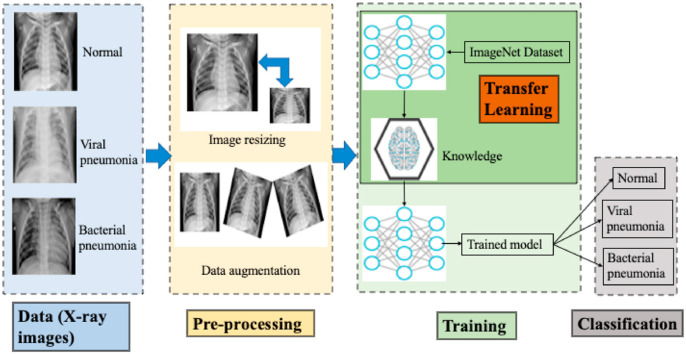
Fig. 1.
Overall workflow summary of all the methods. The first step is the acquisition of the data, and the imaging format can be chest X-ray (CXR) or CT scan. The second step is preprocessing, such as image resizing and data augmentation. Then, the preprocessed data are trained using one of the three methods. The trained model is used for classification and evaluation.
3.1. Transfer learning and fine-tuning approaches
Transfer learning is the go-to method for most of the papers. Pretrained models that are trained on the ImageNet database are used to perform transfer learning. Although the method is the same, different architectures are used in the works [35]. Even if the architectural family is the same, different variants are used. Cross-validation is another technique that is used in some of the methods. In addition, methods with new CNN models are considered, which also utilize the benefits of transfer learning when the dataset is very small.
A comparative study of the available deep learning architectures, namely, MobileNet V2, Inception, Xception, Inception ResNet V2 and VGG19, that use the transfer learning method is performed by Apostolopoulos et al. [35]. Three models - ResNet50, InceptionV3 and Inception-ResNetV2 - are also utilized [36]. Transfer learning on InceptionV3 [37] and AlexNet [38] along with data augmentation is another variation. ResNet18, ResNet50, SqueezeNet [39], and DenseNet-121 [40] are used for transfer learning along with data augmentation methods [41]. Transfer learning using VGG19, DenseNet201, InceptionV3, ResNet152, InceptionResNetV2, Xception, and MobileNetV2 is conducted by Ref. [42]. Transfer learning on ImageNet with VGG16 and ResNet50 [43] by replacing the last layer of both VGG16 and ResNet50 with one global average pooling layer [44] and two fully connected layers is used in Ref. [45].
Transfer learning on AlexNet, ResNet18, DenseNet201 and SqueezeNet is performed by Ref. [46]. Two-class and three-class classification with and without data augmentation is performed with fivefold cross-validation and stochastic gradient descent (SGD) optimization. Fig. 2 illustrates the working principle of [46] stepwise. Similar to Ref. [46], binary and multiclass classification on NASNet-Large, DenseNet169, InceptionV3, ResNet18, and Inception ResNet V2 are implemented by Punn et al. [47]. However [47], uses a weighted class loss function and random oversampling methods to overcome the disproportionate rates in the classes. The class with the ”COVID” label is given higher weight, since it is of higher significance than other classes, using the weighted class loss function. In the random oversampling method, the classes are balanced by increasing the number of samples in the minority class by data augmentation. For denoising, an image mask is created using binary thresholding and subtracted from the original image. Fine-tuning is performed by keeping nontrainable layers as the base model and adding four trainable convolutional layers, one fully connected layer and one classification layer. Transfer learning is also used by Wang et al. [48], but instead of the whole image, region of interests (RoIs)/image patches are provided as input. A total of 195 COVID-positive and 258 COVID-negative image patches are used for training. These image patches are input into a pretrained network for feature extraction, followed by a fully connected classification layer for classification.
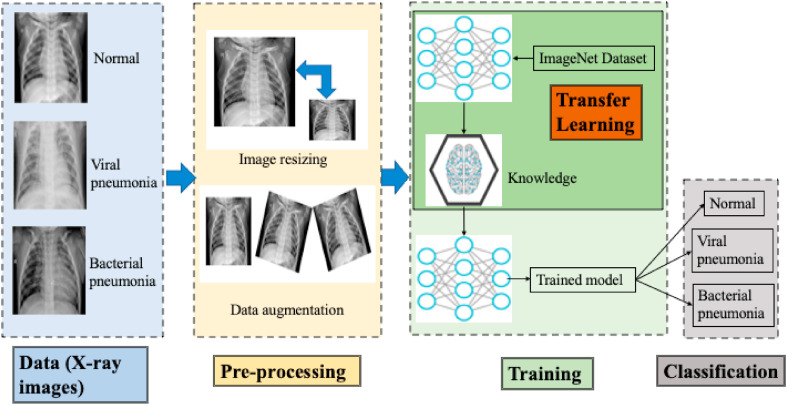
Fig. 2.
Stepwise diagrammatic representation of transfer learning by Chowdhury et al. [46]. The first step is the acquisition of the patients' data from an X-ray imaging machine. Both two-class classification and three-class classification are performed. Second, in the image resizing (preprocessing) step, the input layer of the deep learning method is fit. Data augmentation is performed in one of the experiments. Then, transfer learning is performed on various deep learning architectures. Finally, the trained model is saved, and classification is performed.
Generative adversarial networks (GANs) are used extensively for image reconstruction [49]. Data augmentation is one application of GANs [50]. Since the dataset is small, more data are obtained using a GAN for data augmentation, and the augmented data are split into training and testing sets to train a deep CNN model for binary classification [51]. Three phases are used. First, in the preprocessing phase, the GAN is used for data augmentation. Second, transfer learning on AlexNet, SqueezeNet, GoogleNet, and ResNet18 is performed to train the model. Finally, in the testing phase, the trained model is evaluated.
Along with fine-tuning on the top layers of the CNN, VGG16, VGG19, DenseNet201, Inception_ResNet_V2, Inception_V3, Xception, Resnet50, and MobileNet_V2 architectures, a comparative study is conducted [52]. Three convolutional layers with a filter size of 3 × 3, two max-pooling layers with a filter size of 2 × 2, a fully connected layer and, finally, a classification layer with a sigmoid classifier are proposed. Intensity normalization [53] and contrast limited adaptive histogram equalization (CLAHE) [54] are performed on the images during preprocessing.
First, a dataset is synthesized using a fuzzy color technique. Then, another dataset is created by combining the original and fuzzy color images using the stacking technique. Transfer learning and fine-tuning are performed on the created dataset [55]. Transfer learning on a combination of chest X-ray and CT scan images using the VGG19-CNN, ResNet152 V2, ResNet152 V2 + gated recurrent unit (GRU), and ResNet152 V2 + bidirectional GRU (Bi-GRU) architectures for multiclass classification is performed by Ibrahim et al. [56]. Transfer learning on 3D CT scans using ResNet architectures is also conducted [57]. A machine-learning algorithm-based method is also designed and evaluated for coronavirus detection [58,59].
3.2. Novel architectures
COVID-Net [60] utilizes a new CNN architecture for detecting COVID from CXR images, and an open-source COVID dataset, namely, COVIDx,1 is introduced. COVID-Net can classify CXR images into one of three classes. The architecture is based on lightweight residual projection-expansion projection extension (PEPX) design patterns with two stages of projections, expansions, a depthwise representation and an extension. The authors perform transfer learning by training the CNN architecture initially on the ImageNet dataset and subsequently on the COVIDx dataset.
A model with three parts, namely, a backbone, a classification head and an anomaly detection head, is proposed by Zhang et al. [61]. The pretrained backbone architecture on ImageNet is used to extract high-level features from X-ray images, and these features are fed to the classification and anomaly detection heads to produce a score. A cumulative score for every ’l’ predictions is also used.
COVID-CAPS is a capsule network-based framework for detecting the presence of COVID infection from CXR and CT scan images [62]. One advantage of using a capsule network is that it can perform well even when data are scarce. Transfer learning is also used in this framework. However, this is contrary to other methods of transfer learning on a model that is pretrained with X-ray images from a publicly available dataset 2. This has advantages over the other methods for transfer learning on the ImageNet dataset.
A novel CNN model is proposed by Abbas et al. [63], namely, DeTraC, which consists of three phases: feature extraction, decomposition and class composition. Using the backbone architecture, features from images are obtained. Then, training using the SGD optimizer is performed, followed by class composition for classification.
COVIDLite is a novel architecture that uses the depthwise separable convolutional neural network (DSCNN) to classify CXR images for coronavirus detection [64]. A preprocessing step (CLAHE) is used to improve the visibility and enhance the white balance. White balancing is performed to enhance the color fidelity of the images. Fast COVID-19 detector (FCOD) is another variant of the depthwise separable convolutional neural network, which is based on the inception architecture [65]. Using depthwise separable convolutional layers in place of the normal convolutional layer decreases the computational complexity and computation time. Similar to Ref. [65], depthwise separable convolutional layers are used in the XceptionNet architecture by Singh et al. [66].
A novel CNN with one convolutional block with a 16-filter convolutional layer, batch normalization and ReLU activation and two fully connected layers with softmax classification is proposed by Maghdid et al. [67]. The pretrained Alexnet on the ImageNet dataset is compared with the proposed model. A set of tailored CNN models that are based on established architectures is proposed by Ref. [68]. Each detected image can belong to one of three classes, namely, normal, viral pneumonia and bacterial pneumonia. Additionally, an estimator for the infection rate is provided from the predictions.
A custom CNN model that accepts concatenated features from two models (Xception and ResNet50V2) and passes them through a convolutional layer and a classification layer is proposed by Ref. [69]. Similarly, deep features are extracted from MobileNet as the base model, and they are input into a global pooling layer and a fully connected layer. Then, the feature vector is input into the classifier for classification by Ref. [70]. Three types of techniques are tested, namely, fine-tuning, transfer learning and training from scratch. As in Refs. [69,70], a deep convolutional neural network architecture, namely, CoroNet [71], is used to classify X-rays into four classes: normal, bacterial pneumonia, viral pneumonia and COVID-19 positive. The architecture is based on Xception as the base; however, a dropout and two fully connected layers are used. The Darknet-19 [72] based architecture, which is used for general object detection, is called the DarkCovid net [73]. It uses fewer layers than Darknet-19 with average pooling and softmax for classification, and transfer learning on the ImageNet dataset is performed.
A four-phase method for COVID-19 detection is implemented by Ozyurt [74]. The feature extraction technique is emphasized by using techniques such as exemplar-based pyramid feature generation, ReliefF, and iterative principal component analysis (PCA) analysis. The final stage is classification using a deep neural network (DNN) and an artificial neural network (ANN). CovXNet is a novel CNN architecture with depthwise convolutional layers [75]. Not only is this novel architecture trained from scratch but also different modifications, such as transfer learning and fine-tuning, are designed to compare the performances of various methods. Both binary classification and multiclass classification are performed on chest X-rays by unique CNN architectures without transfer learning by Karakanis et al. [76].
3.3. Other approaches
A pretrained model is used to extract the deep features of the images of a prepared custom dataset [77]. Then, the extracted deep features are input into a linear support vector machine (SVM) and OneVsAll SVM classifier for classification. Eleven established model architectures that are pretrained on the ImageNet dataset [28] are used to extract the deep features: AlexNet, DenseNet201, GoogleNet, InceptionV3, ResNet18, ResNet50, ResNet101, VGG16, VGG19, XceptionNet, and InceptionResNetV2.
A slightly different approach is applied by the authors for the classification of X-ray images [77]. Similar to Ref. [77]., features are extracted from three networks, namely, VGG-16, GoogleNet and ResNet-50 [78], for the classification of CT images. The features are fused, and to reduce the redundancy of the features, the t-test method is used to rank the features based on frequency. The final constructed feature vector is input into a binary SVM classifier for classification. A depthwise separable convolution neural network (DWS-CNN) is used to extract the features from the patient's X-ray images. The extracted features are input into a deep support vector machine (DSVM) for classification. Data acquisition occurs through Internet of things (IoT)-enabled devices. The raw data are passed through a Gaussian filter before feature extraction and classification [79]. A pretrained VGG16 network is used, and the output is upsampled to a depthwise separable convolutional network, which is followed by a shallow 3D CNN block and spatial pyramid pooling for COVID-19 detection [80].
A hierarchical classification method in place of flat classification is another proposed variation [81]. Hierarchical classification considers the relationships between classes, conducts local classification and trains models to perform the classification. Since the dataset is small even after customization, to avoid underfitting or overfitting of the model, the available data are expanded using data augmentation techniques. The EfficentNet [82] architecture family is used as the base model for the classification, which is extended by adding batch normalization and dropout, followed by three fully connected layers and classification using softmax. Additionally, instead of training from scratch, transfer learning on ImageNet dataset weights is carried out.
ResNet50 is used as the base model for classifying the image into three classes: normal, bacterial pneumonia and viral pneumonia [83]. If the prediction is viral, the image is input into DenseNet169 to further classify it as COVID or not. This is similar to hierarchical classification except that a single model is used for the full overflow, in contrast to Ref. [81]. Global average pooling (GAP) and SE-Structure are used to increase the performance of the model. Contrast limited adaptive histogram equalization (CLAHE) and the MoEx structure that is formed from normalization are used for image enhancement to help increase the accuracy. A gradient class activation map (Grad-CAM) is used for visualization to help doctors [84]. U-Net is used to segment the lung in the image, which is also provided as input to the DenseNet model. A workflow that is similar to Ref. [83] is proposed by Gozes et al. [85]. First, lung segmentation using U-Net is performed to extract the ROIs. The ROIs are provided as input for classification, and Grad-CAM is used for visualization.
A preprocessing step, which includes contrast and edge enhancement using histogram equalization (HGE), application of the Perona–Malik filter (PMF), and elimination of noise by unsharp masking edge enhancement, is conducted before the detection of coronavirus infection [86]. This preprocessing can help the model learn and generalize better. An ensemble-based method is employed for detection by training the VGG, ResNet, and DenseNet architectures. An ensemble of the best model predictions is used to obtain the final prediction.
COVID-MobileXpert is a deep learning-based hardware-friendly model with a knowledge transfer and distillation framework [87]. DenseNet-121 is used by the Attending Physician (AP) and Resident Fellow (RF) networks, and MobileNetv2, ShuffleNetV2 and SqueezeNet are used by the Medical Student (MS) network. The MS network has been designed to facilitate the deployment of the model on devices. Transfer learning is conducted on the AP and RF networks, and the RF network is used to train the MS network through knowledge distillation.
An ensemble method with three steps, namely, feature extraction using Alexnet, feature selection using trial and error and classification using the SVM algorithm, is performed. The results are compared with those of other deep learning methods, and the proposed solution has higher overall accuracy [88]. A multitask method is proposed by Rahman et al. [89], along with a new dataset, for image enhancement, segmentation, and classification.
Fig. 3 presents an overview summary of all the methods that are currently available for this application.
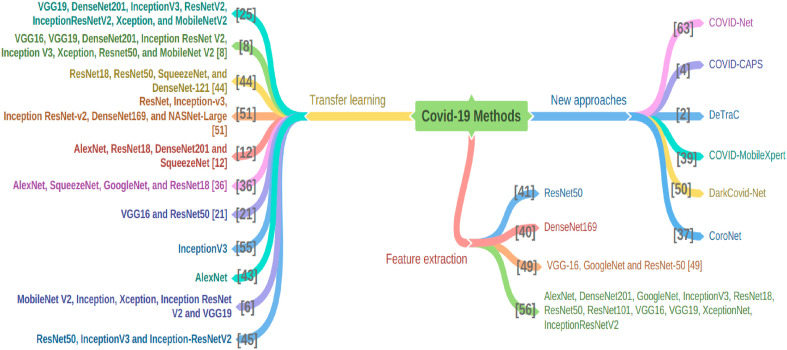
Fig. 3.
Methods and approaches. The surveyed literature works are grouped into three categories, namely, transfer learning and fine-tuning, novel architectures, and other approaches. Three branches are included in the figure, and the methods under each category are listed.
4. Datasets
The size of the data is the key factor for the performance of any deep learning model. However, since COVID-19 is a recent disease, only a limited number of datasets are publicly available. There is a repository of COVID-positive lung X-ray images that is constantly updated [90], solely for classification purposes. It also contains metadata and annotations of the lung segments. Additionally, this repository contains only a limited number of non-COVID images. Another commonly used dataset in this context is from Kaggle.2 Dr. Paul Mooney created a lung image dataset with 5,863 pediatric images under three classes (normal, viral pneumonia and bacterial pneumonia).3 Apart from these, the RSNA Pneumonia Detection Challenge dataset,4 SIRM datasets,5 Covid Chest X-ray dataset [92], and Chexpert dataset [93] are notable datasets that are used for COVID classification. Another important consideration is that some of the methods use binary classes (COVID+ and COVID-) whereas others use more than two classes (normal, COVID, viral pneumonia and bacterial pneumonia) for classification.
The COVIDx datasets that are introduced in COVID-Net [60] include 13,975 CXR images across 13,870 patient cases that have been selected and combined from publicly available datasets. The dataset consists of images in three classes, namely, normal, non-COVID infection and COVID infection. A detailed study and the steps for generating the dataset can be found in Ref. [94]. A custom COVID-Xray-5k dataset is built with 2,031 training images and 3,040 test images [41]. This dataset is a combination of COVID + images from the COVID Chest X-ray dataset [92] and ChexPert [93].6
Two datasets are used in Ref. [35] to evaluate transfer learning on various models. A combination of normal, COVID and bacterial pneumonia images from various sources, such as [90], and [95], are combined into one dataset with 504 normal images, 700 bacterial pneumonia images and 224 confirmed COVID images. However, to fine-tune and improve the performance of the models, another class, namely, viral pneumonia, is added to create another dataset. Dataset 2 consists of 504 normal images, 224 confirmed COVID images, 400 bacterial pneumonia infection images and 314 viral pneumonia infection images. A black background is added, and the images are rescaled to dimensions of 200 × 266. Even after all these efforts, the number of samples in the dataset is small, and the classes are not balanced with the minimum number of images for confirmed COVID cases.
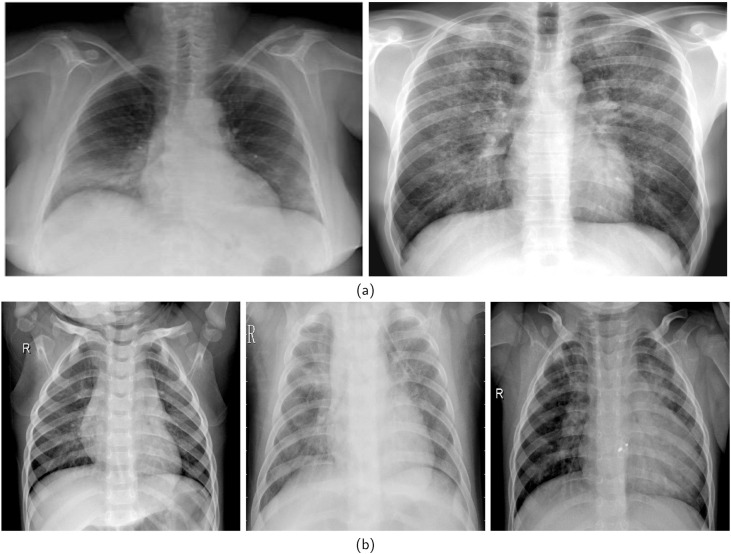
A few images that represent each class from the most commonly used datasets, namely [90,96], are presented in Fig. 4 [90]. has COVID+ and COVID-images, while [96] has normal, bacterial pneumonia, and viral pneumonia images. Table 1 summarizes in detail the most commonly used publicly available datasets.
Fig. 4.
(a). Example images from two classes, namely, COVID+ and COVID-, from the [90] dataset. (b). Example images from three classes, namely, normal, viral pneumonia and bacterial pneumonia, from the [91] dataset.
Table 1.
Summary of major publicly available datasets.
| Dataset | Size | Type | Classes |
|---|---|---|---|
| cohen | Regularly updated | Chest X-ray and CT images | 5 |
| Paul Mooney | 5856 | Chest X-ray | 3 |
| Kaggle | 97 | Chest X-ray and CT images | 2 |
| COVIDx | 104,009 | CT images | 3 |
| ChestXray-8 | 108,948 | Chest x-ray images | 3 |
| CheXpert | 224,316 | Chest radiographs | 5 |
| Kaggle 2 | 2909 | Chest X-ray images | 4 |
Similarly [35,77], develop two datasets for model training and testing. The first dataset consists of 25 COVID + images, excluding MERS, ARDS and SARS, and 25 COVID-images. The second dataset consists of 133 COVID + images, which include Middle East respiratory syndrome (MERS), acute respiratory distress syndrome (ARDS), and SARS images and 133 COVID-images from Refs. [90,97]. Four datasets are used for experiments in Ref. [38], namely, [90,92,93,98]. The experiments are conducted separately, and the classes and the images are combined. Three datasets, namely [90,92,99], are used in Ref. [81]. Multiple classes other than pneumonia are used, which include thorax diseases with COVID + images from Refs. [90,99,100]; pneumonia images from Ref. [101]; and other thoracic disease images from Ref. [92]. To add diversity, data augmentation is conducted.
A dataset with a total of 1300 images, namely, 310 normal, 330 bacterial pneumonia, 327 viral pneumonia and 284 COVID images, is used in Ref. [71]. COVID-positive images are obtained from Ref. [90], and normal, bacterial and viral pneumonia images are obtained from Ref. [91].
In [46], four datasets are combined. COVID images are collected from Refs. [90,100,101] by the authors of [46]. Normal and viral pneumonia images are collected from Ref. [91]. A two-class dataset is created.
Images from 5 sources are combined in Ref. [67]. [73] uses normal and pneumonia images from Ref. [92] and COVID images from Ref. [90]. To obtain a balanced dataset, only 500 random images from Ref. [92] for both classes are selected. Fivefold cross-validation is conducted with two experiments – binary and three-class classification..7
[68] uses [91] with images from normal, bacterial pneumonia and viral pneumonia for training, and testing is performed on COVID images from Ref. [90]. It is assumed that any infections that are caused by COVID-19 are due to viruses; thus, the model has to predict the COVID-positive images under the viral pneumonia class. Covid images from Ref. [90] and pneumonia, no findings and normal images from Ref. [99] are used to form a dataset in Ref. [47].
[36] uses 50 COVID infection images from Ref. [90], 50 normal healthy images from Ref. [91], 100 COVID images from Ref. [90], and 1431 pneumonia infection images from Ref. [92] in Ref. [61]. A total of 130 COVID-19 and 130 normal X-ray images from Refs. [90,96,97] are used in Ref. [37]. A dataset that combines [90,91] is used in Ref. [69]..8 Eighty normal images from Refs. [102,103] and 116 images from Ref. [90], along with data augmentation, are used in Ref. [63]. In Ref. [51], 624 images with two classes [101] are used [101]. is used in Ref. [52] for binary classification using data augmentation [42]. uses [90,104] for COVID and normal images, respectively. In Ref. [42], 25 normal and 25 COVID images are used [90,100,105]. are used to obtain 135 COVID images, and 320 pneumonia images from Ref. [106] are collected to form the dataset in Ref. [43]. To balance the dataset, only 102 images from both classes are considered, and 10-fold cross-validation is performed as the dataset is small.
First [92], is used to train COVID-CAPS. Then, transfer learning is performed on COVIDx [94] in COVID-CAPS [62]. The COVIDx dataset is used in Ref. [86]. Balanced and unbalanced datasets are considered for experiments. CXR images and noisy snapshots of the lung images are the inputs that are used in Ref. [87]. Normal and pneumonia images are obtained from Ref. [18], and COVID images are obtained from Ref. [90]. Microsoft Office Lens is used to capture snapshots of the images on the PC screen to create the noisy snapshot dataset. The captured images are RGB images, which are converted to 8-bit grayscale images [78]. uses 53 COVID images from Ref. [100]. Two patch datasets are obtained from these 53 images by selecting the COVID-infected and noninfected regions in the CT images. Two patch sizes are considered: 16 × 16 and 32 × 32. A total of 3000 patches from COVID infection images and 3000 no-finding patches are used to form the dataset.
A few images that represent each class from the most commonly used datasets, namely [90,96], are presented in Fig. 4 [90]. uses COVID+ and COVID-images, and [96] uses normal, bacterial and viral pneumonia images. Table 2 summarizes the primarily used publicly available datasets.
Table 2.
Comparative analysis of the methods in terms of accuracy, precision/PPV, recall/sensitivity, specificity, NPV and F1-score.
| Method | Model/Backbone | Accuracy | Precision | Sensitivity | Specificity | NPV | F1-score |
|---|---|---|---|---|---|---|---|
| [35] | VGG19 | 98.75 | – | 92.85 | 98.75 | – | – |
| MobileNetV2 | 97.40 | – | 99.10 | 97.09 | – | – | |
| Inception | 86.13 | – | 12.94 | 99.70 | – | – | |
| Xception | 85.57 | – | 0.08 | 99.99 | – | – | |
| InceptionResNetV2 | 84.38 | – | 0.01 | 99.83 | – | – | |
| [36] | InceptionV3 | 95.4 | 73.4 | 90.6 | 96.0 | – | 81.1 |
| ResNet50 | 96.1 | 76.5 | 91.8 | 96.6 | – | 83.5 | |
| ResNet101 | 96.1 | 84.2 | 78.3 | 98.2 | – | 81.2 | |
| ResNet152 | 93.9 | 74.8 | 65.4 | 97.3 | – | 69.8 | |
| InceptionResNetV2 | 94.2 | 67.7 | 83.5 | 95.4 | – | 74.8 | |
| [37] | InceptionV3 | 100 | 100 | 100 | 100 | 100 | 100 |
| [41] | ResNet18 | – | – | 98.0 | 90.7 | – | – |
| ResNet50 | – | – | 98.0 | 89.6 | – | – | |
| SqueezeNet | – | – | 98.0 | 92.9 | – | – | |
| DenseNet121 | – | – | 98.0 | 75.1 | – | – | |
| [42] | VGG19 | 90.0 | 83.0 | 100 | – | – | 91.0 |
| DenseNet201 | 90.0 | 83.0 | 100 | – | – | 91.0 | |
| ResNetV2 | 70.0 | 100 | 40.0 | – | – | 57.0 | |
| InceptionResNetV2 | 80.0 | 100 | 60.0 | – | – | 75.0 | |
| XceptionNet | 80.0 | 100 | 60.0 | – | – | 75.0 | |
| MobileNetV2 | 60.0 | 100 | 20.0 | – | – | 33.0 | |
| [46] | VGG19 | 99.6 | 99.2 | 98.6 | 99.8 | – | 98.9 |
| ResNet18 | 99.6 | 99.6 | 99.6 | 99.3 | – | 99.6 | |
| DenseNet201 | 99.7 | 99.7 | 99.7 | 99.55 | – | 99.7 | |
| SqueezeNet | 99.4 | 99.4 | 99.4 | 99.84 | – | 98.4 | |
| MobileNetV2 | 99.65 | 99.65 | 99.65 | 99.26 | – | 99.65 | |
| ResNet101 | 99.6 | 99.6 | 99.6 | 99.31 | – | 99.6 | |
| InceptionV3 | 99.40 | 98.80 | 98.33 | 99.7 | – | 98.56 | |
| [47] | ResNet | 89.0 | 67.0 | 89.0 | 85.0 | – | 76.0 |
| InceptionV3 | 88.0 | 90.0 | 88.0 | 90.0 | – | 85.0 | |
| InceptionResNetV2 | 95.0 | 97.0 | 96.0 | 94 | – | 96.0 | |
| DenseNet169 | 92.0 | 94.0 | 96.0 | 95.0 | – | 95.0 | |
| NASNetLarge | 98.0 | 95.0 | 91.0 | 98.0 | – | 98.0 | |
| [48] | Inception | 89.5 | 71.0 | 88.0 | 87.0 | 95.0 | 77.0 |
| [51] | AlexNet | 96.1 | 96.52 | 95.37 | – | – | 95.94 |
| ResNet18 | 99.0 | 98.97 | 98.97 | – | – | 98.97 | |
| GoogLeNet | 96.8 | 98.63 | 98.31 | – | – | 94.46 | |
| SqueezeNet | 97.8 | 93.6 | 95.88 | – | – | 98.47 | |
| [52] | CNN | 84.18 | 94.05 | 78.33 | 93.07 | – | 85.66 |
| VGG16 | 86.26 | 87.73 | 85.22 | 87.36 | – | 86.46 | |
| VGG19 | 85.94 | 80.39 | 90.43 | 82.35 | – | 85.11 | |
| ResNet50 | 96.61 | 98.46 | 94.92 | 98.43 | – | 96.67 | |
| MobileNetV2 | 96.27 | 98.06 | 94.61 | 98.02 | – | 96.30 | |
| InceptionV3 | 94.59 | 93.75 | 95.35 | 93.85 | – | 94.54 | |
| InceptionResNetV2 | 96.09 | 98.61 | 93.88 | 98.53 | – | 96.19 | |
| DenseNet201 | 93.66 | 99.01 | 89.44 | 98.89 | – | 93.98 | |
| Xception | 83.14 | 95.77 | 76.45 | 94.34 | – | 85.03 | |
| [56] | VGG19+CNN | 98.05 | 98.43 | 98.05 | 99.5 | 99.3 | 98.24 |
| ResNet152V2 | 95.31 | 95.31 | 95.31 | 98.4 | 98.4 | 95.31 | |
| ResNet152V2+GRU | 96.09 | 96.06 | 96.09 | 98.7 | 98.7 | 96.09 | |
| ResNet152V2+Bi-GRU | 93.36 | 93.35 | 93.16 | 97.8 | 97.8 | 93.26 |
5. Evaluation
As in any other classification tasks, the metrics that are used to evaluate the models are accuracy and precision, which are also called positive prediction value (PPV) and negative prediction value (NPV), respectively; specificity; recall, which is also called sensitivity; and F1-score; these are the most commonly used measures. To calculate these measures, four main metrics are used: (a) correctly identified diseased cases (true positives, TP), (b) incorrectly classified diseased cases (false negatives, FN), (c) correctly identified healthy cases (true negatives, TN), and (d), incorrectly classified healthy cases (false positives, FP). The equations for calculating accuracy, specificity and sensitivity are presented in 1, 5, and 4.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
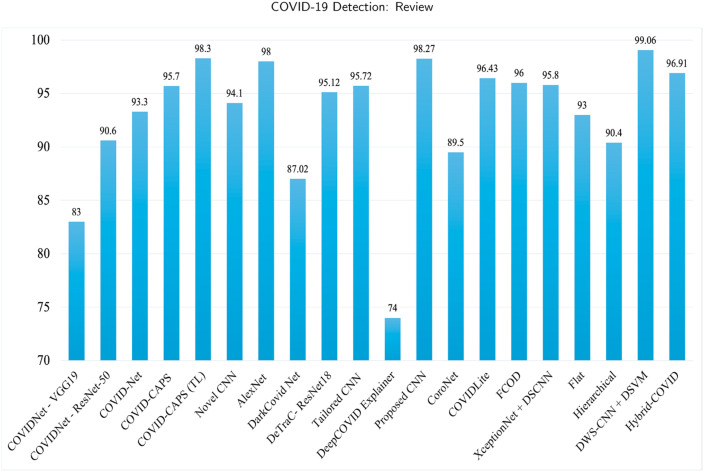
Table 1 summarizes the methods and the accuracies that are realized in various papers. Additionally, methods for binary classification are presented in the table. Fig. 5 presents a detailed comparison of the results of novel architectures and other approaches.
Fig. 5.
Graphical representation of the accuracy results for novel architectures and other approaches.
The receiver operating characteristic (ROC) curve and area under the ROC curve (AUC) are the other evaluation metrics that are commonly used. The ROC curve is used to show the performance of the proposed model by plotting the true positive rate (TPR), which is also called the recall, against the false-positive rate (FPR), at various thresholds. The equation for calculating FPR is presented in Equation (7). Lowering the classification threshold results in the classification of more items as positive, thereby increasing both the number of false positives and the number of true positives. AUC is an aggregate measure for evaluating a model at various possible thresholds. It is the two-dimensional area under the ROC curve between (0,0) and (1,1). AUC is the probability that the model ranks a random positive example higher than a random negative example.
| (7) |
Accuracy, by default, is the common metric that is used by almost every method in the study except in Refs. [7,38,41,70,86], and [87]. Specificity and sensitivity are the measures used in Refs. [7,35,37,41,46,48,52,[60], [61], [62], [63],70,73,77,78], and [85]. Precision/PPV, recall and f1-score are used in Refs. [37,42,[46], [47], [48],51,52,60,73,78,81,83,86], and [85]. Additionally, a statistical analysis among the models is performed using the false-positive rate, F1, MCC and kappa measures in Ref. [77]. ROC-AUCs are used to measure the model performance in Refs. [38,41,46,47,62,83], and [60]. [69] uses the accuracy of all classes, accuracy of each class, precision, recall and specificity. Overall accuracy, classwise precision, recall and F-measure are the measures that are used in Ref. [71]. CPU (%), memory (MB), energy and AUC are used in Ref. [87]. The Matthews Correlation Coefficient (MCC) [107] is the extra metric that is used in Ref. [78].
6. Discussion and future direction
From Table 2, Salman et al. [37] realize the best performance in terms of accuracy, precision, specificity, sensitivity, NPV and F1-score, with values of 100%. This method uses InceptionV3 as the model with transfer learning. However, the use of the same InceptionV3 architecture in Refs. [36,46,47], and [52] did not produce the same results as in Ref. [37]. It is observed that Salman et al. use data augmentation with two classes of 130 images and 260 images in total. The models that produce the second- and third-best results are Denset201 and MobileNet V2 in Ref. [46], which are designed by Chowdhury et al. The second and third best results are not far from the first result of 100%. DenseNet201 achieves 99.7% accuracy, which is the second-best result. MobileNetV2 realizes an accuracy of 99.65%, which is only 0.05% less than the second-best accuracy and 0.35% less than the best accuracy results.
Apart from accuracy, other evaluation metrics are used, namely, precision, recall/sensitivity, specificity, and F1-score. Hemdan et al. [42] produce a precision of 100%. Similar to accuracy, Denset201 produces the second-best result with 99.7% precision, and MobileNetV2 has the third-best precision of 99.65%. DenseNet201 and MobileNetV2 produce the second-best results in terms of sensitivity and F1-score. The sensitivity and F1-score values for DenseNet201 and MobileNetV2 are 99.7% and 99.65%, respectively. However, DensetNet201 and MobileNet V2 do not produce results with higher specificity. In terms of specificity, SqueezeNet realizes the second-best value of 99.84%, and VGG19 produces the third-best values of 99.8%, with a mere difference of 0.04%. NPV values are not used by many methods. The best NPV value of 100% is produced by Inception V3 in Ref. [37]. Better results are produced by the combination of VGG19 and CNN, which realizes 99.3%, and the combination of ResNet152 V2 and GRU, which realizes 98.7% [56].
In summary, most of the methods utilize transfer learning on established architectures for the classification of lung images. Even if novel architectures are proposed, due to scarcity of available image data, transfer learning on the ImageNet dataset is considered [60,71]. Different network architectures are used by different methods. Out of all of them, Inception, DenseNet, MobileNet, SqueezeNet and the VGG family outperform the other families.
To effectively detect coronavirus infection, an easy, fast, and accurate application that can be deployed in hand-held devices has to be developed. Most of the architectures that are used in the literature have many layers and, hence, huge numbers of parameters to store and compute. The ResNet50 architecture has 53 convolutions and one fully connected layer with over 23 million trainable parameters [108]. performed a detailed analysis on the memory requirements of each model before and after deployment in a hand-held device chip. The memory requirement of the ResNet model is so large that it is expensive and impractical to deploy the trained model on a mobile device. Memory is compromised in the place of accuracy. The feasibility and portability of the application for the detection of coronavirus is affected.
Accuracy is the common metric that is used to evaluate the performances of models. VGG19 shows satisfactory performance with an accuracy of 98.75%. In addition, VGG19 has fewer parameters and a shallower model, which makes it easily deployable even in mobile applications and mobile devices. Although some of the other approaches, such as deep feature extraction [78] and hierarchical classification [81], have been tested, they did not achieve better performance in comparison.
As discussed earlier, deep learning methods need large amounts of data to perform well. Although most of the methods have tried to overcome the shortage of data with various data augmentation methods, there is no proof of real-time detection. There is no proven evidence on the effectiveness of data augmentation in real-life and live images for the detection of coronavirus. Creating a public dataset with possible classes requires help from medical experts, which is time-consuming. Since the availability of public datasets is low, studies have tailored custom datasets by combining two or three repositories based on the application. The popular representations are [90] for COVID images and [96] for normal, bacterial and viral pneumonia images.
A preprocessing step for resizing the input image to fit the architecture is conducted before training and testing the model. Careful consideration must be taken when dealing with medical images. Medical images are easily prone to noise, and this noise has to be removed before passing them to the model; otherwise, the model will learn the noise [109]. This may affect the performance of the model. An effective preprocessing step for removing artifacts and noise is essential for improving the model performance.
The major advantage in using the deep learning models is the ease of using them without any requirements for manually picking the features. However, in the case of medical images, the selection and use of features were of higher importance than any other tasks. The features that are selected by the deep learning models are not interpretable by medical professionals, and hence, the reliability is not certain; hence, it is unclear how the application can help them.
The privacy and security of confidential materials such as X-ray images, patient information and other details are of the utmost importance.
In the future, more publicly available datasets with lung images can be collected and constructed for future use. Without the availability of quality data, the performance of the deep learning models cannot be improved. Other research directions include constructing and annotating data and providing metadata information.
7. Conclusion
The COVID-19 pandemic is a novel pandemic that is caused by the coronavirus, and the only preventive measures that are available thus far are social distancing and early detection. For early detection and prevention of spread, deep learning models are trained to detect and classify lung images. Since the spread of the COVID-19 pandemic started recently in the last quarter of 2019, limited data are available for training deep learning models. To overcome this scarcity, researchers created custom datasets by combining many repositories. Transfer learning on established architectures, novel architectures with transfer learning on the ImageNet dataset, and other approaches, such as deep feature extraction using a deep learning architecture and hierarchical classification methods, are the methods that are available in the study. Among these available methods, transfer learning performs the best, and out of all the architectures, InceptionV3, DenseNet201, and MobileNetV2 realize higher accuracy, while SqueezeNet and VGG19 show better specificity. Although vaccine drives are occurring all around the world, supply chain logistics and fear of the vaccine are some of the major issues. The RT–PCR test that is currently used for the detection of coronavirus is expensive, time-consuming, and less sensitive. Chest X-rays, CT scans, and ultrasound images of the lungs are primarily considered for detecting coronavirus detection by health care officials. Deep learning methods can facilitate coronavirus detection using images at early stages. The best results of 100% accuracy, 100% precision, 100% specificity, 100% sensitivity, 100% NPV, and 100% F1-score show the higher reliability of the deep learning methods.
Declaration of competing interest
None declared.
Acknowledgement
This publication was supported by Qatar University COVID19 Emergency Response Grant (QUERG-CENG-2020-1). The findings achieved herein are solely the responsibility of the authors. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the Qatar University.
Biographies

Nandhini Subramanian received master's degree in computing from Qatar University, Doha, and bachelor's degree in Electrical and Electronics Engineering from the PSG College of Technology, India. Her interests include computer vision, artificial intelligence, machine learning and cloud computing. She is currently working as Research Assistant with Dr. Somaya Al-Maadeed in Qatar University, Doha. She is the winner of the national-level AI competition (Qatar) at the student level.

Omar Elharrouss received a master's degree in 2013 from the Faculty of Sciences, Dhar El Mehraz, Fez, Morocco. In 2017, he received a Ph.D. from the LIIAN Laboratory at USMBA-Fez University. His research interests include pattern recognition, image processing, and computer vision.
Somaya Al-Maadeed received a Ph.D. degree in computer science from Nottingham, U.K., in 2004. She is the Coordinator of the Computer Vision and AI Research Group. She enjoys excellent collaboration with national and international institutions and industry. She is a principal investigator of several funded research projects that involve approximately five million people. She has published extensively in the field of pattern recognition and delivered workshops on teaching programming for undergraduate students. She has attended workshops that were related to higher education strategy, assessment methods, and interactive teaching. In 2015, she was elected as the IEEE Chair for the Qatar Section.

Muhammad E. H. Chowdhury received B.Sc. and M.Sc. degrees from the Department of Electrical and Electronics Engineering, University of Dhaka, Bangladesh, and a Ph.D. degree from the University of Nottingham, U.K., in 2014. He worked as a Postdoctoral Research Fellow and a Hermes Fellow with the Sir Peter Mansfield Imaging Centre, University of Nottingham. He is currently working as an Assistant Professor with the Department of Electrical Engineering, Qatar University. Before joining Qatar University, he worked in several universities in Bangladesh. He has two patents and published approximately 80 peer-reviewed journal articles, conference papers, and four book chapters. His current research interests include biomedical instrumentation, signal processing, wearable sensors, medical image analysis, machine learning, embedded system design, and simultaneous EEG/fMRI. He is currently the recipient of several NPRP and UREP grants from QNRF and internal grants from Qatar University, and he conducts academic and government projects. He has been involved in EPSRC, ISIF, and EPSRC-ACC grants, along with various national and international projects. He has worked as a consultant for the project entitled Driver Distraction Management Using Sensor Data Clouds (2013–2014), Information Society Innovation Fund (ISIF) Asia. He is an Active Member of British Radiology, the Institute of Physics, ISMRM, and HBM. He received the ISIF Asia Community Choice Award 2013 for a project entitled Design and Develop-ment of a Precision Agriculture Information System for Bangladesh. He recently received the COVID-19 Dataset Award and National AI Com-petition awards for his contribution to the fight against COVID-19. He is serving as an Associate Editor for IEEE Access and a Topic Editor and Review Editor for Frontiers in Neuroscience.
Footnotes
References
- 1.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh R., Sarsaiya S., Singh T.A., Singh T., Pandey L.K., Pandey P.K., Khare N., Sobin F., Sikarwar R., Gupta M.K. Corona virus (covid-19) symptoms prevention and treatment: a short review. J. Drug Deliv. Therapeut. 2021;11(2-S):118–120. [Google Scholar]
- 3.Zhou L., Wu Z., Li Z., Zhang Y., McGoogan J.M., Li Q., Dong X., Ren R., Feng L., Qi X., et al. One hundred days of coronavirus disease 2019 prevention and control in China. Clin. Infect. Dis. 2021;72(2):332–339. doi: 10.1093/cid/ciaa725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samaranayake L.P., Seneviratne C.J., Fakhruddin K.S. Coronavirus disease 2019 (covid-19) vaccines: a concise review. Oral Dis. 2021:1–11. doi: 10.1111/odi.13916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rehman A., Saba T., Tariq U., Ayesha N. Deep learning-based covid-19 detection using ct and x-ray images: current analytics and comparisons. IT Professional. 2021;23(3):63–68. doi: 10.1109/MITP.2020.3036820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.economist.com. 2020. https://www.economist.com/graphic-detail/2020/03/20/coronavirus-research-is-being-published-at-a-furious-pace Academic articles. [Google Scholar]
- 7.Hassaballah M., Awad A.I. CRC Press; 2020. Deep Learning in Computer Vision: Principles and Applications. [Google Scholar]
- 8.Ciaparrone G., Sánchez F.L., Tabik S., Troiano L., Tagliaferri R., Herrera F. Deep learning in video multi-object tracking: a survey. Neurocomputing. 2020;381:61–88. [Google Scholar]
- 9.Asadi-Aghbolaghi M., Clapes A., Bellantonio M., Escalante H.J., Ponce-López V., Baró X., Guyon I., Kasaei S., Escalera S. 2017 12th IEEE International Conference on Automatic Face & Gesture Recognition (FG 2017) IEEE; 2017. A survey on deep learning based approaches for action and gesture recognition in image sequences; pp. 476–483. [Google Scholar]
- 10.Masi I., Wu Y., Hassner T., Natarajan P. 2018 31st SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI) IEEE; 2018. Deep face recognition: a survey; pp. 471–478. [Google Scholar]
- 11.Subramanian N., Elharrouss O., Al-Maadeed S., Bouridane A. IEEE Access; 2021. Image Steganography: A Review of the Recent Advances. [Google Scholar]
- 12.Elharrouss O., Almaadeed N., Al-Maadeed S. 2020 IEEE International Conference on Informatics, IoT, and Enabling Technologies (ICIoT) 2020. An image steganography approach based on k-least significant bits (k-lsb) pp. 131–135. [DOI] [Google Scholar]
- 13.Subramanian N., Elharrouss O., Al-Maadeed S., Bouridane A. End-to-end image steganography using deep convolutional autoencoders. IEEE Access. 2021 doi: 10.1109/ACCESS.2021.3113953. 1–1. [DOI] [Google Scholar]
- 14.Ulhaq A., Khan A., Gomes D., Pau M. Computer vision for covid-19 control: a survey. arXiv preprint arXiv:2004. 2020 doi: 10.1109/ACCESS.2020.3027685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19. IEEE.Rev. Biomed. Eng. 2020;14:1–15. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 16.Gu J., Wang Z., Kuen J., Ma L., Shahroudy A., Shuai B., Liu T., Wang X., Wang G., Cai J., et al. Recent advances in convolutional neural networks. Pattern Recogn. 2018;77:354–377. [Google Scholar]
- 17.Kunhoth J., Karkar A., Al-Maadeed S., Al-Attiyah A. Comparative analysis of computer-vision and ble technology based indoor navigation systems for people with visual impairments. Int. J. Health Geogr. 2019;18(1):1–18. doi: 10.1186/s12942-019-0193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [Google Scholar]
- 19.Nielsen M.A. vol. 25. Determination press; San Francisco, CA: 2015. (Neural Networks and Deep Learning). [Google Scholar]
- 20.Sarıgül M., Ozyildirim B.M., Avci M. Differential convolutional neural network. Neural Network. 2019;116:279–287. doi: 10.1016/j.neunet.2019.04.025. [DOI] [PubMed] [Google Scholar]
- 21.Fakhrou A., Kunhoth J., Al Maadeed S. Multimedia Tools and Applications; 2021. Smartphone-based Food Recognition System Using Multiple Deep Cnn Models; pp. 1–22. [Google Scholar]
- 22.Ioffe S., Szegedy C. International Conference on Machine Learning. PMLR; 2015. Batch normalization: accelerating deep network training by reducing internal covariate shift; pp. 448–456. [Google Scholar]
- 23.Zhao H., Gallo O., Frosio I., Kautz J. 2015. Loss Functions for Neural Networks for Image Processing. arXiv preprint arXiv:1511.08861. [Google Scholar]
- 24.Chakraborty S., Mondal R., Singh P.K., Sarkar R., Bhattacharjee D. Transfer learning with fine tuning for human action recognition from still images. Multimed. Tool. Appl. 2021;80(13):20547–20578. [Google Scholar]
- 25.Cai Z., Peng C. 2021 13th International Conference on Knowledge and Smart Technology (KST) IEEE; 2021. A study on training fine-tuning of convolutional neural networks; pp. 84–89. [Google Scholar]
- 26.Simonyan K., Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv. 2014:1409–1556. https://arxiv.org/abs/1409.1556 [Google Scholar]
- 27.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., Berg A.C., Fei-Fei L. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115(3):211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 28.Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L. ImageNet: A Large-Scale Hierarchical Image Database. 2009. CVPR09. [Google Scholar]
- 29.Szegedy C., Liu W., Jia Y., Sermanet P., Reed S., Anguelov D., Erhan D., Vanhoucke V., Rabinovich A. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015. Going deeper with convolutions; pp. 1–9. [Google Scholar]
- 30.Szegedy C., Ioffe S., Vanhoucke V., Alemi A.A. Thirty-first AAAI Conference on Artificial Intelligence. 2017. Inception-v4, inception-resnet and the impact of residual connections on learning. [Google Scholar]
- 31.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 32.Huang G., Liu Z., van der Maaten L., Weinberger K.Q. 2016. Densely Connected Convolutional Networks. arXiv:1608.06993. [Google Scholar]
- 33.Howard A.G., Zhu M., Chen B., Kalenichenko D., Wang W., Weyand T., Andreetto M., Adam H., Mobilenets . 2017. Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv preprint arXiv:1704.04861 arXiv:1704.04861. [Google Scholar]
- 34.Chollet F. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Xception: deep learning with depthwise separable convolutions; pp. 1251–1258. [Google Scholar]
- 35.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng.Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021:1–14. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salman F.M., Abu-Naser S.S., Alajrami E., Abu-Nasser B.S., Alashqar B.A. Covid-19 detection using artificial intelligence. Int. J. Acad. Eng. Res.(IJAER) 2020;4:18–25. [Google Scholar]
- 38.Maguolo G., Nanni L. A critic evaluation of methods for covid-19 automatic detection from x-ray images. arXiv preprint arXiv:2004. 2020:12823. doi: 10.1016/j.inffus.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iandola F.N., Han S., Moskewicz M.W., Ashraf K., Dally W.J., Keutzer K. Squeezenet: Alexnet-level accuracy with 50x fewer parameters and< 0.5 mb model size. arXiv preprint arXiv:1602.07360. 2016 [Google Scholar]
- 40.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 41.Minaee S., Kafieh R., Sonka M., Yazdani S., Soufi G.J. Deep-covid: predicting covid-19 from chest x-ray images using deep transfer learning. Med. Image Anal. 2020;65 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hemdan E.E.-D., Shouman M.A., Karar M.E. Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv preprint arXiv:2003. 2020:11055. [Google Scholar]
- 43.Hall L.O., Paul R., Goldgof D.B., Goldgof G.M. Finding covid-19 from chest x-rays using deep learning on a small dataset. arXiv preprint arXiv:2004. 2020 [Google Scholar]
- 44.Lin M., Chen Q., Yan S. Network in network. arXiv preprint arXiv:1312.4400. 2013 [Google Scholar]
- 45.Ma W., Lu J. An equivalence of fully connected layer and convolutional layer. arXiv preprint arXiv:1712.01252. 2017 [Google Scholar]
- 46.Chowdhury M.E., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Al Emadi N., et al. Can ai help in screening viral and covid-19 pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- 47.Punn N.S., Agarwal S. Automated diagnosis of covid-19 with limited posteroanterior chest x-ray images using fine-tuned deep neural networks. Appl. Intell. 2021;51(5):2689–2702. doi: 10.1007/s10489-020-01900-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., Cai M., Yang J., Li Y., Meng X., et al. A deep learning algorithm using ct images to screen for corona virus disease (covid-19) Eur. Radiol. 2021:1–9. doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., Courville A., Bengio Y. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 2014;27 [Google Scholar]
- 50.Creswell A., White T., Dumoulin V., Arulkumaran K., Sengupta B., Bharath A.A. Generative adversarial networks: an overview. IEEE Signal Process. Mag. 2018;35(1):53–65. [Google Scholar]
- 51.Khalifa N.E.M., Taha M.H.N., Hassanien A.E., Elghamrawy S. Detection of coronavirus (covid-19) associated pneumonia based on generative adversarial networks and a fine-tuned deep transfer learning model using chest x-ray dataset. arXiv preprint arXiv:2004. 2020 [Google Scholar]
- 52.El Asnaoui K., Chawki Y., Idri A. Artificial Intelligence and Blockchain for Future Cybersecurity Applications. Springer; 2021. Automated methods for detection and classification pneumonia based on x-ray images using deep learning; pp. 257–284. [Google Scholar]
- 53.Sintorn I.-M., Bischof L., Jackway P., Haggarty S., Buckley M. Gradient based intensity normalization. J. Microsc. 2010;240(3):249–258. doi: 10.1111/j.1365-2818.2010.03415.x. [DOI] [PubMed] [Google Scholar]
- 54.Yadav G., Maheshwari S., Agarwal A. 2014 International Conference on Advances in Computing, Communications and Informatics (ICACCI) IEEE; 2014. Contrast limited adaptive histogram equalization based enhancement for real time video system; pp. 2392–2397. [Google Scholar]
- 55.Toğaçar M., Ergen B., Cömert Z. Covid-19 detection using deep learning models to exploit social mimic optimization and structured chest x-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ibrahim D.M., Elshennawy N.M., Sarhan A.M. Deep-chest: multi-classification deep learning model for diagnosing covid-19, pneumonia, and lung cancer chest diseases. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Serte S., Demirel H. Deep learning for diagnosis of covid-19 using 3d ct scans. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alves M.A., Castro G.Z., Oliveira B.A.S., Ferreira L.A., Ramírez J.A., Silva R., Guimarães F.G. Explaining machine learning based diagnosis of covid-19 from routine blood tests with decision trees and criteria graphs. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shiri I., Sorouri M., Geramifar P., Nazari M., Abdollahi M., Salimi Y., Khosravi B., Askari D., Aghaghazvini L., Hajianfar G., et al. Machine learning-based prognostic modeling using clinical data and quantitative radiomic features from chest ct images in covid-19 patients. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L., Wong A. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. arXiv preprint arXiv:2003. 2020 doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J., Xie Y., Li Y., Shen C., Xia Y. Covid-19 screening on chest x-ray images using deep learning based anomaly detection. arXiv preprint arXiv:2003. 2020:12338. [Google Scholar]
- 62.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. Covid-caps: a capsule network-based framework for identification of covid-19 cases from x-ray images. Pattern Recogn. Lett. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of covid-19 in chest x-ray images using detrac deep convolutional neural network. arXiv preprint arXiv:2003. 2020:13815. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siddhartha M., Santra A. Covidlite: a depth-wise separable deep neural network with white balance and clahe for detection of covid-19. arXiv preprint arXiv:2006. 2020:13873. [Google Scholar]
- 65.Panahi A.H., Rafiei A., Rezaee A. Fcod: fast covid-19 detector based on deep learning techniques. Informatics in Medicine Unlocked. 2021;22 doi: 10.1016/j.imu.2020.100506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh K.K., Siddhartha M., Singh A. Diagnosis of coronavirus disease (covid-19) from chest x-ray images using modified xceptionnet. Rom. J. Inf. Sci. Technol. 2020;23(657):91–115. [Google Scholar]
- 67.Maghdid H.S., Asaad A.T., Ghafoor K.Z., Sadiq A.S., Khan M.K. Diagnosing covid-19 pneumonia from x-ray and ct images using deep learning and transfer learning algorithms. arXiv preprint arXiv:2004. 2020 00038. [Google Scholar]
- 68.Hammoudi K., Benhabiles H., Melkemi M., Dornaika F., Arganda-Carreras I., Collard D., Scherpereel A. Deep learning on chest x-ray images to detect and evaluate pneumonia cases at the era of covid-19. arXiv preprint arXiv:2004. 2020 doi: 10.1007/s10916-021-01745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rahimzadeh M., Attar A. A new modified deep convolutional neural network for detecting covid-19 from x-ray images. arXiv e-prints (2020) arXiv. 2004 doi: 10.1016/j.imu.2020.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Apostolopoulos I.D., Aznaouridis S.I., Tzani M.A. Extracting possibly representative covid-19 biomarkers from x-ray images with deep learning approach and image data related to pulmonary diseases. J. Med. Biol. Eng. 2020:1. doi: 10.1007/s40846-020-00529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khan A.I., Shah J.L., Bhat M.M. Coronet: a deep neural network for detection and diagnosis of covid-19 from chest x-ray images. Comput. Methods Progr. Biomed. 2020 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Redmon J., Farhadi A. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Yolo9000: better, faster, stronger; pp. 7263–7271. [Google Scholar]
- 73.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ozyurt F., Tuncer T., Subasi A. An automated covid-19 detection based on fused dynamic exemplar pyramid feature extraction and hybrid feature selection using deep learning. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mahmud T., Rahman M.A., Fattah S.A. Covxnet: a multi-dilation convolutional neural network for automatic covid-19 and other pneumonia detection from chest x-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020;122 doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karakanis S., Leontidis G. Lightweight deep learning models for detecting covid-19 from chest x-ray images. Comput. Biol. Med. 2021;130 doi: 10.1016/j.compbiomed.2020.104181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sethy P.K., Behera S.K. Detection of coronavirus disease (covid-19) based on deep features. Preprints 2020030300. 2020 2020. [Google Scholar]
- 78.Ozkaya U., Ozturk S., Barstugan M. Coronavirus (covid-19) classification using deep features fusion and ranking technique. arXiv preprint arXiv:2004. 2020 [Google Scholar]
- 79.Le D.-N., Parvathy V.S., Gupta D., Khanna A., Rodrigues J.J., Shankar K. Iot enabled depthwise separable convolution neural network with deep support vector machine for covid-19 diagnosis and classification. Int. J.Mach.Learn.Cybern. 2021:1–14. doi: 10.1007/s13042-020-01248-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bayoudh K., Hamdaoui F., Mtibaa A. Hybrid-covid: a novel hybrid 2d/3d cnn based on cross-domain adaptation approach for covid-19 screening from chest x-ray images. Phys. Eng.Sci. Med. 2020;43(4):1415–1431. doi: 10.1007/s13246-020-00957-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luz E.J.d.S., Silva P.L., Silva R., Silva L., Moreira G., Menotti D. Towards an effective and efficient deep learning model for covid-19 patterns detection in x-ray images. CoRR. 2020:1–14. [Google Scholar]
- 82.Tan M., Le Q. International Conference on Machine Learning. PMLR; 2019. Efficientnet: rethinking model scaling for convolutional neural networks; pp. 6105–6114. [Google Scholar]
- 83.Lv D., Qi W., Li Y., Sun L., Wang Y. A cascade network for detecting covid-19 using chest x-rays. arXiv preprint arXiv:2005. 2020 01468. [Google Scholar]
- 84.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proceedings of the IEEE International Conference on Computer Vision. 2017. Grad-cam: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- 85.Gozes O., Frid-Adar M., Sagie N., Zhang H., Ji W., Greenspan H. Coronavirus detection and analysis on chest ct with deep learning. arXiv preprint arXiv:2004.02640. 2020 [Google Scholar]
- 86.Karim M., Döhmen T., Rebholz-Schuhmann D., Decker S., Cochez M., Beyan O., et al. Deepcovidexplainer: explainable covid-19 predictions based on chest x-ray images. arXiv preprint arXiv:2004. 2020:4582. [Google Scholar]
- 87.Li X., Li C., Zhu D. Covid-mobilexpert: on-device covid-19 screening using snapshots of chest x-ray. arXiv preprint arXiv:2004. 2020:3042. [Google Scholar]
- 88.Jin W., Dong S., Dong C., Ye X. Hybrid ensemble model for differential diagnosis between covid-19 and common viral pneumonia by chest x-ray radiograph. Comput. Biol. Med. 2021;131 doi: 10.1016/j.compbiomed.2021.104252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rahman T., Khandakar A., Qiblawey Y., Tahir A., Kiranyaz S., Kashem S.B.A., Islam M.T., Al Maadeed S., Zughaier S.M., Khan M.S., et al. Exploring the effect of image enhancement techniques on covid-19 detection using chest x-ray images. Comput. Biol. Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cohen J.P., Morrison P., Dao L., Roth K., Duong T.Q., Ghassemi M. Covid-19 image data collection: prospective predictions are the future. arXiv preprint arXiv:2006.11988. 2020 [Google Scholar]
- 91.chest-xray pneumonia, Kaggle lung pneumonia dataset2. 2020. https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
- 92.Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Jul 2017. Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. [DOI] [Google Scholar]
- 93.Irvin J., Rajpurkar P., Ko M., Yu Y., Ciurea-Ilcus S., Chute C., Marklund H., Haghgoo B., Ball R., Shpanskaya K., Seekins J., Mong D.A., Halabi S.S., Sandberg J.K., Jones R., Larson D.B., Langlotz C.P., Patel B.N., Lungren M.P., Ng A.Y. Chexpert: a large chest radiograph dataset with uncertainty labels and expert comparison. arXiv:1901. 2019:7031. [Google Scholar]
- 94.COVID-NET Covidx dataset. 2020. https://github.com/lindawangg/COVID-Net
- 95.Kermany D.S., Goldbaum M., Cai W., Valentim C.C., Liang H., Baxter S.L., McKeown A., Yang G., Wu X., Yan F., Dong J., Prasadha M.K., Pei J., Ting M.Y., Zhu J., Li C., Hewett S., Dong J., Ziyar I., Shi A., Zhang R., Zheng L., Hou R., Shi W., Fu X., Duan Y., Huu V.A., Wen C., Zhang E.D., Zhang C.L., Li O., Wang X., Singer M.A., Sun X., Xu J., Tafreshi A., Lewis M.A., Xia H., Zhang K. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. http://www.sciencedirect.com/science/article/pii/S0092867418301545 e9. URL. [DOI] [PubMed] [Google Scholar]
- 96.Kaggle, Kaggle lung pneumonia dataset. 2020. https://www.kaggle.com
- 97.NIH . 2020. Nih Dataset.https://openi.nlm.nih.gov/ [Google Scholar]
- 98.Kaggle Kaggle lung pneumonia dataset1. 2020. https://www.kaggle.com/andrewmvd/convid19-x-rays
- 99.RSNA Rsna pneumonia detection challenge dataset. 2020. https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data
- 100.SIRM, S.i. s. o. m. a. i. radiology . 2020. covid-19 Database.https://www.sirm.org/category/senza-categoria/covid-19/ 2020. [Google Scholar]
- 101.Kermany D., Zhang K., Goldbaum M. Labeled optical coherence tomography (oct) and chest x-ray images for classification. Mendeley data. 2018;2 [Google Scholar]
- 102.Candemir S., Jaeger S., Palaniappan K., Musco J.P., Singh R.K., Xue Z., Karargyris A., Antani S., Thoma G., McDonald C.J. Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imag. 2013;33(2):577–590. doi: 10.1109/TMI.2013.2290491. [DOI] [PubMed] [Google Scholar]
- 103.Jaeger S., Karargyris A., Candemir S., Folio L., Siegelman J., Callaghan F., Xue Z., Palaniappan K., Singh R.K., Antani S., et al. Automatic tuberculosis screening using chest radiographs. IEEE Trans. Med. Imag. 2013;33(2):233–245. doi: 10.1109/TMI.2013.2284099. [DOI] [PubMed] [Google Scholar]
- 104.Adrian Medical images. 2020. https://www.pyimagesearch.com/category/medical/
- 105.Radiopedia Radiopedia. 2020. https://radiopaedia.org/
- 106.Nih chestxrays nih-chestxrays. 2020. https://www.kaggle.com/nih-chestxrays/data
- 107.Chicco D., Jurman G. The advantages of the matthews correlation coefficient (mcc) over f1 score and accuracy in binary classification evaluation. BMC Genom. 2020;21(1):1–13. doi: 10.1186/s12864-019-6413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Siu K., Stuart D.M., Mahmoud M., Moshovos A. 2018 IEEE International Symposium on Workload Characterization (IISWC) IEEE; 2018. Memory requirements for convolutional neural network hardware accelerators; pp. 111–121. [Google Scholar]
- 109.Rawat S., Rana K., Kumar V. A novel complex-valued convolutional neural network for medical image denoising. Biomed. Signal Process Control. 2021;69 [Google Scholar]