Abstract
We evaluated the sensitivity of a DNA amplification test for the detection of Mycobacterium avium in blood samples using different blood components and different DNA extraction methods. M. avium-inoculated blood samples were processed to obtain separate blood components: peripheral blood mononuclear cells (PBMCs), polymorphonuclear cells (PMNCs), and whole-blood sodium dodecyl sulfate (SDS)-lysate pellets. The sensitivity for the detection of the lowest mycobacterial load (1 CFU/ml) was significantly greater (P < 0.01) with DNA extracted from SDS-lysate pellets than with DNA extracted from PBMCs or PMNCs. Subsequently, DNA extraction methods based on guanidine NaOH, and proteinase were compared. The sensitivity of the guanidine-based method was significantly greater (P < 0.01) than those of the others.
In the last 2 decades, the impact of the AIDS epidemic and the diffusion of immunosuppressive conditions have resulted in a substantial increase in the incidence of disseminated Mycobacterium avium complex (MAC) infections (5, 8, 13, 14, 21, 22, 31). The diagnosis of disseminated MAC infection is often based on suggestive clinical signs and symptoms (2, 5, 11, 16), but in only 12% of the cases it is confirmed by a positive blood culture (6). The culture of mycobacteria from blood takes from 2 to 4 weeks after the culture inoculation (1, 2, 16); thus, there is a considerable need to develop more-rapid diagnostic tests on blood samples. Several studies proposing different amplification protocols have been published so far (4, 7, 9, 16, 17, 20, 29), but the standardization of a reliable amplification method for disseminated MAC infection diagnosis has not been achieved yet. The aim of this study was to analyze the sensitivity of a PCR-based method for the detection of M. avium in blood samples by using on different blood components or different DNA extraction methods.
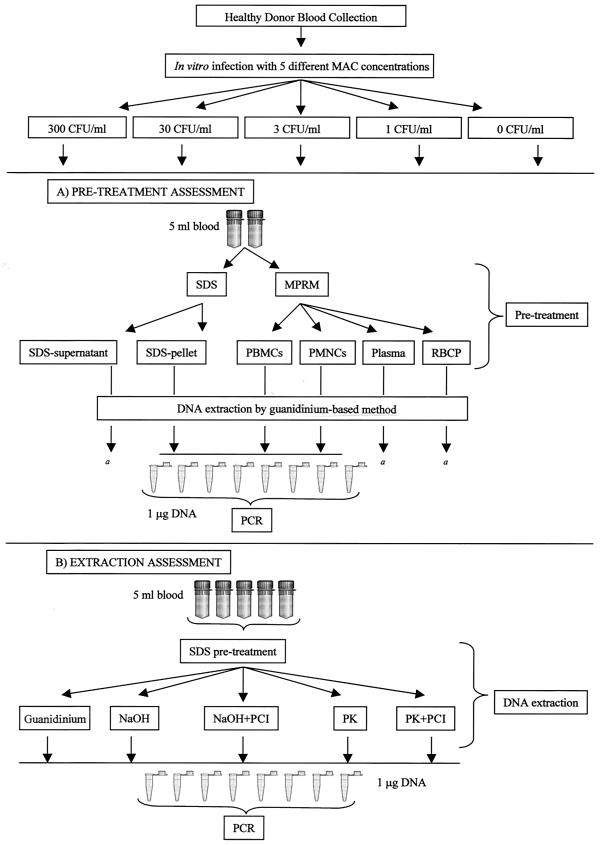
First, we evaluated the performance of the assay by using different blood components (experiment A). Fifty milliliters of peripheral blood was drawn from a healthy donor (sodium citrate at 3.8% was the anticoagulant) and divided into five parts (10 ml each); four parts were inoculated in vitro with four different M. avium bacterial loads (300, 30, 3, and 1 CFU/ml), and the remaining part was utilized as a negative control (Fig. 1A).
FIG. 1.
(A) Whole peripheral blood (50 ml) was divided into five parts (10 ml each). Four parts were inoculated in vitro with four different M. avium bacterial loads (300, 30, 3, and 1 CFU/ml), and the remaining part was utilized as a negative control. Each part was further subdivided to obtain two aliquots (5 ml each). These aliquots underwent one of two different pretreatments (lysis by SDS or cell separation by gradient centrifugation) to obtain three separate blood components: SDS-lysate pellet, PBMCs, and PMNCs. The guanidine-based method was used for DNA extraction. PCR amplification was performed with 1 μg of DNA per replicate, and eight replicates were amplified for each DNA sample. a, the presence of M. avium in SDS-lysate supernatants, plasma layers, and red blood cell pellets (RBCP) (after M-PRM separation) from 300-CFU/ml blood samples was also investigated; PCR analysis was performed for the totality of DNA extracted from these components. (B) Whole peripheral blood (125 ml) was divided into five parts (25 ml each). Four parts were inoculated in vitro with four different M. avium bacterial loads, and the remaining part was utilized as a negative control. Each part was further subdivided to obtain five aliquots (5 ml each). These aliquots underwent pretreatment with SDS. Three DNA extraction methods (guanidine-based method, NaOH-based method with or without purification, and PK-based method with or without purification) were used for DNA extraction. PCR amplification was performed with 1 μg of DNA per replicate, and eight replicates were amplified for each DNA sample. PCI, phenol-chloroform-isoamylalcohol.
For blood sample inoculation, an M. avium isotonic saline suspension was prepared from a clinical isolate identified as M. avium by standard microbiological and biochemical tests (18). The isolate was grown at 35°C and 5% CO2 on Lowenstein-Jensen medium supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC) enrichment medium (Becton Dickinson, Rutherford, N.J.). The suspension was homogenized and sonicated in a sonicating water bath (50-Hz Cole-Parmer sonicator) for 3 min at room temperature. The mycobacterial concentration was determined on the basis of optical density (OD) and adjusted to 3 × 107 mycobacteria/ml. The suspension was serially diluted (1:103, 1:104, 1:105, and 0.33:105), and four isotonic saline volumes (10 ml each) were inoculated with 100 μl of the dilutions. Triplicate 100-μl samples of the inoculated isotonic saline volumes were plated on agar medium (Middlebrook 7H10 plus 10% OADC; Becton Dickinson), and the colonies were counted after 14 and 28 days of incubation at 37°C. The concentrations of the isotonic saline volumes were 300 (coefficient of variation, ±10%), 30 (coefficient of variation, ±10%), 3 (coefficient of variation, ±5%), and 1 CFU/ml (coefficient of variation, ±7%), respectively (coefficients of variation refer to mycobacterial CFU counts of three controls per concentration). The serial dilutions of the mycobacterial suspension were utilized to inoculate blood samples (100 μl of dilutions/10 ml of blood in experiment A and 250 μl of dilutions/25 ml of blood in experiment B). Inoculated blood samples were incubated at 37°C and 5% CO2 for 1 h on a rotator to allow phagocytosis and then subdivided in aliquots of 5 ml each. The range of bacterial load of inoculated blood samples corresponds to bacterial titers in disseminated MAC infections in the majority of patients at the time of diagnosis (from 1 to 300 CFU/ml) (6, 12, 15, 20).
The 5-ml aliquots underwent two different pretreatments to obtain three separate blood components. (i) Gradient separation by Mono-Poly resolving medium (M-PRM; density coefficient, 1.114 ± 0.002; Flow Laboratories Inc., McLean, Va.) was used to collect peripheral blood mononuclear cells (PBMCs) and polymorphonuclear cells (PMNCs) according to the manufacturer's instructions (upper layers of plasma and red blood cell pellets were also separately collected). (ii) Lysis by sodium dodecyl sulfate (SDS) (final concentration, 1%) and subsequent centrifugation (4,000 × g at 5°C for 30 min) was used to obtain lysate-blood pellets, as previously described (10) (supernatants were also collected). DNA extraction was performed using a guanidine-based method (Easy-DNA kit; Invitrogen BV, Leek, The Netherlands) according to the manufacturer's instructions; OD was used to determine the DNA concentration. The concentrations of extracted DNA from PBMCs, PMNCs, and SDS-lysate pellets were normalized at 100 ng/μl by adding nuclease-free H2O. Each DNA sample was amplified by utilizing 1 μg of DNA per PCR replicate and performing eight PCR replications per sample (a total of 8 μg of DNA per sample underwent PCR amplification) (Fig. 1A). Amplification and a hybridization assay were performed to identify the M. avium target sequence according to a previously described protocol (3, 23, 24). The PCR-hybridization colorimetric reaction was read at 450 nm using a spectrophotometer to determine positive and negative replicates: OD values higher than 0.4 were considered positive (24). DNA samples were amplified for the β-globin gene sequence to assess the presence of PCR inhibitors and the integrity of the template DNA (25). Each PCR-hybridization run was performed using positive (10 fg of DNA from M. avium ATCC 15769) and negative (human DNA and nuclease-free H2O) controls and sterilization procedures in accordance with guidelines for maintaining contamination-free conditions to prevent false-positive results (19). PCR-hybridization analysis was also performed on the totality of DNA (extracted by the guanidine method) from SDS-lysate supernatants of 300-CFU/ml blood samples and from plasma layers and red blood cell pellets which were collected after M-PRM separation of 300-CFU/ml blood samples. After evaluation of the pretreatment methods, three DNA extraction methods were compared (experiment B). Five 25-ml aliquots of peripheral blood, inoculated with M. avium, were obtained as described above. Each part was further subdivided to obtain five aliquots (5 ml each). These aliquots underwent pretreatment with SDS to obtain SDS-lysate pellets. Three DNA extraction methods were used: (i) a guanidine-based method as described above; (ii) an NaOH-based method as previously described (20), with or without DNA purification by a phenol-chloroform-isoamyl alcohol (PCI) protocol (27); and (iii) a proteinase K (PK)-based method as previously described (30), with or without PCI DNA purification (Fig. 1B). OD was used to determine DNA concentration.
Both experiments A and B were repeated three times in three separate sessions to evaluate the reproducibility of tests. The proportions of positive replicates of the eight-PCR-replicate set of three separate experiments were calculated, and the sensitivity of each single replicate was calculated. Student's unpaired t test was used to compare the proportions of positive replicates.
M. avium PCR-hybridization analysis performed on DNA from PBMCs, PMNCs, and SDS-lysate pellets of noninoculated blood aliquots was negative for all 72 PCR replicates, while β-globin gene PCR yielded 100% positive replicates, ruling out PCR inhibitors or lack of DNA integrity.
The results of PCR-hybridization analysis on DNA extracted from PBMCs showed that the M. avium target sequence was detectable on 300-, 30-, and 3-CFU/ml samples; the sensitivities of single PCR replicates were 41, 20, and 8%, respectively (Table 1). PCR-hybridization results for DNA extracted from PMNCs showed an increased sensitivity of single PCR replicates in 300- and 30-CFU/ml samples (62 and 45%, respectively) compared to the sensitivity of those in PBMC samples; the same sensitivity was evident from 3-CFU/ml samples (8%). PCR-hybridization analysis failed to detect the target sequence from samples with the lowest bacterial load (1 CFU/ml) in DNA extracted from both PMNCs and PBMCs in three separate experiments.
TABLE 1.
Experiment A: variability of sensitivity depending on different pretreatments and blood components
| Pretreatment | Blood component | No. of PCR replicates/ expta | Result for a mycobacterial concentrationb (CFU/ml) of:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 300
|
30
|
3
|
1
|
|||||||||||
| No. of pos replicates/ expt | Proportion of pos replicates/ expt | Sensitivity/ replicate (%) | No. of pos, replicates/ expt | Proportion of pos replicates expt | Sensitivity/ replicate (%) | No. of pos replicates/ expt | Proportion of pos replicates/ expt | Sensitivity/ replicate (%) | No. of pos replicates/ expt | Proportion of pos replicates/ expt | Sensitivity/ replicate (%) | |||
| SDS 1% | Lysate pellet | 8 | 8 | 1.00 | 87 | 4 | 0.50 | 50 | 5 | 0.62 | 50 | 2 | 0.25 | 20 |
| 8 | 6 | 0.75 | 4 | 0.50 | 4 | 0.50 | 2 | 0.25 | ||||||
| 8 | 7 | 0.87 | 4 | 0.50 | 3 | 0.37 | 1 | 0.12 | ||||||
| M-PRM | PMNC | 8 | 6 | 0.75 | 62 | 4 | 0.50 | 45 | 1 | 0.12 | 8 | 0 | 0.00 | 0 |
| 8 | 4 | 0.50 | 4 | 0.50 | 1 | 0.12 | 0 | 0.00 | ||||||
| 8 | 5 | 0.62 | 3 | 0.37 | 0 | 0.00 | 0 | 0.00 | ||||||
| M-PRM | PBMC | 8 | 4 | 0.50 | 41 | 2 | 0.25 | 20 | 1 | 0.12 | 8 | 0 | 0.00 | 0 |
| 8 | 3 | 0.37 | 2 | 0.25 | 1 | 0.12 | 0 | 0.00 | ||||||
| 8 | 3 | 0.37 | 1 | 0.12 | 0 | 0.00 | 0 | 0.00 | ||||||
Total of three experiments.
Pos, positive.
As regards the data obtained by DNA extracted from SDS-lysate pellets, the sensitivity of single PCR replicates was significantly greater (P < 0.01) than the sensitivities of replicates in PMNC and PBMC samples (87% for 300 CFU/ml, 50% for 30 CFU/ml, 50% for 3 CFU/ml, and 20% for 1 CFU/ml) and the PCR-hybridization assay detected the target sequence even from samples with the lowest bacterial load (1 CFU/ml).
No target DNA sequence was detectable in SDS-lysate supernatants of 300-CFU/ml blood samples, thus showing that M. avium DNA was absent from or present in small amounts in SDS-lysate supernatants. Similarly, PCR analysis was performed on DNA from plasma layers and red blood cell pellets which were collected after M-PRM separation of 300-CFU/ml blood samples, and no target sequence was detectable. Thus, we assumed that all mycobacteria were adherent to blood leukocytes or phagocytosed by monocytes or PMNCs. The β-globin gene PCR control showed that no inhibitors were present in any of the DNA samples tested.
Subsequently, the comparison of the sensitivities of the three different DNA extraction methods analyzed showed that the NaOH-based method allowed for the detection of M. avium DNA in samples with concentrations as low as 3 CFU/ml, with single-replicate sensitivities of 8% when PCI-purified DNA was analyzed and 4% when not-purified DNA was analyzed (Table 2). PK extraction allowed target detection solely in 300-CFU/ml samples, with single-replicate sensitivity of 16% for PCI-purified DNA; target detection failed in 30-, 3-, and 1-CFU/ml samples. Both NaOH-based and PK-based methods showed sensitivities significantly lower than that for the guanidine-based DNA extraction technique (P < 0.01), which yielded a positive detection even at the lowest bacterial load (1-CFU/ml samples), with single-replicate sensitivity of 20%, in three separate experiments (Table 2).
TABLE 2.
Experiment B: variability of sensitivity depending on different extraction methods
| DNA extraction methoda | No. of PCR replicates/ exptb | Result for a mycobacterial concentrationc (CFU/ml) of:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 300
|
30
|
3
|
1
|
||||||||||
| No. of pos replicates/ expt | Proportion of pos replicates/ expt | Sensitivity/ replicates (%) | No. of pos replicates/ expt | Proportion of pos replicates/ expt | Sensitivity/ replicate (%) | No. of pos replicates/ expt | Proportion of pos replicates/ expt | Sensitivity/ replicates expt | No. of pos replicates/ expt | Proportion of pos replicates/ expt | Sensitivity/ replicate (%) | ||
| Guanidine | 8 | 8 | 1.00 | 91 | 5 | 0.62 | 54 | 4 | 10.50 | 45 | 2 | 0.25 | 20 |
| 8 | 6 | 0.75 | 4 | 0.50 | 4 | 0.50 | 2 | 0.25 | |||||
| 8 | 8 | 1.00 | 4 | 0.50 | 3 | 0.37 | 1 | 0.12 | |||||
| NaOH (PCI-purified DNA) | 8 | 6 | 0.75 | 58 | 2 | 0.25 | 20 | 1 | 0.12 | 8 | 0 | 0.00 | 0 |
| 8 | 4 | 0.50 | 2 | 0.25 | 1 | 0.12 | 0 | 0.00 | |||||
| 8 | 4 | 0.50 | 1 | 0.12 | 0 | 0.00 | 0 | 0.00 | |||||
| NaOH (not-purified DNA) | 8 | 2 | 0.25 | 20 | 2 | 0.25 | 12 | 1 | 0.12 | 4 | 0 | 0.00 | 0 |
| 8 | 2 | 0.25 | 1 | 0.12 | 0 | 0.00 | 0 | 0.00 | |||||
| 8 | 1 | 0.12 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | |||||
| PK (PCI-purified DNA) | 8 | 2 | 0.25 | 16 | 0 | 0.00 | 0 | 0 | 0.00 | 0 | 0 | 0.00 | 0 |
| 8 | 1 | 0.12 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | |||||
| 8 | 1 | 0.12 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | |||||
| PK (not-purified DNA) | 8 | 2 | 0.25 | 12 | 0 | 0.00 | 0 | 0 | 0.00 | 0 | 0 | 0.00 | 0 |
| 8 | 1 | 0.12 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | |||||
| 8 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | |||||
DNA was extracted from SDS-lysate pellets by three methods; a DNA purification step was provided by kit instructions when the guanidine method was used.
Total of three experiments.
Pos, positive.
Several PCR-based methods for the diagnosis of MAC bacteremia are described in the literature, and different authors report sensitivities ranging from 56 to 100% (4, 7, 9, 16, 17, 20, 29). The sensitivity depends on different factors: volume of sample and number of organisms per milliliter of blood, procedures for the treatment of blood samples, extraction methods, total amount of DNA utilized for PCR analysis, and the PCR assay detection limit. Furthermore, when applied to DNA extracted from blood samples, PCR amplification is frequently hampered by the presence of inhibitors, and false-positive results, due to contamination, may reduce the usefulness of PCR methods in routine diagnosis.
We tested different pretreatment techniques to evaluate the blood component-related sensitivity of the PCR assay. Indeed, the sensitivity was apparently related to the analyzed blood components. Our results showed a significantly greater sensitivity (P < 0.01) for the PCR assay when it was done on DNA extracted from SDS-lysate pellets. This result could be explained by the specimen preparation procedures. We could lyse most of the human cells by SDS, preserving the integrity of mycobacteria, given the resistance of the mycobacterial cell wall (10, 25, 26, 29). Compared to the other pretreatment methods, this technique allowed an increase in the mycobacterial DNA/total DNA ratio of about 5- to 10-fold, with a consequent proportional rise in the probability of detecting target DNA. For this purpose, SDS-lysate pellet preparation was highly efficient, as demonstrated by a PCR-hybridization assay done on SDS-lysate supernatants that showed the absence of M. avium DNA in this component. Moreover, the recovery of PMNCs and PBMCs (either by M-PRM or Ficoll-Hypaque) is less than 100% efficient; this inefficiency could be correlated with the lower sensitivities of these methods.
The better sensitivity of guanidine-based DNA extraction could be explained by the finding that the method using guanidine isothiocyanate has a greater ability than the other methods to lyse mycobacterial cell walls.
In conclusion, our study shows that the sensitivity of the PCR method for detection of M. avium in blood samples depends on the pretreatment extraction methods utilized and strictly correlates to the DNA extraction techniques performed.
Acknowledgments
We thank Bianca Ghisi for computer counseling, Mario Corbellino and Stefano Rusconi for their valuable help, Elizabeth Kaplan and Alan Michael Rosen for their professional assistance, Maura Mezzetti for statistical analysis, and Mauro Moroni for helpful discussion.
This work was supported by a grant from the Italian National Institute of Health, 2nd National Tuberculosis Project.
REFERENCES
- 1.Anargyros P, Astill D S J, Lim I S L. Comparison of improved BACTEC and Lowenstein-Jensen media for culture of mycobacteria from clinical specimens. J Clin Microbiol. 1990;28:1288–1291. doi: 10.1128/jcm.28.6.1288-1291.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson C A, Ellner J J. Mycobacterium avium complex infection and AIDS: advances in theory and practice. Clin Infect Dis. 1993;17:7–20. doi: 10.1093/clinids/17.1.7. [DOI] [PubMed] [Google Scholar]
- 3.Böddinghaus B, Rogall T, Flohr T, Blöcker H, Böttger E C. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogner J R, Rusch-Gerdes S, Mertenskotter T, Loch O, Emminger C, Baumgarten R, Brockmeyer N H, Brockhaus W, Jablonowski H, Stoehr A, Roth A, Albrecht H, Roth K, Tschauder S, Dietrich M. Patterns of Mycobacterium avium culture and PCR positivity in immunodeficient HIV-infected patients: progression from localized to systematic disease. German AIDS Study Group. Scand J Infect Dis. 1997;29:579–584. doi: 10.3109/00365549709035898. [DOI] [PubMed] [Google Scholar]
- 5.Chaisson R E, Moore R D, Richman D D, Keruly J, Creagh T. Incidence and natural history of Mycobacterium avium complex infections in patients with advanced human immunodeficiency virus disease treated with zidovudine. Am Rev Respir Dis. 1992;146:285–289. doi: 10.1164/ajrccm/146.2.285. [DOI] [PubMed] [Google Scholar]
- 6.Chin D P, Reingold A L, Horsburgh C R, Yajko D M, Hadley W K, Elkin E P, Stone E N, Simon E M, Gonzalez P L, Ostroff S M, Jacobson M A, Hopewell P C. Predicting Mycobacterium avium complex bacteremia in patients infected with human immunodeficiency virus: a prospectively validated model. Clin Infect Dis. 1994;19:668–674. doi: 10.1093/clinids/19.4.668. [DOI] [PubMed] [Google Scholar]
- 7.De Francesco M A, Colombrita D, Pinsi G, Gargiulo F, Caligaris S, Bertelli D, Martinelli F, Gao J, Turano A. Detection and identification of Mycobacterium avium in the blood of AIDS patients by polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 1996;15:551–555. doi: 10.1007/BF01709362. [DOI] [PubMed] [Google Scholar]
- 8.Ellner J J, Goldberger M J, Parenti D M. Mycobacterium avium infection and AIDS: a therapeutic dilemma in rapid evolution. J Infect Dis. 1991;163:1326–1335. doi: 10.1093/infdis/163.6.1326. [DOI] [PubMed] [Google Scholar]
- 9.Folgueira L, Delgado R, Palenque E, Aguado J M, Noriega A R. Rapid diagnosis of Mycobacterium tuberculosis bacteremia by PCR. J Clin Microbiol. 1996;34:512–515. doi: 10.1128/jcm.34.3.512-515.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamboa F, Fernandez G, Padilla E, Manterola J M, Lonca J, Cardona P J, Matas L, Ausina V. Comparative evaluation of initial and new versions of the Gen-Probe amplified Mycobacterium tuberculosis direct test for direct detection of Mycobacterium tuberculosis in respiratory and nonrespiratory specimens. J Clin Microbiol. 1998;36:684–689. doi: 10.1128/jcm.36.3.684-689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Havlik J A, Horsburgh C R, Metchock B, Williams P P, Fann S A, Thompson S E. Disseminated Mycobacterium avium complex infection: clinical identification and epidemiologic trends. J Infect Dis. 1992;165:577–580. doi: 10.1093/infdis/165.3.577. [DOI] [PubMed] [Google Scholar]
- 12.Havlir D, Kemper C A, Deresinski S C. Reproducibility of lysis-centrifugation cultures for quantification of Mycobacterium avium complex bacteremia. J Clin Microbiol. 1993;31:1794–1798. doi: 10.1128/jcm.31.7.1794-1798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawkins C C, Gold J W M, Whimbey E, Kiehn T E, Brannon P, Cammarata R, Brown A E, Armstrong D. Mycobacterium avium complex infections in patients with AIDS. Ann Intern Med. 1986;105:184–188. doi: 10.7326/0003-4819-105-2-184. [DOI] [PubMed] [Google Scholar]
- 14.Horsburgh C R. Mycobacterium avium complex infection in the acquired immunodeficiency syndrome. N Engl J Med. 1991;324:1332–1338. doi: 10.1056/NEJM199105093241906. [DOI] [PubMed] [Google Scholar]
- 15.Horsburgh C R, Metchock B, Gordon S M, Havlik J A, Mcgowan J E, Thompson S E. Predictors of survival in patients with AIDS and disseminated Mycobacterium avium complex disease. J Infect Dis. 1994;170:573–577. doi: 10.1093/infdis/170.3.573. [DOI] [PubMed] [Google Scholar]
- 16.Inderlied C B, Kemper C A, Bermudez L E M. The Mycobacterium avium complex. Clin Microbiol Rev. 1993;6:266–310. doi: 10.1128/cmr.6.3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iralu J V, Sritharan V K, Pieciak W S, Wirth D F, Maguire J H, Barker R H. Diagnosis of Mycobacterium avium bacteremia by polymerase chain reaction. J Clin Microbiol. 1993;31:1811–1814. doi: 10.1128/jcm.31.7.1811-1814.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkins P A, Pattyn S R, Portels F. Diagnostic bacteriology. In: Ratledge C, Stanford J, editors. The biology of the mycobacteria. London, England: Academic Press Ltd.; 1982. pp. 441–476. [Google Scholar]
- 19.Kent P T, Kubica G. Public health mycobacteriology. A guide for the level III laboratory. Atlanta, Ga: Centers for Disease Control; 1985. [Google Scholar]
- 20.MacGregor R R, Dreyer K, Herman S, Hocknell P K, Nghiem L, Tevere V J, Williams A L. Use of PCR in detection of Mycobacterium avium complex (MAC) bacteremia: sensitivity of the assay and effect of treatment for MAC infection on concentrations of human immunodeficiency virus in plasma. J Clin Microbiol. 1999;37:90–94. doi: 10.1128/jcm.37.1.90-94.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masur H the Public Health Task Force on Prophylaxis and Therapy for Mycobacterium avium. Special report. Recommendations on prophylaxis and therapy for disseminated Mycobacterium avium complex disease in patients infected with the human immunodeficiency virus. N Engl J Med. 1993;329:898–905. doi: 10.1056/NEJM199309163291228. [DOI] [PubMed] [Google Scholar]
- 22.Nightingale S D, Byrd L T, Southern P M, Jockusch J D, Cal S X, Winne B A. Incidence of Mycobacterium avium-intracellulare complex bacteremia in human immunodeficiency virus-positive patients. J Infect Dis. 1992;165:1082–1085. doi: 10.1093/infdis/165.6.1082. [DOI] [PubMed] [Google Scholar]
- 23.Oggioni M R, Fattorini L, Li B, De Milito A, Zazzi M, Pozzi G, Orefici G, Valensin P E. Identification of Mycobacterium tuberculosis complex, Mycobacterium avium and Mycobacterium intracellulare by selective nested polymerase chain reaction. Mol Cell Probes. 1995;9:321–326. doi: 10.1016/s0890-8508(95)91604-0. [DOI] [PubMed] [Google Scholar]
- 24.Rossi M C, Gori A, Zehender G, Marchetti G, Ferrario G, De Maddalena C, Catozzi L, Bandera A, Degli Esposti A, Franzetti F. A PCR-colorimetric microwell plate hybridization assay for detection of Mycobacterium tuberculosis and M. avium from culture samples and Ziehl-Neelsen-positive smears. J Clin Microbiol. 2000;38:1772–1776. doi: 10.1128/jcm.38.5.1772-1776.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnheim N. Enzymatic amplification of β-globin genomic sequences and restriction site analysis of sickle cell anemia. Science. 1985;230:1350–1357. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 26.Salfinger M, Kafader F M. Comparison of two pretreatment methods for the detection of mycobacteria of BACTEC and Lowenstein-Jensen slants. J Microbiol Methods. 1987;6:315–321. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. E.3–E.4. [Google Scholar]
- 28.Sjobring U, Mecklenburg M, Andersen A B, Miürner H. Polymerase chain reaction for detection of Mycobacterium tuberculosis. J Clin Microbiol. 1990;28:2200–2204. doi: 10.1128/jcm.28.10.2200-2204.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stauffer F, Haber H, Rieger A, Mutschlechner R, Hasenberger P, Tevere V J, Young K K. Genus level identification of mycobacteria from clinical specimens by using an easy-to-handle Mycobacterium-specific PCR assay. J Clin Microbiol. 1998;36:614–617. doi: 10.1128/jcm.36.3.614-617.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallis J M, Hannah J B. Mycobacterium avium complex infection in patients with AIDS. A clinicopathologic study. Chest. 1988;93:926–932. doi: 10.1378/chest.93.5.926. [DOI] [PubMed] [Google Scholar]
- 32.Wayne L G. Cultivation of Mycobacterium tuberculosis for research purposes. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C.: American Society for Microbiology; 1994. pp. 73–83. [Google Scholar]