Abstract
Background
Gastric cancer (GC) is one of the most commonly diagnosed malignancies of the human digestive tract, and currently there is a dearth of effective biomarkers for this disease.
Methods
MiR-598 expression levels were analyzed by the cancer genome atlas (TCGA database) mining and verified in GC patient plasma using real-time reverse transcription polymerase chain reaction (RT-PCR) assay. We used the GEPIA and UALCAN databases and immunohistochemistry (IHC) to analyze SOX4 expression. The MTT assay assessed MNK28 and SGC7901 cell proliferation after transfection with miR-596 plasmids. The analytical tools, Functional Enrichment Analysis Tool (FunRich), Database of Immune Cell Expression (DICE) and Tumor IMmune Estimation Resource (TIMER) were used to analyze correlations between SOX4 and immune responses. Furthermore, a Kaplan Meier plotter database explored correlations between miR-596, SOX4 and overall patient survival.
Results
Data from TCGA and RT-PCR indicated that miR-598 was lowly expressed in GC patients. The miRWalk database showed that SOX4 was the target genes of miR-596 and also revealed that miR-596 bound directly to SOX4. MiR-596 over-expression further depressed GC cell proliferation. In addition, the FunRich database showed that SOX4 was involved in immune responses, and was further shown to be differentially expressed in CD4+ T cells by DICE. Specifically, TIMER indicated that high expression of SOX4 was negatively correlated with infiltrating CD4+ T cells in stomach adenocarcinoma (STAD). Moreover, high expression of miR-596 and low expression of SOX4 prolonged the overall survival (OS) of GC patients.
Conclusions
Our study reveals a crucial role for miR-596 in tumor-associated immune infiltration and predicting prognoses in GC patients.
Keywords: Bioinformatics analysis, gastric cancer (GC), immune infiltration, miR-596, SOX4
Introduction
Gastric cancer (GC) is one of the most commonly diagnosed malignancies of the human digestive tract, with a poor prognosis and a five-year survival rate of 20–40% (1). Currently, patients with early-stage GC receive effective treatments with curative clinical outcomes, whereas patients with moderate- or late-stage GC lack effective forms of treatment (2,3). Thus, it is essential to determine novel GC biomarkers.
MicroRNAs (miRNAs) are a class of single stranded noncoding RNAs, consisting of 19–25 nucleotides (4). These molecules play critical biological roles in human pathological and physiological processes, through a range of molecular mechanisms (5,6). Also, aberrant miRNA expression leads to a variety of diseases such as cancer, by interacting with 3’-untranslated regions of target mRNAs, and potentially dysregulating protein translation (7). Increasing evidence shows that miRNAs are involved in a variety of biological processes, including proliferation, apoptosis, migration and differentiation, which suggests miRNAs are potential and promising targets for cancer therapeutics (8). Recently, miR-596 was reported as dysregulated in multiple human cancers, such as hepatocellular, gastric, cervical and oral cancer (9-12). In oral cancer, high expression of miR-596 suppresses tumor cell migration and invasion (12). Similarly, the high expression of this molecule also contributes to survival benefits of cervical cancer patients (11). Interestingly, miR-596 also acts as a tumor suppressor in GC and is upregulated by promoter demethylation (12). However, little is known about how miR-596 is associated with immune infiltration in GC (10).
SOX4 is an essential developmental transcription factor, and plays a key role in mediating immune cells (13). Moreover, SOX4 regulates progenitor development, multiple developmental pathways, and is also associated with several cancers (14). SOX4 overexpression promotes cell proliferation in rectal cancer, non-small cell lung cancer and breast cancer (15-17). Recently, studies have indicated that miR-381, miR-138, miR-30a-5p, miR-211 and miR-30a-5p inhibit cell migration and invasion in human GC by down regulating SOX4 (18-21). In this study, we identified low expression of miR-596 in GC by analyzing three GC miRNA microarray datasets, and serum from GC patients. As predicted by bioinformatics, miR-596 directly targets SOX4 that had connection with tumor infiltrating immune cells in GC.
Methods
SGF-7901 and MNK28 cell culture
SGC-7901 and MNK28 cells were purchased from the China Center for Type Culture Collection (Wuhan, China), and cultured in complete RPMI-1640 medium (R0883, Sigma-Aldrich) (10% fetal bovine serum (FBS) (FND500, Thermo Fisher Scientific, USA), 100 U/mL streptomycin (15140122, Thermo Fisher Scientific) at 37 °C in a humidified atmosphere containing 5% CO2.
Quantitative real-time PCR and bioinformatic analysis
The analysis of miR-596 expression in real-time qRT-PCR assay was performed as previously described (18,19). Total RNA was extracted using the RNeasy Mini Kit (Qiagen) and real-time qRT-PCR was performed as previously reported (18,19). The primers for miRNAs were synthesized by Sangon Biotech (Shanghai, China). The primers for SOX4 were: 5'-GAGGAGGAAGAGGGTAGACAG-3' and 5'-CAACATCAATAACAACAATCACAGG-3'. The primers for miR-596 were: 5'-AAGCCTGCCCGGCTC-3' and 5'-CAGTGCAGGGTCCGAGGTAT-3'. The primers for U6 were: 5'-CTCGCTTCGGCAGCACATATACT-3' and 5'-ACGCTTCACGAATTTGCGTGTC-3'. The following parameters were used: 95 °C for 3 min followed by 39 cycles of 95 °C for 10 s, and 60 °C for 30 s. The relative expression levels of miR-596 were normalized to the internal control, U6 using the 2−ΔΔCt cycle threshold method.
Cell transfection and cell proliferation assay
PEGFP-N1-miR-596 and an empty PEGFP-N1 (NC) were purchased from Shanghai Genechem Co. Ltd. SGC-7901 and MNK28 cells were seeded 1×106 in 6-well culture plates (657160, Corning, NYK, USA) and transfected PEGFP-N1-miR-596 and Empty PEGFP-N1 with Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA, USA) following manufacturer’s instructions. SGC-7901 and MNK28 cells were also seeded in 96-well culture plates (3690, Corning, NYC, USA) at a density of 1×104 cells/ml in complete RPMI. Following incubation for 24 h at 37 °C, cells were transfected with PEGFP-N1-miR-596 for 12, 24 and 48 h. After these periods, 5 mg/mL MTT (10 µL) was added to each well, and cells were further incubated at 37 °C for 3 hours. Then, DMSO (150 µL) was added to each well, and the optical density was determined at 490 nm using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Tissue immunohistochemistry (IHC)
Primary GC tissue and corresponding non-tumor-adjacent gastric tissue were collected from the Biobank of Zhejiang Cancer Hospital. Prior to participating, each patient signed an informed consent sheet. This study was approved by the Medical Ethics Committee, Zhejiang Medical College. The immunohistochemical staining kit was purchased from Beijing Zhongshan Golden Bridge Biotechnology Co. Ltd. In brief, paraffin sections were successively deparaffinized, rehydrated, and boiled for antigen retrieval. Then, a primary SOX4 antibody (Invitrogen) (1:1,000) was added to each section for 2 h at room temperature. After sections were gently washed three times in PBS, 400 µL histochemical polymer enhancer was added for 20 minutes at room temperature. After gently washing three times in PBS, the secondary antibodies were added and incubated for 20 min, followed by washing, DAB staining, counterstaining, and mounting.
Exploration of differentially expressed miRNAs
RNA sequencing (RNA-Seq) data were derived from gene expression omnibus (GSE23739). We analyzed 112 adjacent normal tissues and 412 gastric tumor tissues, with available miRNASeqs. We explored differentially and significantly expressed miRNAs between normal and cancer tissue using R package version 0.10.2.1 for R statistics software. By analyzing miRNA differential expression, we identified miR-596. Heatmap is plotted using pheatmap R package (version 0.7.7). The source code of pheatmap package was slightly modified to improve the layout and to add some features.
Functional enrichment analysis
SOX4 and target gene functional enrichment were analyzed by FunRich (http://www.funrich.org/). FunRich is a stand-alone software tool for the functional enrichment and interaction network analysis of genes and proteins. Results can be depicted by Venn, bar, column, pie and doughnut charts. The FunRich database currently supports enrichment analysis of the following: biological processes, cellular components, molecular functions, protein domains, biological pathways, transcription factors and clinical synopsis phenotypic terms. In this study, we imported the name of SOX4 into the gene enrichment component of FunRich software, and analyzed its biological functions.
MiRNA-target interactions
MiRWalk is an improved version of the previous database, and stores predicted data obtained with a machine learning algorithm, including experimentally verified miRNA-target interactions (PMID: 25055920).
Database of immune cell expression
The DICE (https://dice-database.org/landing) (Database of Immune Cell Expression, Expression quantitative trait loci (eQTLs) and Epigenomics) identifies the expression quantitative trait loci for 12,254 unique genes, which represents 61% of all protein-coding genes expressed in different cell types. Strikingly, a large fraction (41%) of these genes shows strong cis-associations with genotypes in a single cell type. It also shows that biological sex is associated with major differences in immune cell gene expression in a highly cell-specific manner. In this study, we analyzed SOX4 expression in different immune cells.
Tumor IMmune Estimation Resource (TIMER) analysis
TIMER (https://cistrome.shinyapps.io/timer/) is a comprehensive resource for the systematic analysis of immune infiltrates across different cancer types. It infers the abundance of tumor infiltrating immune cells from gene expression profiles. Across 32 cancer types, there are 10,897 samples in the TIMER database that estimate immune infiltrates. SOX4 expression levels in GC patients (n=415) were displayed using log2 RSEM.
SCNAs in the TIMER database are defined by GISTIC 2.0, including deep deletion (−2), arm-level deletion (−1), diploid/normal (0), arm-level gain (1), and high amplification (2). Box plots show the distribution of each immune subset at each copy number status, in selected cancer types. The infiltration level for each SCNA category is compared with the normal, using two-sided Wilcoxon rank sum tests. In this study, we analyzed various SOX4 mutations in different immune cells.
Survival analysis
Overall survival (OS) was calculated using the Kaplan-Meier (KM) Plotter (http://www.kmplot.com) database, which evaluates the impact of 54,675 genes on survival time of cancer patients. Approximately 10,188 cancer samples were analyzed in this program, including 1,648 ovarian, 4,142 breast, 1,065 GC and 2,437 lung sample microarray expression profiles. We also analyzed the effects of miR-596 [high (n=299) vs. low (n=132) expression] on the OS of GC patients, using a log-rank P-value and a hazard ratio (HR), with 95% confidence intervals (CI).
The Biomedical Informatics Institute (http://bioinfo.henu.edu.cn/Index.html) studies human cancers, genetic diseases and the development of biomedical databases/software using high-throughput and high dimension data. The goal of this database is to molecularly subtype diseases, and prognostic markers, and evaluates the impact of 54,675 genes on survival time of cancer patients. In this study, we analyzed the effects of SOX4 differential expression [high (n=826) vs. low (n=277) expression] on the OS of GC patients, with a log-rank P value and hazard ratio (HR).
Statistical analysis
Statistical analyses were performed using SPSS 19.0 software (IBM, Armonk, NY, USA). Differences between groups were compared using median analysis, and all data were presented as the median. P values <0.05 were considered statistically significant.
Results
MiR-596 is under expressed in GC patients
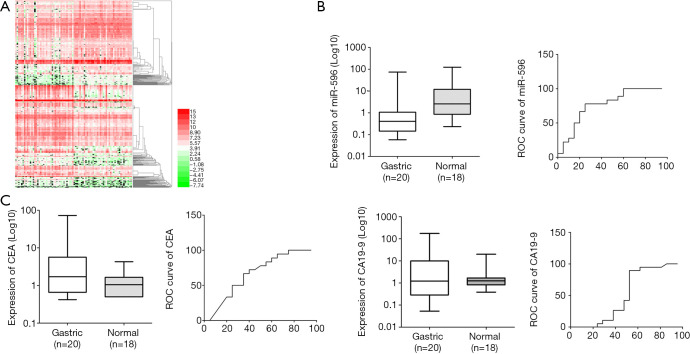
We performed a genome-wide screen in blood samples from GC patients and identified a series of abnormally expressed miRNAs (over-expressed or under-expressed) (Figure 1A). We then analyzed miR-596 serum expression levels and two gastrointestinal cancer markers carcinoembryonic antigen (CEA) and carbohydrate antigen 199 (CA19-9) in GC patients and healthy individuals. We observed that CEA and CA19-9 serum expression was higher, while miR-596 expression was lower in GC patients when compared to healthy individuals (Figure 1B,C). We also compared the area under curve of miR-596 with CEA and CA19-9 and found that miR-596 generated higher sensitivity and specificity.
Figure 1.
Low expression of miR-596 in GC. (A) Heatmap of miRNA differential expression in GC; ascending normalized expression levels are colored from green to red. (B) RT-PCR analysis of miR-596 levels in serum from GC patients (n=20), when compared with healthy individuals (n=18). Expression levels were normalized to U6. ROC curve of miR-596. (P<0.05). (C) High CEA and CA19-9 expression levels in serum samples from GC patients. ROC curve analysis of CEA and CA19-9 (P<0.05). GC, gastric cancer; CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 199.
SOX4 is highly expressed in GC
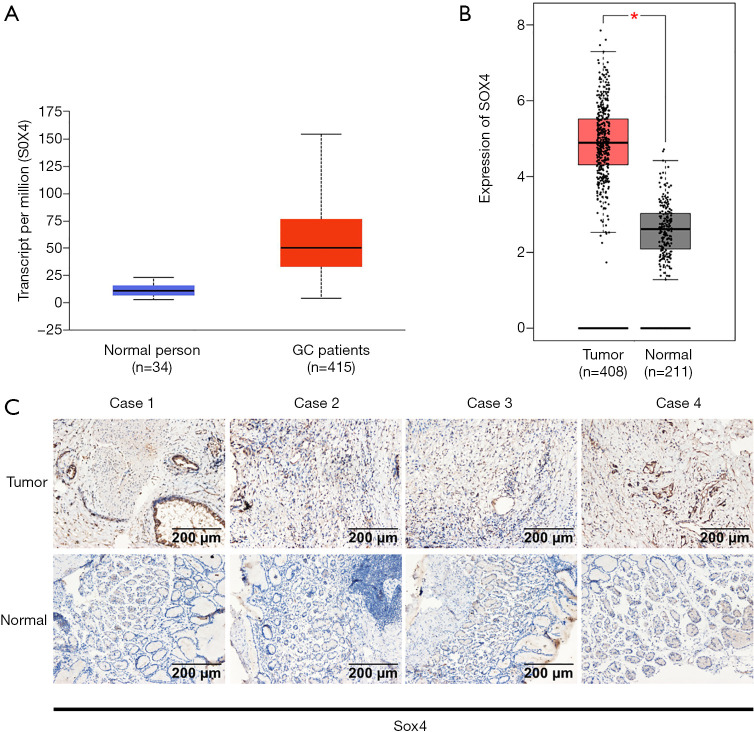
We analyzed the UALCAN and GEPIA databases and found that SOX4 expression was higher in primary gastric tumor samples, when compared to normal samples (Figure 2A,B). After this, we analyzed 10 pairs of gastric tumor tissues and corresponding adjacent non-tumor gastric tissues to verify SOX4 expression levels. IHC staining showed that SOX4 was abundantly and uniformly expressed in tumor samples, but was significantly down-regulated in all adjacent non-tumor gastric tissues (Figure 2C).
Figure 2.
High SOX4 expression levels in GC. (A,B) High SOX4 levels in gastric primary tumors according to UALCAN [primary tumor (n) =415; normal (n) =34] and GEPIA databases [tumor (n) =408; normal (n) =211]. (C) SOX4 immunohistochemistry in gastric tumor and adjacent non-tumor gastric tissue. *, P<0.05. GC, gastric cancer.
MiR-596 negatively regulates SOX4 expression
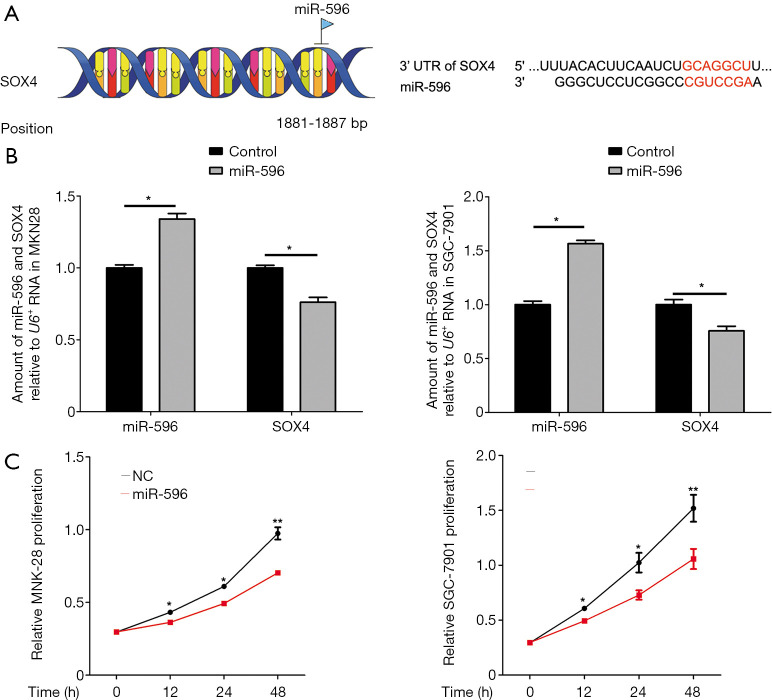
The interaction between miR-596 and SOX4 was also investigated. From the miRWalk database, miR-596 bound directly to the 1881-1887 base pair site of SOX4 (Figure 3A). RT-PCR analysis revealed that SOX4 expression was significantly decreased in PEGFP-N1-miR-596 transfected MNK28 and SGC7901 cells, when compared with NC transfected cells (P<0.05, Figure 3B). And then, MTT assay showed that the proliferation ability of over-expression of miR-596 in MNK28 and SGC-7901 cells was significantly reduced on 12, 24 and 48 h compared with that of the control cells (Figure 3C).
Figure 3.
miR-596 negatively regulates SOX4 expression. (A) Potential binding sites between miR-596 and SOX4. (B) RT-PCR analysis of SOX4 mRNA levels relative to U6. Relative SOX4 levels in MNK28 and SGC7901 cells were arbitrarily designated as 1. Each column represents the mean ± SD from three biological repeats. (C) MNK28 and SGC-7901 cell viability. Cells were transfected with PEGFP-N1-miR-596 and NC, and analyzed by MTT assay at 12, 24 and 48 h. *, P<0.05; **, P<0.01.
SOX4 is highly expressed in CD4+ T cells
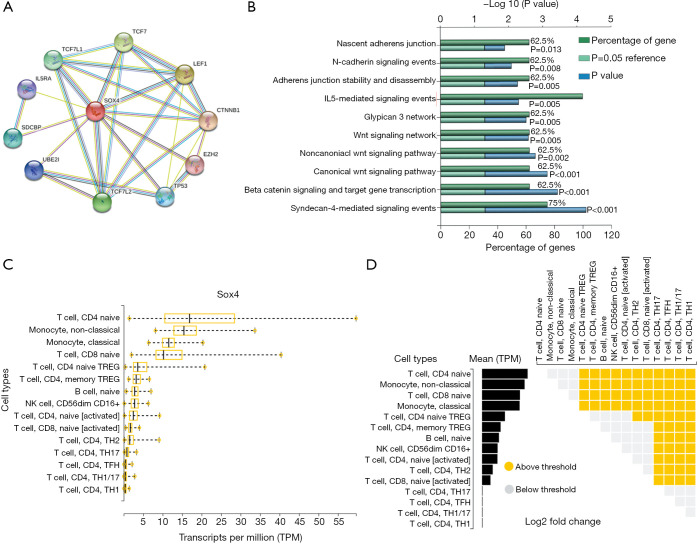
Next, we analyzed SOX4 target genes using string (Figure 4A). Functional enrichment analysis in the FunRich database showed that SOX4 function and target genes were mainly focused on interleukin 5 (IL5) mediated signaling (Figure 4B). We also analyzed SOX4 expression in several immune cells; according to the DICE database, SOX4 was highly expressed in CD8+ T and CD4+ T cells (P=0.005, Figure 4C,D).
Figure 4.
SOX4 expression in immune cells. (A) Target genes of SOX4. (B) SOX4 and target genes focus on the immune response. (C,D) SOX4 is highly expressed in CD4+ T and CD8+ T cells.
SOX4 expression is correlated with immune infiltration in GC
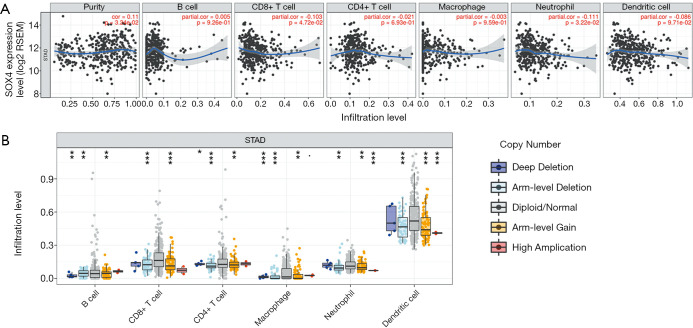
We investigated whether SOX4 expression was correlated with immune infiltration levels in GC, using the TIMER database. As shown (Figure 5A), SOX4 expression was significantly positively correlated with tumor purity in STAD, and negatively correlated with CD8+ T and CD4+ cells. We also investigated infiltration levels of different somatic copy number alterations for SOX4, and found diploid/normal SOX4 was significantly highly expressed in CD8+ T and CD4+ T cells (Figure 5B).
Figure 5.
Correlation of SOX4 expression with immune infiltration levels in stomach adenocarcinoma (STAD). (A) SOX4 expression is significantly negatively associated with tumor purity, but no significant correlations with infiltrating CD8+ T and CD4+ T cell levels were determined (n=415). (B) Significant highly expression of normal SOX4 in CD8+ T and CD4+ T cells.
MiR-596 and SOX4 are connected with GC patient survival
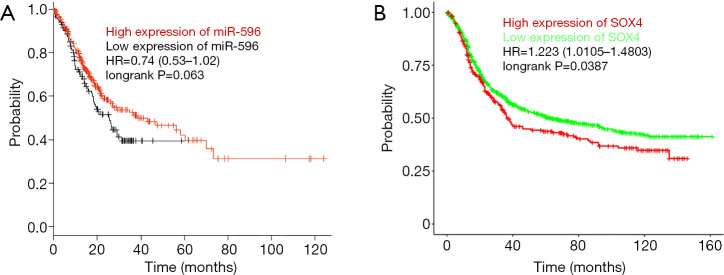
Based on the Kaplan-Meier Plotter database, miR-596 and SOX4 were assessed for associations with GC patient prognoses. We observed that high levels of miR-596 expression and low SOX4 expression levels were significantly associated with a longer OS in GC patients (Figure 6A,B).
Figure 6.
Differential expression of miR-596 and SOX4 as prognostic markers for the OS of GC patients. (A) High miR-596 levels prolong GC patients (n=431) survival. (B) Low SOX4 levels prolong GC patients (n=1,103) survival. GC, gastric cancer; OS, overall survival.
Discussion
MiRNAs are accepted as important noncoding RNAs, and are involved in the development and progression of several cancers (22). Recently, increasing numbers of miRNAs have been identified as important GC therapeutic targets (23). In this study, we explored the expression, prognosis, and function of miR-596 in GC patients, and found that miR-596 was down-regulated in these patients, when compared to healthy individuals. MiR-596 overexpression decreased SOX4 expression. In addition, miR-596 was confirmed as playing an important role in GC development and progression, such that low expression could lead to poor prognoses in GC patients. In agreement with our data/results, miR-596 was under-expressed in hepatocellular, cervical and oral cancer, and was associated with an unfavorable prognosis in hepatocellular, cervical and oral patient groups (12). A recent study reported that miR-596 also acts as a tumor suppressor in GC and is upregulated by promoter demethylation. However, the precise mechanism whereby miR-596 is implicated in GC requires further exploration.
Recently studies have indicated that SOX4 overexpression plays important roles in multiple cancers (14). SOX4 activation contributes to prostate cancer prognosis and progression (15). The miR-138/SOX4 axis is involved in sorafenib mediated reverse sorafenib induced changes in cholangiocarcinoma cell apoptosis and viability (24). Moreover, high expression of SOX4 is associated with poor clinical outcomes in colon cancer and acute lymphoblastic leukemia (ALL) patients (25). In this study, miR-596 appeared to downregulate SOX4 expression, with significant negative correlations for SOX4 with CD8+ T and CD4+ cells in GC patients. Thus, miR-596 high expression and SOX4 low expression were significantly associated with a longer OS in GC patients. Our preliminary results have uncovered negative regulatory correlations between miR-596 and SOX4 in GC, however the precise mechanisms whereby miR-596 mediates SOX4 expression requires further exploration.
Conclusions
We show that miR-596 is a negative regulator of SOX4. As predicted by bioinformatics, miR-596 directly targets SOX4 that had connection with tumor infiltrating immune cells in GC.
Acknowledgments
Funding: This study was supported by the Zhejiang Province Public Welfare Technology Application Research Project (No. 2020362827), Zhejiang Province Public Welfare Technology Application Research Project (No. 2017198118) and Zhejiang Province Public Welfare Technology Application Research Project (No. 2018KY294).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.02). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Ajani JA, D'Amico TA, Almhanna K, et al. Gastric Cancer, Version 3.2016, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2016;14:1286-312. 10.6004/jnccn.2016.0137 [DOI] [PubMed] [Google Scholar]
- 3.Fock KM. Review article: the epidemiology and prevention of gastric cancer. Aliment Pharmacol Ther 2014;40:250-60. 10.1111/apt.12814 [DOI] [PubMed] [Google Scholar]
- 4.Liu SP, Fu RH, Yu HH, et al. MicroRNAs regulation modulated self-renewal and lineage differentiation of stem cells. Cell Transplant 2009;18:1039-45. 10.3727/096368909X471224 [DOI] [PubMed] [Google Scholar]
- 5.Eyking A, Reis H, Frank M, et al. MiR-205 and MiR-373 are associated with aggressive human mucinous colorectal cancer. PLoS One 2016;11:e0156871. 10.1371/journal.pone.0156871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Musilova K, Mraz M. MicroRNAs in B-cell lymphomas: how a complex biology gets more complex. Leukemia 2015;29:1004-17. 10.1038/leu.2014.351 [DOI] [PubMed] [Google Scholar]
- 7.Nielsen BS, Jorgensen S, Fog JU, et al. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis 2011;28:27-38. 10.1007/s10585-010-9355-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capula M, Mantini G, Funel N, et al. New avenues in pancreatic cancer: exploiting microRNAs as predictive biomarkers and new approaches to target aberrant metabolism. Expert Rev Clin Pharmacol 2019;12:1081-90. 10.1080/17512433.2019.1693256 [DOI] [PubMed] [Google Scholar]
- 9.Fen H, Hongmin Z, Wei W, et al. RHPN1-AS1 drives the progression of hepatocellular carcinoma via regulating miR-596/IGF2BP2 axis. Curr Pharm Des 2020;25:4630-40. 10.2174/1381612825666191105104549 [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, Dai DQ. MicroRNA-596 acts as a tumor suppressor in gastric cancer and is upregulated by promotor demethylation. World J Gastroenterol 2019;25:1224-37. 10.3748/wjg.v25.i10.1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang YW, Kuo CT, Chen JH, et al. Hypermethylation of miR-203 in endometrial carcinomas. Gynecol Oncol 2014;133:340-5. 10.1016/j.ygyno.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endo H, Muramatsu T, Furuta M, et al. Potential of tumor-suppressive miR-596 targeting LGALS3BP as a therapeutic agent in oral cancer. Carcinogenesis 2013;34:560-9. 10.1093/carcin/bgs376 [DOI] [PubMed] [Google Scholar]
- 13.Mehta A, Mann M, Zhao JL, et al. The microRNA-212/132 cluster regulates B cell development by targeting Sox4. J Exp Med 2015;212:1679-92. 10.1084/jem.20150489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanieh H, Ahmed EA, Vishnubalaji R, et al. SOX4: Epigenetic regulation and role in tumorigenesis. Semin Cancer Biol 2019. [Epub ahead of print]. 10.1016/j.semcancer.2019.06.022 [DOI] [PubMed] [Google Scholar]
- 15.Ha Thi HT, Kim HY, Kim YM, et al. MicroRNA-130a modulates a radiosensitivity of rectal cancer by targeting SOX4. Neoplasia 2019;21:882-92. 10.1016/j.neo.2019.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Lin X, Jiang W, et al. lncRNA LEF1-AS1 Promotes Malignancy in Non-Small-Cell Lung Cancer by Modulating the miR-489/SOX4 Axis. DNA Cell Biol 2019;38:1013-21. 10.1089/dna.2019.4717 [DOI] [PubMed] [Google Scholar]
- 17.Song Z, Zhang X, Lin Y, et al. LINC01133 inhibits breast cancer invasion and metastasis by negatively regulating SOX4 expression through EZH2. J Cell Mol Med 2019;23:7554-65. 10.1111/jcmm.14625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang M, Huang S, Long D, et al. MiR-381 inhibits migration and invasion in human gastric carcinoma through downregulatedting SOX4. Oncol Lett 2017;14:3760-6. 10.3892/ol.2017.6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Y, Zhang J, Ye B, et al. miR-138 mediates sorafenib-induced cell survival and is associated with poor prognosis in cholangiocarcinoma cells. Clin Exp Pharmacol Physiol 2020;47:459-65. 10.1111/1440-1681.13205 [DOI] [PubMed] [Google Scholar]
- 20.Quan X, Li X, Yin Z, et al. p53/miR-30a-5p/ SOX4 feedback loop mediates cellular proliferation, apoptosis, and migration of non-small-cell lung cancer. J Cell Physiol 2019;234:22884-95. 10.1002/jcp.28851 [DOI] [PubMed] [Google Scholar]
- 21.Wang CY, Hua L, Sun J, et al. MiR-211 inhibits cell proliferation and invasion of gastric cancer by down-regulating SOX4. Int J Clin Exp Pathol 2015;8:14013-20. [PMC free article] [PubMed] [Google Scholar]
- 22.Bahreyni-Toossi MT, Dolat E, Khanbabaei H, et al. microRNAs: Potential glioblastoma radiosensitizer by targeting radiation-related molecular pathways. Mutat Res 2019;816-818:111679. 10.1016/j.mrfmmm.2019.111679 [DOI] [PubMed] [Google Scholar]
- 23.Bhat SA, Majid S, Rehman MU. Scenario and future prospects of microRNAs in gastric cancer: A review. Iran J Basic Med Sci 2019;22:345-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vervoort SJ, van Boxtel R, Coffer PJ, et al. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene 2013;32:3397-409. 10.1038/onc.2012.506 [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Ju HL, Yuan XY, et al. SOX4 is a potential prognostic factor in human cancers: a systematic review and meta-analysis. Clin Transl Oncol 2016;18:65-72. 10.1007/s12094-015-1337-4 [DOI] [PubMed] [Google Scholar]