Abstract
Background
The aim was to explore the interaction among chemokine (C-X-C motif) ligand (CXCL) 1/2/8 expressions, and their associations with clinicopathologic features and survival profiles in non-small cell lung cancer (NSCLC) patients.
Methods
The tumor tissue specimens from 232 primary NSCLC patients with TNM stage I-IIIA underwent resection were obtained and the expressions of CXCL1, CXCL2 and CXCL8 were measured by immunohistochemical assay. Disease-free survival (DFS) and overall survival (OS) were calculated according to survival data.
Results
There were 117(50.4%) CXCL1 low expression patients versus (vs.) 115 (49.6%) CXCL1 high expression patients, 107(46.1%) CXCL2 low expression patients vs. 125 (53.9%) CXCL2 high expression patients, 93 (40.1%) CXCL8 low expression patients vs. 139 (59.9%) CXCL8 high expression patients. Meanwhile, CXCL1 expression was positively correlated with CXCL2 expression and CXCL8 expression; CXCL2 expression was also positively correlated with CXCL8 expression. For tumor features, CXCL1, CXCL2 and CXCL8 were positively correlated with lymph node (LYN) metastasis and TNM stage, but not correlated with differentiation, tumor size or carcinoembryonic antigen (CEA) level. For prognosis, CXCL1 high expression was associated with worse DFS and OS, so did CXCL2 high expression, while there was no correlation of CXCL8 with DFS or OS; Multivariate Cox’s regression disclosed that high expression of CXCL1, but not CXCL2 or CXCL8, was an independent factor predicting shorter DFS and OS.
Conclusions
An inter-correlation is observed among CXCL1, CXCL2 and CXCL8 expressions, and they show diversified potential as biomarkers for tumor features and survival profiles in NSCLC patients.
Keywords: Chemokine (C-X-C motif) ligand 1, chemokine (C-X-C motif) ligand 2, chemokine (C-X-C motif) ligand 8, non-small cell lung cancer (NSCLC), survival profiles
Introduction
Lung cancer is the most commonly diagnosed cancer and the leading cause of cancer deaths (1). According to the global cancer statistics 2018, it causes approximately 2,093,876 new cases (11.6% of the total new cancer cases) and 1,761,007 deaths (18.4% of the total cancer deaths) worldwide (1). Non-small cell lung cancer (NSCLC), as the most common histologic subtype of lung cancer, accounts on nearly 85% of lung cancer and contributes to high morbidity and mortality (2,3). Current balance of benefits and risks suggests that completed surgical resection is an optimum treatment for NSCLC patients in early stage, while most of NSCLC patients have been in advanced stage at the first diagnosis, and over 65% of them present with local or distant metastasis, who lose the best time of surgical resection (2,4). In addition, although obvious improvements have been achieved in various treatments for NSCLC (including chemotherapy, radiotherapy as well as immunological therapy) and these treatments have effects on release symptoms and delay disease progression, the 5-year overall survival (OS) rate of NSCLC patients still range from 15% to 25% (2,4). Thus, the exploration of potential biomarkers supervising disease progression may provide opportunities for disease management and improve prognosis in NSCLC patients.
Chemokines (C-X-C motif) (CXC) are a group of 17 α-chemokines that exert physiological and pathological functions, and many CXC chemokines [such as CXC ligand (CXCL) 1, CXCL2 and CXCL8] not only play critical roles in tumor microenvironment alone, but also interact with each other to promote neutrophils’ secretion of several pro-inflammatory, angiogenic and immunoregulatory factors, thereby contributing to tumor progression (5-8). Regarding lung cancer, several previous studies have shown that CXCL1, CXCL2 and CXCL8 participant in the microenvironment of lung cancer through participating in multiple mechanisms, including recruitment of tumor-associated neutrophils (9), involving in anlotinib resistance (10), or affecting tumor cell proliferation and angiogenesis (11). Apart from their roles in cancer pathology, CXCL1, CXCL2 and CXCL8 have been considered as tumor promoters in cancer patients (12-14). For example, CXCL1 is positively related to tumor invasion, tumor-node-metastasis (TNM) stage, tumor size and lymph node metastasis in gastric cancer patients (12); CXCL2 is positively associated with infiltration extent of primary tumor and lymph node metastasis in colorectal cancer (CRC) (13); CXCL8 is positively correlated with depth of tumor invasion and C-reactive protein (CRP) concentrations in esophageal cancer (OC) (14). More importantly, a previous report explores the clinicopathological significance of CXCR2 ligands (including CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL7, and CXCL8) in gastric cancer patients, and reveals that among the CXCR2 ligands, CXCL7 and CXCL1 server as important roles in the malignant progression of gastric cancer by regulating CXCR2 signaling (6). Whereas little is known about the clinical implication of CXCL1/2/8 in NSCLC patients. To solve this problem, we conducted this study with aim to explore the interaction among CXCL1/2/8 expressions, and their associations with clinical characteristics and prognosis in NSCLC patients. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2539).
Methods
Patients
We retrospectively reviewed 232 primary NSCLC patients who underwent resection in our hospital between January 2011 and December 2013. The inclusion criteria were: (I) histopathological proof of NSCLC; (II) age above 18 years; (III) received resection, and tumor tissues excised from resection were well preserved; (IV) clinical features were completely recorded; (V) follow-up data were complete and available. The exclusion criteria included: (I) TNM stage IIIB or IV; (II) relapsed NSCLC; (III) received chemotherapy, radiotherapy or other anti-cancer therapies before resection; (IV) history of hematologic malignancies or other solid tumors; (V) pregnant or lactating women. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Shanghai Chest Hospital, Shanghai Jiao Tong University (KSY1672). The written informed consents were acquired from the patients or their family members.
Data and sample collection
Clinicopathologic features including demographics, complications and tumor features were collected from electronic medical records. Preoperative carcinoembryonic antigen (CEA) level in the serum of patients was also collected, which was determined by electrochemiluminescence immunoassay (ECLIA) using RocheElecsys601 automatic immune analyzer (Roche diagnostics, Mannheim, Germany) and corresponding test kits. And the Formalin-fixed and paraffin-embedded tumor tissue specimens were obtained from the store room of pathology department.
Immunohistochemical (IHC) assay
The expressions of CXCL1, CXCL2 and CXCL8 in tissue specimens were measured by IHC. All antibodies used in IHC were purchased from Thermo Fisher Scientific (Waltham, Massacsetts, USA), which included Rabbit CXCL1 Polyclonal Antibody (1:20), Rabbit CXCL2 Monoclonal Antibody (1:20), Rabbit CXCL8 Polyclonal Antibody (1:200) and Goat anti-Rabbit IgG (H+L) (HRP) Antibody (1:10,000). All procedures were carried out referring to a previous study (15). The IHC score was assessed by a semi-quantitative scoring method as previously described (16), and the total IHC score was ranging from 0 to 12. The IHC score >3 was defined as high expression, while the IHC score ≤3 was defined as low expression.
Follow-up
Survival data were extracted from follow-up records, and the last follow-up date was December 31, 2018. According to the survival data, the disease-free survival (DFS) and OS were calculated. The DFS was defined as the duration from resection to disease relapse, disease progression or death, and OS was defined as the duration from resection to death.
Statistical analysis
Statistical analyses were performed using SPSS 22.0 software (IBM, Chicago, IL, USA), and figures were plotted using GraphPad Prism 7.01 software (GraphPad Software, San Diego, California, USA). Correlation analysis was determined by Chi-square test or Spearman’s rank correlation test. DFS and OS were displayed using Kaplan-Meier (K-M) curve. Comparison of DFS and OS between two groups was determined by Log-rank test. Factors predicting DFS and OS were analyzed by univariate and forward stepwise multivariate Cox’s proportional hazard regression model. P value <0.05 was considered significant.
Results
Clinicopathological characteristics in NSCLC patients
The mean age was 61.1±10.6 years of 232 NSCLC patients [including 68 (29.3%) female and 164 (70.7%) males]. The number of patients with history of smoke, history of drink, hypertension, hyperlipidemia and diabetes was 128 (55.2%), 97 (41.8%), 89 (38.4%), 74 (31.9%) and 43 (18.5%) respectively. For tumor features, there were 34 (14.7%), 137 (59.0%) and 61 (26.3%) patients with well, moderate and poor differentiation; the mean value of tumor size was 5.2±2.0 cm, and there were 141 (60.8%) patients with tumor size ≤5 cm, but 91 (39.2%) patients with tumor size >5 cm; 88 (37.9%) patients had lymph node (LYN) metastasis. As to TNM stage, the number of patients with stage I, stage II and stage III was 76 (32.8%), 67 (28.9%) and 89 (38.3%), respectively. For carcinoembryonic antigen (CEA) level, the median value was 6.8 (3.1–27.3) ng/mL, and there were 137 (59.1%) patients with abnormal CEA level and 95 (40.9%) patients with normal CEA level (Table 1).
Table 1. Clinicopathologic characteristics.
| Items | NSCLC patients (N=232) |
|---|---|
| Age (years), mean ± SD | 61.1±10.6 |
| ≤60 years, No. (%) | 115 (49.6) |
| >60 years, No. (%) | 117 (50.4) |
| Gender, No. (%) | |
| Female | 68 (29.3) |
| Male | 164 (70.7) |
| History of smoke, No. (%) | |
| No | 104 (44.8) |
| Yes | 128 (55.2) |
| History of drink, No. (%) | |
| No | 135 (58.2) |
| Yes | 97 (41.8) |
| Hypertension, No. (%) | |
| No | 143 (61.6) |
| Yes | 89 (38.4) |
| Hyperlipidemia, No. (%) | |
| No | 158 (68.1) |
| Yes | 74 (31.9) |
| Diabetes, No. (%) | |
| No | 189 (81.5) |
| Yes | 43 (18.5) |
| Differentiation, No. (%) | |
| Well | 34 (14.7) |
| Moderate | 137 (59.0) |
| Poor | 61 (26.3) |
| Tumor size (cm), mean ± SD | 5.2±2.0 |
| ≤5 cm, No. (%) | 141 (60.8) |
| >5 cm, No. (%) | 91 (39.2) |
| LYN metastasis, No. (%) | |
| No | 144 (62.1) |
| Yes | 88 (37.9) |
| TNM stage, No. (%) | |
| I | 76 (32.8) |
| II | 67 (28.9) |
| III | 89 (38.3) |
| CEA (ng/mL) level, median (IQR) | 6.8 (3.1–27.3) |
| Normal*, No. (%) | 95 (40.9) |
| Abnormal*, No. (%) | 137 (59.1) |
*, normal CEA level ≤5 ng/mL and abnormal CEA level >5 ng/mL. NSCLC, non-small cell lung carcinoma; SD, standard deviation; LYN, lymph node; CEA, carcinoembryonic antigen; IQR, interquartile range.
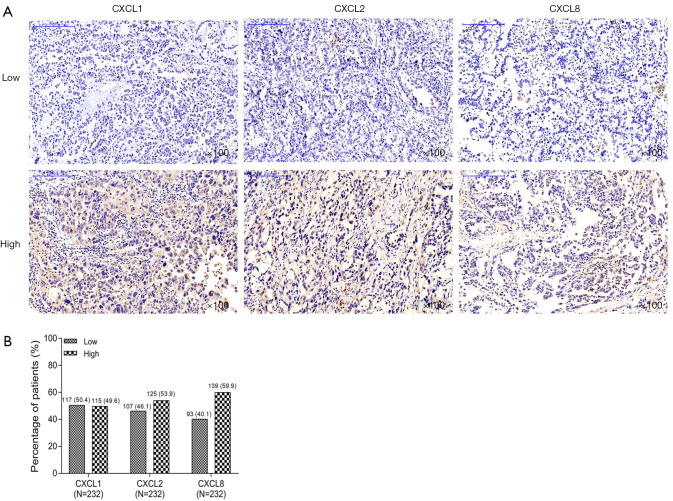
CXCL1, CXCL2 and CXCL8 expressions in NSCLC patients
The examples of CXCL1, CXCL2 and CXCL8 staining results by IHC were shown in Figure 1A. For CXCL1, there were 117 (50.4%) patients with CXCL1 low expression and 115 (49.6%) patients with CXCL1 high expression (Figure 1B). For CXCL2, there were 107 (46.1%) patients with CXCL2 low expression and 125 (53.9%) patients with CXCL2 high expression. For CXCL8, there were 93 (40.1%) patients with CXCL8 low expression and 139 (59.9%) patients with CXCL8 high expression.
Figure 1.
CXCL1, CXCL2 and CXCL8 expressions. (A) Example samples of CXCL1, CXCL2 and CXCL8 staining results by immunohistochemical assay in NSCLC patients; (B) the percentage of NSCLC patients with high expression and low expression of CXCL1, CXCL2 and CXCL8, respectively. NSCLC, non-small cell lung cancer; CXCL, chemokine (C-X-C motif) ligand.
Association among CXCL1, CXCL2 and CXCL8 expressions in NSCLC patients
CXCL1 expression was positively correlated with CXCL2 expression (P<0.001) and CXCL8 expression (P<0.001) (Table 2). Meanwhile, CXCL2 expression was also positively correlated with CXCL8 expression (P<0.001).
Table 2. Correlations among CXCL1, CXCL2 and CXCL8 expressions.
| Items | CXCL1 expression | CXCL2 expression | |||||
|---|---|---|---|---|---|---|---|
| Low | High | P value | Low | High | P value | ||
| CXCL2 expression, No. (%) | <0.001 | – | |||||
| Low | 85 (36.6) | 22 (9.5) | – | – | |||
| High | 32 (13.8) | 93 (40.1) | – | – | |||
| CXCL8 expression, No. (%) | <0.001 | <0.001 | |||||
| Low | 85 (36.6) | 8 (3.4) | 73 (31.5) | 20 (8.6) | |||
| High | 32 (13.8) | 107 (46.1) | 34 (14.7) | 105 (45.3) | |||
CXCL, chemokine (C-X-C motif) ligand.
Association of CXCL1, CXCL2 and CXCL8 expressions with tumor features in NSCLC patients
For CXCL1, it was positively correlated with LYN metastasis (P=0.002) and TNM stage (P=0.010), while there was no correlation of CXCL1 expression with differentiation (P=0.126), tumor size (P=0.295) or CEA level (P=0.275) (Table 3). For CXCL2, it was positively correlated with LYN metastasis (P=0.039) and TNM stage (P=0.008), while no correlation was found of CXCL2 expression with differentiation (P=0.638), tumor size (P=0.423) or CEA level (P=0.827). For CXCL8, it was also positively correlated with LYN metastasis (P=0.005) and TNM stage (P=0.004), while presented with a trend to be correlated with differentiation (P=0.064) but with no statistical significance; meanwhile, there was no correlation of CXCL8 expression with tumor size (P=0.219) or CEA level (P=0.180). In addition, we further explored the correlation of CXCL1/CXCL2/CXCL8 with lymphocytes, and we found no correlation of CXCL1 (P=0.963), CXCL2 (P=0.209), CXCL8 (P=0.434) with lymphocyte count in NSCLC patients (Table S1). The possible explanation might be that lymphocytes counts in blood sample reflect the whole-body conditions, thus, their associations with tumor CXCL1, 2, 8 might be weak.
Table 3. Correlation of CXCL1/CXCL2/CXCL8 with tumor features.
| Items | CXCL1 | CXCL2 | CXCL8 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | P | Low | High | P | Low | High | P | |||
| Differentiation, No. (%) | 0.126 | 0.638 | 0.064 | ||||||||
| Well | 22 (18.8) | 12 (10.4) | 18 (16.8) | 16 (12.8) | 18 (19.4) | 16 (11.5) | |||||
| Moderate | 67 (57.3) | 70 (60.9) | 61 (57.0) | 76 (60.8) | 55 (59.1) | 82 (59.0) | |||||
| Poor | 28 (23.9) | 33 (28.7) | 28 (26.2) | 33 (26.4) | 20 (21.5) | 41 (29.5) | |||||
| Tumor size, No. (%) | 0.295 | 0.423 | 0.219 | ||||||||
| ≤5 cm | 75 (64.1) | 66 (57.4) | 68 (63.6) | 73 (58.4) | 61 (65.6) | 80 (57.6) | |||||
| >5 cm | 42 (35.9) | 49 (42.6) | 39 (36.4) | 52 (41.6) | 32 (34.4) | 59 (42.4) | |||||
| LYN metastasis, No. (%) | 0.002 | 0.039 | 0.005 | ||||||||
| No | 84 (71.8) | 60 (52.2) | 74 (69.2) | 70 (56.0) | 68 (73.1) | 76 (54.7) | |||||
| Yes | 33 (28.2) | 55 (47.8) | 33 (30.8) | 55 (44.0) | 25 (26.9) | 63 (45.3) | |||||
| TNM stage, No. (%) | 0.010 | 0.008 | 0.004 | ||||||||
| I | 47 (40.2) | 29 (25.2) | 45 (42.0) | 31 (24.8) | 39 (41.9) | 37 (26.6) | |||||
| II | 33 (28.2) | 34 (29.6) | 28 (26.2) | 39 (31.2) | 28 (30.1) | 39 (28.1) | |||||
| III | 37 (31.6) | 52 (45.2) | 34 (31.8) | 55 (44.0) | 26 (28.0) | 63 (45.3) | |||||
| CEA level, No. (%) | 0.275 | 0.827 | 0.180 | ||||||||
| Normal* | 52 (44.4) | 43 (37.4) | 43 (40.2) | 52 (41.6) | 43 (46.2) | 52 (37.4) | |||||
| Abnormal* | 65 (55.6) | 72 (62.6) | 64 (59.8) | 73 (58.4) | 50 (53.8) | 87 (62.6) | |||||
*, normal CEA level ≤5 ng/mL and abnormal CEA level >5 ng/mL. CXCL, chemokine (C-X-C motif) ligand; LYN, lymph node; CEA, carcinoembryonic antigen.
Association of CXCL1, CXCL2 and CXCL8 with survival profiles in NSCLC patients
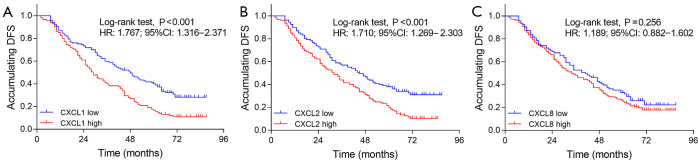
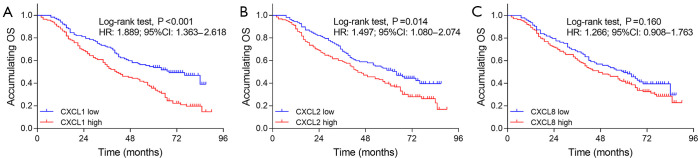
The high expressions of CXCL1 (P<0.001) (Figure 2A) and CXCL2 (P<0.001) (Figure 2B) were correlated with worse DFS, while no correlation was found of CXCL8 (P=0.256) (Figure 2C) with DFS in NSCLC patients. In addition, high expressions of CXCL1 (P<0.001) (Figure 3A) and CXCL2 (P=0.014) (Figure 3B) were correlated with poor OS, while there was no correlation of CXCL8 (P=0.160) (Figure 3C) with OS in NSCLC patients.
Figure 2.
Correlation of CXCL1, CXCL2 and CXCL8 with DFS. (A) Correlation of CXCL1 with DFS in NSCLC patients; (B) correlation of CXCL2 with DFS in NSCLC patients; C: Correlation of CXCL8 with DFS in NSCLC patients. CXCL, chemokine (C-X-C motif) ligand; NSCLC, non-small cell lung cancer; DFS, disease-free survival.
Figure 3.
Correlation of CXCL1, CXCL2 and CXCL8 with OS. (A) Correlation of CXCL1 with OS in NSCLC patients; (B) correlation of CXCL2 with OS in NSCLC patients; (C) correlation of CXCL8 with OS in NSCLC patients. CXCL, chemokine (C-X-C motif) ligand; NSCLC, non-small cell lung cancer; OS, overall survival.
Predictive factors for DFS in NSCLC patients
Based on univariate Cox’s regression analysis, CXCL1 high expression (P<0.001) and CXCL2 high expression (P<0.001) were associated with shorter DFS, and male (P=0.038), poor differentiation (P=0.038), larger tumor size (P=0.013), LYN metastasis (P<0.001), increased TNM stage (P<0.001) as well as abnormal CEA level (P=0.024) were also correlated with poor DFS in NSCLC patients (Table 4). Forward stepwise multivariate Cox’s regression disclosed that CXCL1 high expression (P=0.003) was an independent factor predicting shorter DFS, and male (P=0.012) as well as LYN metastasis (P<0.001) also independently predicted worse DFS in NSCLC patients.
Table 4. Analysis of factors predicting DFS.
| Items | Cox’s regression model | |||
|---|---|---|---|---|
| P value | HR | 95% CI | ||
| Lower | Higher | |||
| Univariate Cox’s regression | ||||
| CXCL1 high | <0.001 | 1.767 | 1.316 | 2.371 |
| CXCL2 high | <0.001 | 1.710 | 1.269 | 2.303 |
| CXCL8 high | 0.256 | 1.189 | 0.882 | 1.602 |
| Age (>60 years) | 0.086 | 1.291 | 0.965 | 1.728 |
| Male | 0.038 | 1.429 | 1.020 | 2.001 |
| History smoke | 0.527 | 0.910 | 0.680 | 1.218 |
| History drink | 0.297 | 1.168 | 0.872 | 1.565 |
| Hypertension | 0.628 | 0.929 | 0.689 | 1.253 |
| Hyperlipidemia | 0.406 | 0.874 | 0.637 | 1.200 |
| Diabetes | 0.222 | 0.786 | 0.534 | 1.157 |
| Poor differentiation | 0.038 | 1.250 | 1.013 | 1.542 |
| Tumor size | 0.013 | 1.453 | 1.083 | 1.951 |
| LYN metastasis | <0.001 | 2.571 | 1.909 | 3.464 |
| TNM stage | <0.001 | 1.499 | 1.257 | 1.788 |
| Abnormal CEA level* | 0.024 | 1.409 | 1.046 | 1.897 |
| Forward stepwise multivariate Cox’s regression | ||||
| CXCL1 high | 0.003 | 1.582 | 1.174 | 2.133 |
| Male | 0.012 | 1.546 | 1.101 | 2.169 |
| LYN metastasis | <0.001 | 2.536 | 1.873 | 3.433 |
Factors with P value less than 0.05 in univariate Cox’s regression were included in forward stepwise multivariate Cox’s regression. *, normal CEA level ≤5 ng/mL and abnormal CEA level >5 ng/mL. DFS, disease-free survival; HR, hazard ratio; CI, confidence interval; CXCL, chemokine (C-X-C motif) ligand; LYN, lymph node; CEA, carcinoembryonic antigen.
Predictive value of factors for OS in NSCLC patients
Univariate Cox’s regression analysis displayed that CXCL1 high expression (P<0.001) and CXCL2 high expression (P=0.015) were associated with poor OS, meanwhile, poor differentiation (P=0.019), larger tumor size (P=0.001), LYN metastasis (P<0.001), increased TNM stage (P<0.001) and abnormal CEA level (P=0.002) were associated with worse OS in NSCLC patients as well (Table 5). According to forward stepwise multivariate Cox’s regression, CXCL1 high expression (P=0.004) independently predicted shorter OS, and poor differentiation (P=0.029), LYN metastasis (P<0.001) as well as abnormal CEA level (P=0.006) were also independent factors predicting worse OS in NSCLC patients.
Table 5. Analysis of factors predicting OS.
| Items | Cox’s regression model | |||
|---|---|---|---|---|
| P value | HR | 95% CI | ||
| Lower | Higher | |||
| Univariate Cox’s regression | ||||
| CXCL1 high | <0.001 | 1.889 | 1.363 | 2.618 |
| CXCL2 high | 0.015 | 1.497 | 1.080 | 2.074 |
| CXCL8 high | 0.164 | 1.266 | 0.908 | 1.763 |
| Age (>60 years) | 0.154 | 1.262 | 0.916 | 1.740 |
| Male | 0.346 | 1.189 | 0.830 | 1.704 |
| History smoke | 0.361 | 0.862 | 0.626 | 1.186 |
| History drink | 0.534 | 1.108 | 0.802 | 1.529 |
| Hypertension | 0.766 | 0.951 | 0.683 | 1.324 |
| Hyperlipidemia | 0.677 | 0.929 | 0.655 | 1.317 |
| Diabetes | 0.340 | 0.814 | 0.533 | 1.243 |
| Poor differentiation | 0.019 | 1.325 | 1.047 | 1.676 |
| Tumor size | 0.001 | 1.700 | 1.233 | 2.344 |
| LYN metastasis | <0.001 | 3.219 | 2.323 | 4.458 |
| TNM stage | <0.001 | 1.485 | 1.223 | 1.804 |
| Abnormal CEA level* | 0.002 | 1.685 | 1.205 | 2.355 |
| Forward stepwise multivariate Cox’s regression | ||||
| CXCL1 high | 0.004 | 1.628 | 1.167 | 2.273 |
| Poor differentiation | 0.029 | 1.328 | 1.029 | 1.714 |
| LYN metastasis | <0.001 | 2.983 | 2.142 | 4.154 |
| Abnormal CEA level* | 0.006 | 1.612 | 1.151 | 2.259 |
Factors with P value less than 0.05 in univariate Cox’s regression were included in forward stepwise multivariate Cox’s regression. *, normal CEA level ≤5 ng/mL and abnormal CEA level >5 ng/mL. OS, overall survival; HR, hazard ratio; CI, confidence interval; CXCL, chemokine (C-X-C motif) ligand; LYN, lymph node; CEA, carcinoembryonic antigen.
Discussion
As common members of the CXC chemokine family, CXCL1, CXCL2 and CXCL8 serve as critical roles in multiple malignancies. For example, one previous study discloses that CXCL1 actives the extracellular signal-regulated kinase (ERK)/matrix metalloproteinase (MMP) 2/9 signaling axis to stimulate cell migration and invasion in ER-negative breast cancer cells (17); In addition, CXCL1 and CXCL2 high expressions attract CD11b+Gr1+myeloid cells into the tumor to produce S100A8/9, thereby causing chemoresistance and metastasis in breast cancer (18); Furthermore, the Ras-dependent secretion of CXCL8 has been reported to induce neo-vascularization to accelerate tumor progression in ovarian cancer (19); Another recent study reveals that CXCL8 is interacted with ING4 (a tumor suppressor gene) to promote tumor growth and vascular density in gliomas (20). In lung cancer, CXCL1 has been reported to interact with miR141-CXCR2 pathway regulates migration of regulatory T cells (Treg) into malignant pleural effusion (MPE) (21). CXCL2 is discovered to obviously offset anlotinib-induced cell migration inhibition and promote invasion of anlotinib-treated cells in NSCLC, indicated that CXCL2 is involved in the resistance in anlotinib resistant NSCLC cells (10). CXCL8/interleukin-8 analogue CXCL8 [3–72] K11R/G31P interacts with CXCR1/2 to suppress tumor cell proliferation and inhibit angiogenesis, thereby restricting lung cancer growth (11). Except for the oncogenic effects of CXCL1, CXCL2 and CXCL8 in several malignancies alone, they also have the obvious interaction with each other in tumor microenvironment. For instance, CXCL1/8-CXCR2 axis is interacted with SMAD4 to attract neutrophils, subsequently promoting the neutrophils’ secretion of matrix metalloproteinases (MMPs) and vascular endothelial growth factor (VEGF) in colorectal cancer (7). In addition, CXCL1, CXCL2 and CXCL8 regulate multiple common genes [such as CXC type chemokine receptor (CXCR)2], suggesting that three of CXCL1, CXCL2 and CXCL8 are able to be interacted with each other via mediating common genes (22-24). Therefore, CXCL1, CXCL2 and CXCL8 not only play important roles in tumor microenvironments alone, but also they are interacted with each other to induce tumor progression.
Apart from the tumor promoter roles of CXCL1, CXCL2 and CXCL8 in cancer pathology, three of them also are discovered as an oncogene in patients with various carcinomas. For instance, one previous study CXCL1 expression is increased and it is positively correlated with T invasion (T2–T4), lymph node metastasis, lymphatic invasion, venous invasion, peritoneal cytology, peritoneal metastasis in gastric cancer patients (24); another study determines that CXCL2 high expression is associated with the multiple tumor numbers in hepatocellular carcinoma patients (25); Meanwhile, CXCL8 is highly expressed in cervical cancer tissues compared with normal cervical tissues, and it is associated with advanced clinical stage, distant metastasis, histological type and histological grade in cervical cancer patients (26). Although CXCL1, CXCL2 and CXCL8 have been reported to be oncogenic roles in patients with several cancers, the clinical implication of CXCL1, CXCL2 and CXCL8 in NSCLC patients is still unclear. To solve this issue, we performed this study that enrolled 232 primary NSCLC patients who underwent resection, and we observed that CXCL1, CXCL2 and CXCL8 had a positive correlation with each other, which might be that there were various common genes regulated by all of CXCL1, CXCL2 and CXCL8 (such as CXCR2), thus, they could be interacted with each other and had a positive association with each other in NSCLC patients (22-24). In order to further explore whether CXCL1, CXCL2 and CXCL8 have clinical significance in NSCLC patients, we assessed the correlation of CXCL1, CXCL2 and CXCL8 with tumor features in NSCLC patients, and we discovered that all CXCL1, CXCL2 and CXCL8 were positively correlated with LYN metastasis and TNM stage in NSCLC patients. The possible explanations were as follows: (I) for CXCL1, it might regulate multiple pathways (including ERK/MMP2/9 signaling axis) to accelerate cell migration and invasion, and then devoted into tumor metastasis and correlated with LYN metastasis in NSCLC patients; (II) for CXCL2, it might be interacted with receptor activator of NF-kappaB ligand to stimulate the cell adhesion and migration, thereby contributed to tumor metastasis in NSCLC patients (27); (III) for CXCL8, it has been considered as a pro-inflammatory chemokine [referred to as interleukin-8 (IL-8)] involving in tumor angiogenesis to promote tumorigenesis and metastasis, thus CXCL8 high expression was related to LYN metastasis in NSCLC patients (19,20). (IV) CXCL1, CXCL2 and CXCL8 were interacted with each other to promote the neutrophils’ secretion of several pro-inflammatory, angiogenic and immunoregulatory factors (including MMPs and VEGF), thereby promoting tumor metastasis in NSCLC patients (7,8,28).
A number of recent studies have been performed to investigate the prognostic value of CXCL1, CXCL2 and CXCL8 in cancer patients. For example, CXCL1 is an independent factor predicting poor OS in gastric cancer patients (24); furthermore, CXCL8 overexpression is reported to be obviously associated with poor DFS and OS in colorectal cancer patients (29) as well as in lung adenocarcinoma patients (30). The prognostic significance of CXCL1, CXCL2 and CXCL8 is still unknown in NSCLC patients. In this study, we discovered that (I) CXCL1 high expression were correlated with shorter DFS as well as OS, and it was an independent factor predicting worse survival profiles, which might be resulted in that CXCL1 not only correlated with deteriorative disease conditions (above-mentioned), thereby indirectly related to worse prognosis in NSCLC patients, but also was interacted with S100A8/9 to increase chemoresistance and then caused worse treatment outcomes, thereby directly independently predicted worse prognosis in NSCLC patients; (II) CXCL2 high expression were associated with shorter DFS and OS, but it could not independently predict DFS and OS, which might be caused by that CXCL2 might be interacted with LYN metastasis to indirectly affect survival profiles in NSCLC patients; (III) CXCL8 was not related to DFS and OS in NSCLC patients, whose discrepancy with other previous data (29,30) would most likely to due to the heterogenicity in human cancers.
Despite of interesting results in this study, the main limitation of the relatively small sample size in this study still existed, further study enrolled more NSCLC patients was necessary. Moreover, the enrolled NSCLC patients in this study were at TNM stage I-IIIA, but not TNM stage IIIB or IV. Thus, the clinical value of CXCL1, CXCL2 and CXCL8 in all NSCLC patients was confused. Furthermore, although the clinical implication of CXCL1, CXCL2 and CXCL8 in NSCLC patients was explored, their detailed mechanism underlying NSCLC pathology was not investigated in this study, further in vitro and in vivo experiments were needed. Besides, the association between CXCL1, CXCL2, CXCL8 and inflammatory factors was not explored, further study should be performed.
In conclusion, an inter-correlation is observed among CXCL1, CXCL2 and CXCL8 expressions, and they show diversified potential as biomarkers for tumor features and survival profiles in NSCLC patients.
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Shanghai Chest Hospital, Shanghai Jiaotong University (KSY1672). The written informed consents were acquired from the patients or their family members.
Footnotes
Reporting Checklist: The authors have completed the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2539).
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2539
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-2539
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2539). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Goldstraw P, Ball D, Jett JR, et al. Non-small-cell lung cancer. Lancet 2011;378:1727-40. 10.1016/S0140-6736(10)62101-0 [DOI] [PubMed] [Google Scholar]
- 3.Remark R, Becker C, Gomez JE, et al. The non-small cell lung cancer immune contexture. A major determinant of tumor characteristics and patient outcome. Am J Respir Crit Care Med 2015;191:377-90. 10.1164/rccm.201409-1671PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ettinger DS, Wood DE, Aisner DL, et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017;15:504-35. 10.6004/jnccn.2017.0050 [DOI] [PubMed] [Google Scholar]
- 5.Boro M, Balaji KN. CXCL1 and CXCL2 Regulate NLRP3 Inflammasome Activation via G-Protein-Coupled Receptor CXCR2. J Immunol 2017;199:1660-71. 10.4049/jimmunol.1700129 [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto Y, Kuroda K, Sera T, et al. The Clinicopathological Significance of the CXCR2 Ligands, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL7, and CXCL8 in Gastric Cancer. Anticancer Res 2019;39:6645-52. 10.21873/anticanres.13879 [DOI] [PubMed] [Google Scholar]
- 7.Ogawa R, Yamamoto T, Hirai H, et al. Loss of SMAD4 Promotes Colorectal Cancer Progression by Recruiting Tu-mor-Associated Neutrophils via the CXCL1/8-CXCR2 Axis. Clin Cancer Res 2019;25:2887-99. 10.1158/1078-0432.CCR-18-3684 [DOI] [PubMed] [Google Scholar]
- 8.Kuang DM, Zhao Q, Wu Y, et al. Peritumoral neutrophils link inflammatory response to disease progression by fostering angiogenesis in hepatocellular carcinoma. J Hepatol 2011;54:948-55. 10.1016/j.jhep.2010.08.041 [DOI] [PubMed] [Google Scholar]
- 9.Yuan M, Zhu H, Xu J, et al. Tumor-Derived CXCL1 Promotes Lung Cancer Growth via Recruitment of Tumor-Associated Neutrophils. J Immunol Res 2016;2016:6530410. 10.1155/2016/6530410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J, Xu W, Qian J, et al. Transcriptome profiling analysis reveals that CXCL2 is involved in anlotinib resistance in human lung cancer cells. BMC Med Genomics 2019;12:38. 10.1186/s12920-019-0482-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan MN, Wang B, Wei J, et al. CXCR1/2 antagonism with CXCL8/Interleukin-8 analogue CXCL8(3-72)K11R/G31P restricts lung cancer growth by inhibiting tumor cell proliferation and suppressing angiogenesis. Oncotarget 2015;6:21315-27. 10.18632/oncotarget.4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang Z, Jiang DP, Xia GG, et al. CXCL1 expression is correlated with Snail expression and affects the prognosis of patients with gastric cancer. Oncol Lett 2015;10:2458-64. 10.3892/ol.2015.3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song HW, Wang GH, Wang XD, et al. Expression and clinical significance of CXCL2 in colorectal cancer. Laboratory Medicine and Clinic 2016;13:1045-8. [Google Scholar]
- 14.Łukaszewicz-Zając M, Paczek S, Muszynski P, et al. Comparison between clinical significance of serum CXCL-8 and classical tumor markers in oesophageal cancer (OC) patients. Clin Exp Med 2019;19:191-9. 10.1007/s10238-019-00548-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei L, Liu Y, Ma Y, et al. C-X-C chemokine receptor 2 correlates with unfavorable prognosis and facilitates ma-lignant cell activities via activating JAK2/STAT3 pathway in non-small cell lung cancer. Cell Cycle 2019;18:3456-71. 10.1080/15384101.2019.1689471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian Y, Zhao K, Yuan L, et al. EIF3B correlates with advanced disease stages and poor prognosis, and it promotes proliferation and inhibits apoptosis in non-small cell lung cancer. Cancer Biomark 2018;23:291-300. 10.3233/CBM-181628 [DOI] [PubMed] [Google Scholar]
- 17.Yang C, Yu H, Chen R, et al. CXCL1 stimulates migration and invasion in ERnegative breast cancer cells via acti-vation of the ERK/MMP2/9 signaling axis. Int J Oncol 2019;55:684-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acharyya S, Oskarsson T, Vanharanta S, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 2012;150:165-78. 10.1016/j.cell.2012.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sparmann A, Bar-Sagi D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angio-genesis. Cancer Cell 2004;6:447-58. 10.1016/j.ccr.2004.09.028 [DOI] [PubMed] [Google Scholar]
- 20.Garkavtsev I, Kozin SV, Chernova O, et al. The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis. Nature 2004;428:328-32. 10.1038/nature02329 [DOI] [PubMed] [Google Scholar]
- 21.Lv M, Xu Y, Tang R, et al. miR141-CXCL1-CXCR2 signaling-induced Treg recruitment regulates metastases and survival of non-small cell lung cancer. Mol Cancer Ther 2014;13:3152-62. 10.1158/1535-7163.MCT-14-0448 [DOI] [PubMed] [Google Scholar]
- 22.Ruffini PA. The CXCL8-CXCR1/2 Axis as a Therapeutic Target in Breast Cancer Stem-Like Cells. Front Oncol 2019;9:40. 10.3389/fonc.2019.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen MC, Baskaran R, Lee NH, et al. CXCL2/CXCR2 axis induces cancer stem cell characteristics in CPT-11-resistant LoVo colon cancer cells via Galphai-2 and Galphaq/11. J Cell Physiol 2019;234:11822-34. 10.1002/jcp.27891 [DOI] [PubMed] [Google Scholar]
- 24.Kasashima H, Yashiro M, Nakamae H, et al. Clinicopathologic significance of the CXCL1-CXCR2 axis in the tumor microenvironment of gastric carcinoma. PLoS One 2017;12:e0178635. 10.1371/journal.pone.0178635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subat S, Mogushi K, Yasen M, et al. Identification of genes and pathways, including the CXCL2 axis, altered by DNA methylation in hepatocellular carcinoma. J Cancer Res Clin Oncol 2019;145:675-84. 10.1007/s00432-018-2824-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan R, Shuai H, Luo X, et al. The clinical and prognostic value of CXCL8 in cervical carcinoma patients: immunohistochemical analysis. Biosci Rep 2017;37:BSR20171021. 10.1042/BSR20171021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ha J, Choi HS, Lee Y, et al. CXC chemokine ligand 2 induced by receptor activator of NF-kappa B ligand enhances osteoclastogenesis. J Immunol 2010;184:4717-24. 10.4049/jimmunol.0902444 [DOI] [PubMed] [Google Scholar]
- 28.Liang J, Piao Y, Holmes L, et al. Neutrophils promote the malignant glioma phenotype through S100A4. Clin Cancer Res 2014;20:187-98. 10.1158/1078-0432.CCR-13-1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao YC, Yang ZB, Cheng XS, et al. CXCL8, overexpressed in colorectal cancer, enhances the resistance of colorectal cancer cells to anoikis. Cancer Lett 2015;361:22-32. 10.1016/j.canlet.2015.02.021 [DOI] [PubMed] [Google Scholar]
- 30.Sunaga N, Kaira K, Tomizawa Y, et al. Clinicopathological and prognostic significance of interleukin-8 expression and its relationship to KRAS mutation in lung adenocarcinoma. Br J Cancer 2014;110:2047-53. 10.1038/bjc.2014.110 [DOI] [PMC free article] [PubMed] [Google Scholar]