Abstract
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) transmissions are occurring rapidly; it is raising the alarm around the globe. Though vaccines are currently available, the evolution and mutations in the SARS-CoV-2 threaten available vaccines' significance. The drugs are still undergoing clinical trials, and certain medications are approved for “emergency use” or as an “off-label” drug during the pandemic. These drugs have been effective yet accommodating side effects, which also can be lethal. Complementary and alternative medicine is highly demanded since it embraces a holistic approach. Since ancient times, natural products have been used as drugs to treat various diseases in the medical field and are still widely practiced. Medicinal plants contain many active compounds that serve as the key to an effective drug design. The Kabasura kudineer and Nilavembu kudineer are the two most widely approved formulations to treat COVID-19. However, the mechanism of these formulations is not well known. The proposed study used a network pharmacology approach to understand the immune-boosting mechanism by the Kabasura kudineer, Nilavembu kudineer, and JACOM in treating COVID-19. The plants and phytochemical chemical compounds in the Kabasura kudineer, Nilavembu kudineer, and JACOM were obtained from the literature. The Swiss target prediction algorithm was used to predict the targets for these phytochemical compounds. The common genes for the COVID-19 infection and the drug targets were identified. The gene–gene interaction network was constructed to understand the interactions between these common genes and enrichment analyses to determine the biological process, molecular functions, cellular functions, pathways involved, etc. Finally, virtual screening and molecular docking studies were performed to identify the most potential targets and significant phytochemical compounds to treat the COVID-19. The present study identified potential targets as ACE, Cathepsin L, Cathepsin B, Cathepsin K, DPP4, EGFR, HDAC2, IL6, RIPK1, and VEGFA. Similarly, betulinic acid, 5″-(2⁗-Hydroxybenzyl) uvarinol, antofine, (S)-1′-methyloctyl caffeate, (Z)-3-phenyl-2-propenal, 7-oxo-10α-cucurbitadienol, and PLX-4720 collectively to be potential treatment agents for COVID-19.
Keywords: COVID-19, SARS-CoV-2, Network pharmacology, Nilavembu kudineer, Kabasura kudineer, JACOM, Traditional Indian Medicine
Abbreviations
- ACE
angiotensin-converting enzyme
- COVID-19
coronavirus disease 2019
- CTSB
Cathepsin B
- CTSL
Cathepsin L
- DPP4
dipeptidyl peptidase-4
- EGFR
epidermal growth factor receptor
- HDAC2
histone deacetylase 2
- IL-6
interleukin 6
- PABPC1
poly(A)-binding protein C1
- POLA
polymerase I
- RIPK1
receptor-interacting serine/threonine-protein kinase 1
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- SIGMAR1
Sigma non-opioid intracellular receptor 1
- TCM
Traditional Chinese Medicine
- TIM
Traditional Indian Medicine
- VEGFA
vascular endothelial growth factor A
- WHO
World Health Organization
1. Introduction
SARS-CoV-2 is a genetic variant of coronavirus known to cause respiratory illness and was reported as a global pandemic on March 11th, 2020 (Cucinotta & Vanelli, 2020). RT-PCR performed detects the infection and predicts the presence of Coronavirus. The virus has a close genetic resemblance with bat coronavirus, implying that it might have arisen from a bat-borne virus. This protocol has been published by World Health Organization (WHO) and used as a guideline for detecting the disease (Corman et al., 2020). Rapidly the viral infection begun to spread, increasing the number of cases in China and various parts of the world resulting from human-to-human transmission through close contact and primarily via respiratory droplets produced via coughs and sneezes (Dhand & Li, 2020). Currently, India stands behind the USA in the second position in the total number of people infected by COVID-19 (Kumar, Kumar, Christopher, & Doss, 2020; Kumar, Kumar, Siva, & Doss, 2020). As of September 6th, 2021, the reported cases are more than 220,563,227, and at least 4,565,483 people have died. The infection begins with the SARS-CoV-2 attaching to the enzyme called ACE2, which again is expressed in cells of various tissues and organs but is particularly abundant in type 2 lung pneumocytes. The people involved are frequently asymptomatic throughout the early stages of incubation, as well as the type I interferon response slows viral replication. With active replication and dissemination of SARS-CoV-2, the disease advances from mild to moderate symptoms such as sore throat, tear, fiber, and muscle pains, leading by both virus-associated respiratory tissue damage and antiviral activity (Cain & Cidlowski, 2020).
Several variants of concern (VOC) have appeared worldwide and seem to pose a significant threat to public health due to enhanced transmissibility or infectivity (Kumar, Shaikh, Kumar, Doss, & Zayed, 2021). Despite efficient vaccines, the coronavirus pandemic elicits widespread concern about discovering critical new variants from California, South Africa, UK, and Brazil (Plante et al., 2021). SARS-CoV-2 variants raise concerns about higher transmission and escaping on both vaccine and natural infection protection, especially in light of concerns about a specific mink strain, which triggered a person's illnesses and the possibility for future alterations. While most mutations are insignificant, the virus occasionally obtains a mutation that offers it an edge over other strains. The virus uses the spike protein to penetrate living cells through the ACE2 receptor. The spike protein's receptor-binding domain (RBD) is the most changeable component of the coronavirus genome (Tai et al., 2020). Mutations in viruses may lead to immune suppression in the host that the host antibodies initiate, necessitating their identification and monitoring for antibody therapy efficacy. The N439K, N440K, Q493K, and E484K spike mutations were prone to immunological escape, and this recent discovery is drawing attention (Srivastava, Banu, Singh, Sowpati, & Mishra, 2021). These mutations have given rise to several lineages. In the absence of direct drugs for the treatment, many efforts have been directed towards developing vaccines against COVID-19. Live attenuated or inactivated viruses, virus-like particles (VLP), protein subunits, viral vector (non-replicating and replicating), DNA, RNA, and nanoparticles, among additional vaccinations, are used in the conduit, each with its own set of advantages and disadvantages. According to the WHO report, these vaccines could only be 50% effective (What Is COVID-19 Vaccine Efficacy? | WHO | Regional Office for Africa, 2021a).
Complementary and alternative medicine is in high demand since it embraces a holistic approach. Since ancient times, natural products have been used as drugs in medical treatment and are still widely practiced. Medicinal plants contain many active compounds, which serve as the key to an effective drug design. Herbal medicine is achieving attention because of its extensive therapeutics like potent antiviral, immunomodulatory, anti-inflammatory, and anti-oxidant properties (Ross, 2009). Traditional Indian Medicine (TIM)—AYUSH and Traditional Chinese medicine (TCM) are ancient yet living traditions. These two traditional medicines are very philosophical and based on experiments, and they seem to be very effective in combatting viral diseases. Using these drugs from their natural origin is the main root of therapy (Patwardhan, Warude, Pushpangadan, & Bhatt, 2005). Pharmaceutical companies have re-established their policies in favor of natural product drug development and discovery. China has effectively endorsed its therapies over the world with a scientific approach. The emerging popularity of TCM can be verified by the rapid upsurge in licensed Chinese medicine providers in the United States. Constant efforts in advancing these therapies in China have set TCM creditable (Youns, Hoheisel, & Efferth, 2010). Worldwide, Siddha is recognized and emerging, and there has been an increase in demand for medicinal plants in India. The growing use of traditional therapies requires more scientific evidence for the principles behind treatments and the effectiveness of medicines. Latest developments in the biological sciences, genomics, and proteomics can justify these therapies (Rathinam et al., 2020).
Several medicines mentioned by Ministry AYUSH, India have been in practice for viral diseases like Chikungunya and Dengue for the past two decades. Herbal formulations such as Kabasura kudineer and Nilavembu kudineer are widely used to treat phlegmatic and hemorrhagic fevers and are approved by the Siddha medicine (Jain et al., 2020; Natarajan et al., 2020). Decoding using in silico studies in Nilavembu kudineer against SARS-CoV-2 spike protein showcases that these medicines can be recognized as a valuable drug to combat COVID-19. Nilavembu kudineer interacts with ACE2 receptor, which serves as the pathogen entrance, and in the outcome, the pathogen cannot enter into the host body. Kabasura kudineer or choornam possesses antiviral solid, anti-bacterial, and immunomodulatory properties. Numerous studies have unveiled the anti-inflammatory properties of Kabasura kudineer. This herbal formulation became reasonably recognized during times of flu due to its therapeutic qualities. In silico docking was performed using Kabasura kudineer against SARS-CoV-2 spike protein. Its rich active Phyto-constituents revealed a favorable outcome that prevented the merging of viral replication binding with viral proteins and inhibited host receptors' binding. This verifies that Kabasura kudineer could be a potential herbal formulation to combat COVID-19 if proven with further preclinical and clinical confirmatory studies (Natarajan et al., 2020).
In our study, medicinal plants used in herbal formulations of Nilavembu kudineer were our primary focus to understand their drug inhibitory potential against SARS-CoV-2. In comparison to modern medicine, Siddha medicine's approach is more holistic. Hence, investigating and ameliorating the effectiveness of Siddha medicine to obtain the solution with the most negligible side effects on immune-compromised patients and the patients with co-morbid conditions (Kiran et al., 2020).
2. Materials and methods
2.1. Genes responsible for COVID-19
The human genes responsible for the COVID-19 disease were collected from the GeneCards database (Rebhan, Chalifa-Caspi, Prilusky, & Lancet, 1997). This database summarizes the current accessible biomedical information, including the human genes, proteins, and relevant diseases. The term “COVID-19” was used as the search term.
2.2. Siddha formulation as the supplement
The standard Siddha formulations such as Kabasura kudineer, Nilavembu kudineer, and JACOM that showed promising treatment results were chosen for this study (Jain et al., 2020; Kiran et al., 2020; Natarajan et al., 2020). The plant sources for these formulations were taken from the previous literature. The list of phytochemical compounds in these plant sources was obtained from the Chemical Entities of Biological Interest (ChEBI) database. This database provides an ontology of molecular entities focused on ‘small’ chemical compounds (Hastings et al., 2013).
2.3. Druglikeness of phytochemical compounds
The drug-likeness of the phytochemical compounds was evaluated using the SwissADME server (Daina, Michielin, & Zoete, 2017). The canonical SMILES format of phytochemical compounds was given as the input in SwissADME. We have considered the number of Lipinski violations given for each compound. The compounds with one or less than one Lipinski violations were selected for further analysis.
2.4. Target prediction for the phytochemical compounds
The various human protein targets for each phytochemical compound were identified using the SwissTargetPrediction (Gfeller et al., 2014). This server predicts bioactive molecules (query molecule) targets based on a blend of 2D and 3D resemblance procedures with known ligands. The top-ranking targets obtained for each phytochemical compound, with a probability of more than 0 was chosen for further analysis. The canonical SMILES format of phytochemical compounds was given as the input in SwissTargetPrediction.
2.5. Common target identification
A Venn diagram was created to find the expected targets for the identified phytochemical compounds and COVID-19 (genes affected by COVID-19). This was achieved using an online tool named Bioinformatics & Evolutionary Genomics. Using the Venn diagram, we identified the targets of phytochemical compounds with the same target as COVID-19 infection. Thus, the typical targets were used for further studies.
2.6. Enrichment analysis
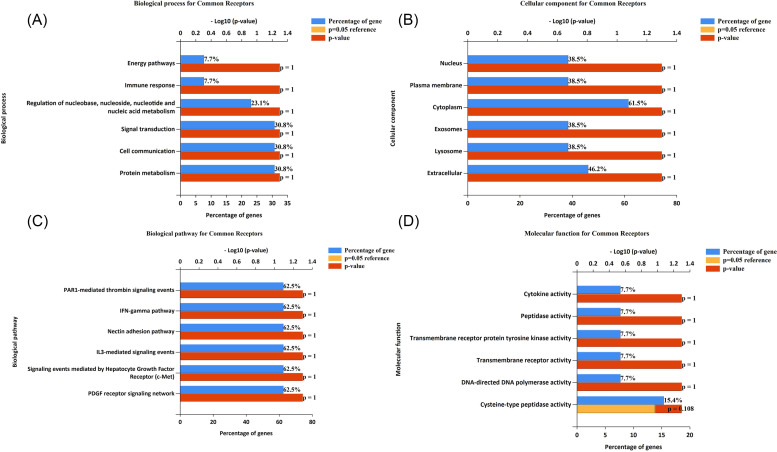
The enrichment analysis was executed using the FunRich (Pathan et al., 2015). FunRich is a standalone package primarily utilized for the enrichment and interaction network analysis of proteins and genes. Enrichment analysis can be performed for biological process (BP), cellular process (CC), biological pathways, and molecular function (MF).
2.7. Pathway analysis
The pathway analysis was performed using the Reactome. Reactome is a comprehensive and well-annotated library of human molecular pathways and reactions (Fabregat et al., 2016).
2.8. Gene–gene interaction analysis
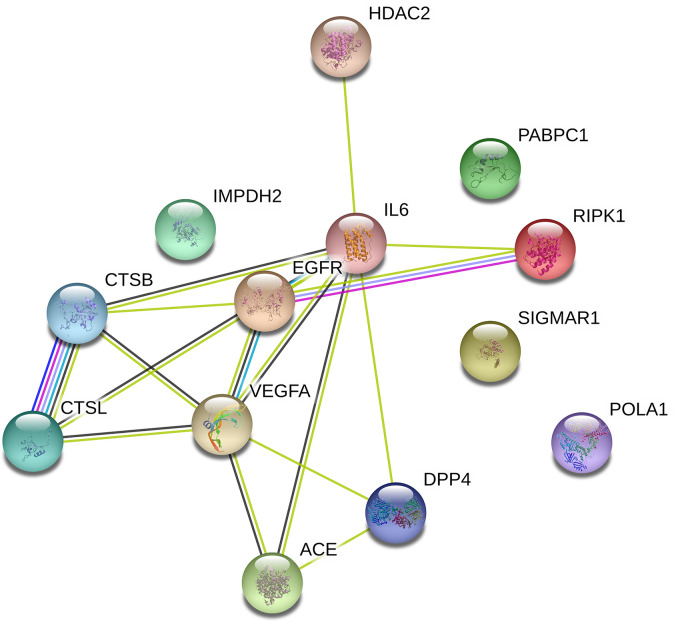
The standard targets obtained from the Venn diagram were subjected to a network analysis study. This was performed using the STRING database with default parameters (Szklarczyk et al., 2019). The STRING database aims to gather, score, and integrate all openly accessible protein–protein interaction information sources and complement these with computational predictions. Thus, this network analysis explains how these genes are biologically linked or overlapped through different pathways and functions.
2.9. Virtual screening and molecular interaction analysis
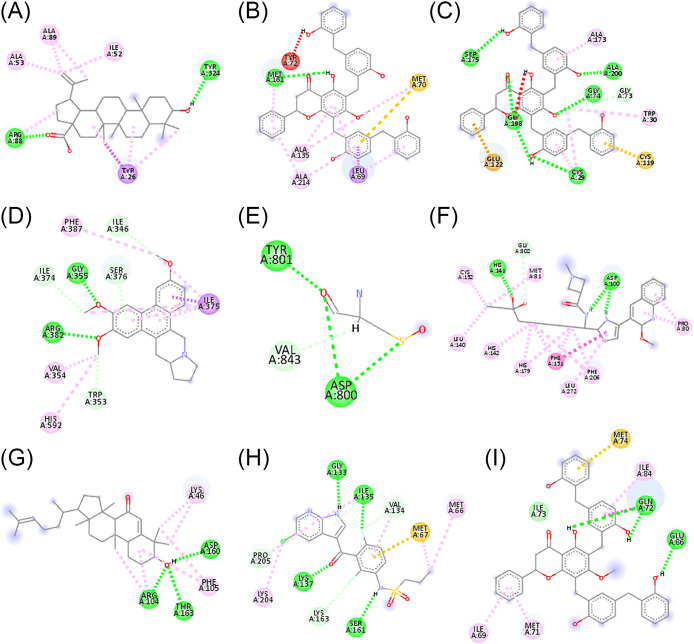
Several identified targets mapped in the gene–gene interaction have been targeted for more than one phytochemical compound. The virtual screening was performed using the AutoDock Vina plugin of the PyRx with the default parameter (Dallakyan & Olson, 2015; Trott & Olson, 2010). The protein structures with PDB IDs 6H5W, 2XU1, 6AY2, 2QT9, 4WKQ, 6WBW, 1ALU, 4ITJ, and 3QTK were taken for the proteins ACE, Cathepsin L, Cathepsin B, Cathepsin K, DPP4, EGFR, HDAC2, IL6, RIPK1, and VEGFA respectively. The compound showing the best affinity was taken for docking using the AutoDock standalone package (Morris et al., 2009). Blind docking protocol was performed using the Lamarckian Genetic Algorithm (Morris et al., 1998). The amino acid interaction with the compounds was visualized using the Discovery Studio.
3. Results and discussions
3.1. Genes responsible for COVID-19
Human genes that are involved in response to any disease are numerous and highly diverse within the genome. A total of 339 genes were obtained. A set of genes were retrieved from the GeneCards database with the keyword COVID. The detailed table with external identifiers is provided in Table 1 .
Table 1.
The list of genes responsible for the COVID-19 with the identifiers obtained from GeneCards database.
| Input term | Symbol | Ensembl | HGNC | NCBI Entrez gene | OMIM | UniProtKB/Swiss-Prot |
|---|---|---|---|---|---|---|
| AKAP8 | AKAP8 | ENSG00000105127 | 378 | 10270 | 604692 | O43823 |
| AKAP8L | AKAP8L | ENSG00000011243 | 29857 | 26993 | 609475 | Q9ULX6 |
| AKAP9 | AKAP9 | ENSG00000127914 | 379 | 10142 | 604001 | Q99996 |
| GGCX | GGCX | ENSG00000115486 | 4247 | 2677 | 137167 | P38435 |
| MDN1 | MDN1 | ENSG00000112159 | 18302 | 23195 | 618200 | Q9NU22 |
| MAT2B | MAT2B | ENSG00000038274 | 6905 | 27430 | 605527 | Q9NZL9 |
| GDF15 | GDF15 | ENSG00000130513 | 30142 | 9518 | 605312 | Q99988 |
| GCC1 | GCC1 | ENSG00000179562 | 19095 | 79571 | 607418 | Q96CN9 |
| GCC2 | GCC2 | ENSG00000135968 | 23218 | 9648 | 612711 | Q8IWJ2 |
| MIPOL1 | MIPOL1 | ENSG00000151338 | 21460 | 145282 | 606850 | Q8TD10 |
| MIB1 | MIB1 | ENSG00000101752 | 21086 | 57534 | 608677 | Q86YT6 |
| GOLGA2 | GOLGA2 | ENSG00000167110 | 4425 | 2801 | 602580 | Q08379 |
| GOLGA3 | GOLGA3 | ENSG00000090615 | 4426 | 2802 | 602581 | Q08378 |
| GNB1 | GNB1 | ENSG00000078369 | 4396 | 2782 | 139380 | P62873 |
| GNG5 | GNG5 | ENSG00000174021 | 4408 | 2787 | 600874 | P63218 |
| GOLGB1 | GOLGB1 | ENSG00000173230 | 4429 | 2804 | 602500 | Q14789 |
| GORASP1 | GORASP1 | ENSG00000114745 | 16769 | 64689 | 606867 | Q9BQQ3 |
| CD14 | CD14 | ENSG00000170458 | 1628 | 929 | 158120 | P08571 |
| CCDC86 | CCDC86 | ENSG00000110104 | 28359 | 79080 | 611293 | Q9H6F5 |
| MAP7D1 | MAP7D1 | ENSG00000116871 | 25514 | 55700 | NA | Q3KQU3 |
| FBXL12 | FBXL12 | ENSG00000127452 | 13611 | 54850 | 609079 | Q9NXK8 |
| ATE1 | ATE1 | ENSG00000107669 | 782 | 11101 | 607103 | O95260 |
| FAM162A | FAM162A | ENSG00000114023 | 17865 | 26355 | 608017 | Q96A26 |
| EXOSC2 | EXOSC2 | ENSG00000130713 | 17097 | 23404 | 602238 | Q13868 |
| EXOSC3 | EXOSC3 | ENSG00000107371 | 17944 | 51010 | 606489 | Q9NQT5 |
| EXOSC5 | EXOSC5 | ENSG00000077348 | 24662 | 56915 | 606492 | Q9NQT4 |
| EXOSC8 | EXOSC8 | ENSG00000120699 | 17035 | 11340 | 606019 | Q96B26 |
| EZH2 | EZH2 | ENSG00000106462 | 3527 | 2146 | 601573 | Q15910 |
| AP2A2 | AP2A2 | ENSG00000183020 | 562 | 161 | 607242 | O94973 |
| AP2M1 | AP2M1 | ENSG00000161203 | 564 | 1173 | 601024 | Q96CW1 |
| AP3B1 | AP3B1 | ENSG00000132842 | 566 | 8546 | 603401 | O00203 |
| ARL6IP6 | ARL6IP6 | ENSG00000177917 | 24048 | 151188 | 616495 | Q8N6S5 |
| ARF6 | ARF6 | ENSG00000165527 | 659 | 382 | 600464 | P62330 |
| ATP13A3 | ATP13A3 | ENSG00000133657 | 24113 | 79572 | 610232 | Q9H7F0 |
| ATP1B1 | ATP1B1 | ENSG00000143153 | 804 | 481 | 182330 | P05026 |
| ATP6AP1 | ATP6AP1 | ENSG00000071553 | 868 | 537 | 300197 | Q15904 |
| ATP6V1A | ATP6V1A | ENSG00000114573 | 851 | 523 | 607027 | P38606 |
| FBLN5 | FBLN5 | ENSG00000140092 | 3602 | 10516 | 604580 | Q9UBX5 |
| FBN1 | FBN1 | ENSG00000166147 | 3603 | 2200 | 134797 | P35555 |
| FBN2 | FBN2 | ENSG00000138829 | 3604 | 2201 | 612570 | P35556 |
| FAM8A1 | FAM8A1 | ENSG00000137414 | 16372 | 51439 | 618409 | Q9UBU6 |
| FAM98A | FAM98A | ENSG00000119812 | 24520 | 25940 | NA | Q8NCA5 |
| ERC1 | ERC1 | ENSG00000082805 | 17072 | 23085 | 607127 | Q8IUD2 |
| ALG11 | ALG11 | ENSG00000253710 | 32456 | 440138 | 613666 | Q2TAA5 |
| ALG5 | ALG5 | ENSG00000120697 | 20266 | 29880 | 604565 | Q9Y673 |
| AGTR2 | AGTR2 | ENSG00000180772 | 338 | 186 | 300034 | P50052 |
| ERGIC1 | ERGIC1 | ENSG00000113719 | 29205 | 57222 | 617946 | Q969X5 |
| LOC117134593 | LOC117134593 | NA | NA | 117134593 | NA | NA |
| LOC117134604 | LOC117134604 | NA | NA | 117134604 | NA | NA |
| LOC117134605 | LOC117134605 | NA | NA | 117134605 | NA | NA |
| LOC117134606 | LOC117134606 | NA | NA | 117134606 | NA | NA |
| LOC117134607 | LOC117134607 | NA | NA | 117134607 | NA | NA |
| LOC117134608 | LOC117134608 | NA | NA | 117134608 | NA | NA |
| LOC117134611 | LOC117134611 | NA | NA | 117134611 | NA | NA |
| LOC117135104 | LOC117135104 | NA | NA | 117135104 | NA | NA |
| LOC117135105 | LOC117135105 | NA | NA | 117135105 | NA | NA |
| LOC117135106 | LOC117135106 | NA | NA | 117135106 | NA | NA |
| ERLEC1 | ERLEC1 | ENSG00000068912 | 25222 | 27248 | 611229 | Q96DZ1 |
| LOC117152610 | LOC117152610 | NA | NA | 117152610 | NA | NA |
| LOC117152611 | LOC117152611 | NA | NA | 117152611 | NA | NA |
| LOC117204000 | LOC117204000 | NA | NA | 117204000 | NA | NA |
| LOC117204001 | LOC117204001 | NA | NA | 117204001 | NA | NA |
| LOC117600004 | LOC117600004 | NA | NA | 117600004 | NA | NA |
| LOC117693187 | LOC117693187 | NA | NA | 117693187 | NA | NA |
| ERO1B | ERO1B | ENSG00000086619 | 14355 | 56605 | 615437 | Q86YB8 |
| ERP44 | ERP44 | ENSG00000023318 | 18311 | 23071 | 609170 | Q9BS26 |
| AGPS | AGPS | ENSG00000018510 | 327 | 8540 | 603051 | O00116 |
| FKBP10 | FKBP10 | ENSG00000141756 | 18169 | 60681 | 607063 | Q96AY3 |
| FKBP15 | FKBP15 | ENSG00000119321 | 23397 | 23307 | 617398 | Q5T1M5 |
| MEPCE | MEPCE | ENSG00000146834 | 20247 | 56257 | 611478 | Q7L2J0 |
| FKBP7 | FKBP7 | ENSG00000079150 | 3723 | 51661 | 607062 | Q9Y680 |
| BCKDK | BCKDK | ENSG00000103507 | 16902 | 10295 | 614901 | O14874 |
| ACE | ACE | ENSG00000159640 | 2707 | 1636 | 106180 | P12821 |
| ACE2 | ACE2 | ENSG00000130234 | 13557 | 59272 | 300335 | Q9BYF1 |
| ADAM9 | ADAM9 | ENSG00000168615 | 216 | 8754 | 602713 | Q13443 |
| ADAMTS1 | ADAMTS1 | ENSG00000154734 | 217 | 9510 | 605174 | Q9UHI8 |
| AAR2 | AAR2 | ENSG00000131043 | 15886 | 25980 | 617365 | Q9Y312 |
| AASS | AASS | ENSG00000008311 | 17366 | 10157 | 605113 | Q9UDR5 |
| AATF | AATF | ENSG00000275700 | 19235 | 26574 | 608463 | Q9NY61 |
| ACSL3 | ACSL3 | ENSG00000123983 | 3570 | 2181 | 602371 | O95573 |
| ACADM | ACADM | ENSG00000117054 | 89 | 34 | 607008 | P11310 |
| BSG | BSG | ENSG00000172270 | 1116 | 682 | 109480 | P35613 |
| FYCO1 | FYCO1 | ENSG00000163820 | 14673 | 79443 | 607182 | Q9BQS8 |
| G3BP1 | G3BP1 | ENSG00000145907 | 30292 | 10146 | 608431 | Q13283 |
| G3BP2 | G3BP2 | ENSG00000138757 | 30291 | 9908 | NA | Q9UN86 |
| BRD2 | BRD2 | ENSG00000204256 | 1103 | 6046 | 601540 | P25440 |
| BRD4 | BRD4 | ENSG00000141867 | 13575 | 23476 | 608749 | O60885 |
| MFGE8 | MFGE8 | ENSG00000140545 | 7036 | 4240 | 602281 | Q08431 |
| BZW2 | BZW2 | ENSG00000136261 | 18808 | 28969 | NA | Q9Y6E2 |
| FOXRED2 | FOXRED2 | ENSG00000100350 | 26264 | 80020 | 613777 | Q8IWF2 |
| FURIN | FURIN | ENSG00000140564 | 8568 | 5045 | 136950 | P09958 |
| C1ORF50 | C1orf50 | ENSG00000164008 | 28795 | 79078 | NA | Q9BV19 |
| LOX | LOX | ENSG00000113083 | 6664 | 4015 | 153455 | P28300 |
| FASTKD5 | FASTKD5 | ENSG00000215251 | 25790 | 60493 | 614272 | Q7L8L6 |
| ATP5MG | ATP5MG | ENSG00000167283 | 14247 | 10632 | 617473 | O75964 |
| CWC27 | CWC27 | ENSG00000153015 | 10664 | 10283 | 617170 | Q6UX04 |
| CYB5B | CYB5B | ENSG00000103018 | 24374 | 80777 | 611964 | O43169 |
| CYB5R3 | CYB5R3 | ENSG00000100243 | 2873 | 1727 | 613213 | P00387 |
| HS2ST1 | HS2ST1 | ENSG00000153936 | 5193 | 9653 | 604844 | Q7LGA3 |
| HS6ST2 | HS6ST2 | ENSG00000171004 | 19133 | 90161 | 300545 | Q96MM7 |
| CTSB | CTSB | ENSG00000164733 | 2527 | 1508 | 116810 | P07858 |
| CTSL | CTSL | ENSG00000135047 | 2537 | 1514 | 116880 | P07711 |
| CRP | CRP | ENSG00000132693 | 2367 | 1401 | 123260 | P02741 |
| HOOK1 | HOOK1 | ENSG00000134709 | 19884 | 51361 | 607820 | Q9UJC3 |
| HEATR3 | HEATR3 | ENSG00000155393 | 26087 | 55027 | 614951 | Q7Z4Q2 |
| HECTD1 | HECTD1 | ENSG00000092148 | 20157 | 25831 | 618649 | Q9ULT8 |
| GLA | GLA | ENSG00000102393 | 4296 | 2717 | 300644 | P06280 |
| GIGYF2 | GIGYF2 | ENSG00000204120 | 11960 | 26058 | 612003 | Q6Y7W6 |
| GFER | GFER | ENSG00000127554 | 4236 | 2671 | 600924 | P55789 |
| ETFA | ETFA | ENSG00000140374 | 3481 | 2108 | 608053 | P13804 |
| ANO6 | ANO6 | ENSG00000177119 | 25240 | 196527 | 608663 | Q4KMQ2 |
| GRPEL1 | GRPEL1 | ENSG00000109519 | 19696 | 80273 | 606173 | Q9HAV7 |
| CDK5RAP2 | CDK5RAP2 | ENSG00000136861 | 18672 | 55755 | 608201 | Q96SN8 |
| GTF2F2 | GTF2F2 | ENSG00000188342 | 4653 | 2963 | 189969 | P13984 |
| CEP112 | CEP112 | ENSG00000154240 | 28514 | 201134 | NA | Q8N8E3 |
| CEP135 | CEP135 | ENSG00000174799 | 29086 | 9662 | 611423 | Q66GS9 |
| CEP250 | CEP250 | ENSG00000126001 | 1859 | 11190 | 609689 | Q9BV73 |
| CEP350 | CEP350 | ENSG00000135837 | 24238 | 9857 | 617870 | Q5VT06 |
| CEP43 | CEP43 | ENSG00000213066 | 17012 | 11116 | 605392 | O95684 |
| CEP68 | CEP68 | ENSG00000011523 | 29076 | 23177 | 616889 | Q76N32 |
| CENPF | CENPF | ENSG00000117724 | 1857 | 1063 | 600236 | P49454 |
| GOLGA7 | GOLGA7 | ENSG00000147533 | 24876 | 51125 | 609453 | Q7Z5G4 |
| HLA-A | HLA-A | ENSG00000206503 | 4931 | 3105 | 142800 | P04439 |
| RHOA | RHOA | ENSG00000067560 | 667 | 387 | 165390 | P61586 |
| HMOX1 | HMOX1 | ENSG00000100292 | 5013 | 3162 | 141250 | P09601 |
| COQ8B | COQ8B | ENSG00000123815 | 19041 | 79934 | 615567 | Q96D53 |
| CLIP4 | CLIP4 | ENSG00000115295 | 26108 | 79745 | NA | Q8N3C7 |
| HLA-DRB1 | HLA-DRB1 | ENSG00000196126 | 4948 | 3123 | 142857 | P01911 |
| CISD3 | CISD3 | ENSG00000277972 | 27578 | 284106 | 611933 | P0C7P0 |
| CIT | CIT | ENSG00000122966 | 1985 | 11113 | 605629 | O14578 |
| CLCC1 | CLCC1 | ENSG00000121940 | 29675 | 23155 | 617539 | Q96S66 |
| RIPK1 | RIPK1 | ENSG00000137275 | 10019 | 8737 | 603453 | Q13546 |
| COLGALT1 | COLGALT1 | ENSG00000130309 | 26182 | 79709 | 617531 | Q8NBJ5 |
| COMT | COMT | ENSG00000093010 | 2228 | 1312 | 116790 | P21964 |
| HDAC2 | HDAC2 | ENSG00000196591 | 4853 | 3066 | 605164 | Q92769 |
| CHPF | CHPF | ENSG00000123989 | 24291 | 79586 | 610405 | Q8IZ52 |
| CHPF2 | CHPF2 | ENSG00000033100 | 29270 | 54480 | 608037 | Q9P2E5 |
| DNAJC11 | DNAJC11 | ENSG00000007923 | 25570 | 55735 | 614827 | Q9NVH1 |
| DNAJC19 | DNAJC19 | ENSG00000205981 | 30528 | 131118 | 608977 | Q96DA6 |
| DNMT1 | DNMT1 | ENSG00000130816 | 2976 | 1786 | 126375 | P26358 |
| MRPS2 | MRPS2 | ENSG00000122140 | 14495 | 51116 | 611971 | Q9Y399 |
| MRPS25 | MRPS25 | ENSG00000131368 | 14511 | 64432 | 611987 | P82663 |
| MRPS27 | MRPS27 | ENSG00000113048 | 14512 | 23107 | 611989 | Q92552 |
| MRPS5 | MRPS5 | ENSG00000144029 | 14498 | 64969 | 611972 | P82675 |
| DPP4 | DPP4 | ENSG00000197635 | 3009 | 1803 | 102720 | P27487 |
| MTCH1 | MTCH1 | ENSG00000137409 | 17586 | 23787 | 610449 | Q9NZJ7 |
| MTARC1 | MTARC1 | ENSG00000186205 | 26189 | 64757 | 614126 | Q5VT66 |
| EDEM3 | EDEM3 | ENSG00000116406 | 16787 | 80267 | 610214 | Q9BZQ6 |
| CD8A | CD8A | ENSG00000153563 | 1706 | 925 | 186910 | P01732 |
| GPT | GPT | ENSG00000167701 | 4552 | 2875 | 138200 | P24298 |
| GPX1 | GPX1 | ENSG00000233276 | 4553 | 2876 | 138320 | P07203 |
| CD209 | CD209 | ENSG00000090659 | 1641 | 30835 | 604672 | Q9NNX6 |
| GRIPAP1 | GRIPAP1 | ENSG00000068400 | 18706 | 56850 | 300408 | Q4V328 |
| GGH | GGH | ENSG00000137563 | 4248 | 8836 | 601509 | Q92820 |
| MOV10 | MOV10 | ENSG00000155363 | 7200 | 4343 | 610742 | Q9HCE1 |
| MPHOSPH10 | MPHOSPH10 | ENSG00000124383 | 7213 | 10199 | 605503 | O00566 |
| DDX10 | DDX10 | ENSG00000178105 | 2735 | 1662 | 601235 | Q13206 |
| DDX21 | DDX21 | ENSG00000165732 | 2744 | 9188 | 606357 | Q9NR30 |
| RNF41 | RNF41 | ENSG00000181852 | 18,401 | 10193 | NA | Q9H4P4 |
| COL6A1 | COL6A1 | ENSG00000142156 | 2211 | 1291 | 120220 | P12109 |
| DCAF7 | DCAF7 | ENSG00000136485 | 30915 | 10238 | 605973 | P61962 |
| DCAKD | DCAKD | ENSG00000172992 | 26238 | 79877 | NA | Q8WVC6 |
| NDUFAF2 | NDUFAF2 | ENSG00000164182 | 28086 | 91942 | 609653 | Q8N183 |
| EIF4E2 | EIF4E2 | ENSG00000135930 | 3293 | 9470 | 605895 | O60573 |
| EIF4H | EIF4H | ENSG00000106682 | 12741 | 7458 | 603431 | Q15056 |
| ELOB | ELOB | ENSG00000103363 | 11619 | 6923 | 600787 | Q15370 |
| ELOC | ELOC | ENSG00000154582 | 11617 | 6921 | 600788 | Q15369 |
| EMC1 | EMC1 | ENSG00000127463 | 28957 | 23065 | 616846 | Q8N766 |
| NEU1 | NEU1 | ENSG00000204386 | 7758 | 4758 | 608272 | Q99519 |
| NEK9 | NEK9 | ENSG00000119638 | 18591 | 91754 | 609798 | Q8TD19 |
| CSNK2A2 | CSNK2A2 | ENSG00000070770 | 2459 | 1459 | 115442 | P19784 |
| CSNK2B | CSNK2B | ENSG00000204435 | 2460 | 1460 | 115441 | P67870 |
| CRTC3 | CRTC3 | ENSG00000140577 | 26148 | 64784 | 608986 | Q6UUV7 |
| PCSK6 | PCSK6 | ENSG00000140479 | 8569 | 5046 | 167405 | P29122 |
| IL17A | IL17A | ENSG00000112115 | 5981 | 3605 | 603149 | Q16552 |
| IL17RA | IL17RA | ENSG00000177663 | 5985 | 23765 | 605461 | Q96F46 |
| IL10 | IL10 | ENSG00000136634 | 5962 | 3586 | 124092 | P22301 |
| IL2RA | IL2RA | ENSG00000134460 | 6008 | 3559 | 147730 | P01589 |
| SPART | SPART | ENSG00000133104 | 18514 | 23111 | 607111 | Q8N0X7 |
| PDZD11 | PDZD11 | ENSG00000120509 | 28034 | 51248 | 300632 | Q5EBL8 |
| PRRC2B | PRRC2B | ENSG00000130723 | 28121 | 84726 | NA | Q5JSZ5 |
| NPTX1 | NPTX1 | ENSG00000171246 | 7952 | 4884 | 602367 | Q15818 |
| SELENOS | SELENOS | ENSG00000131871 | 30396 | 55829 | 607918 | Q9BQE4 |
| NPC2 | NPC2 | ENSG00000119655 | 14537 | 10577 | 601015 | P61916 |
| PABPC1 | PABPC1 | ENSG00000070756 | 8554 | 26986 | 604679 | P11940 |
| PABPC4 | PABPC4 | ENSG00000090621 | 8557 | 8761 | 603407 | Q13310 |
| IDE | IDE | ENSG00000119912 | 5381 | 3416 | 146680 | P14735 |
| PRKACA | PRKACA | ENSG00000072062 | 9380 | 5566 | 601639 | P17612 |
| PCNT | PCNT | ENSG00000160299 | 16068 | 5116 | 605925 | O95613 |
| PRKAR2A | PRKAR2A | ENSG00000114302 | 9391 | 5576 | 176910 | P13861 |
| PDE4DIP | PDE4DIP | ENSG00000178104 | 15580 | 9659 | 608117 | Q5VU43 |
| SAAL1 | SAAL1 | ENSG00000166788 | 25158 | 113174 | NA | Q96ER3 |
| SCARB1 | SCARB1 | ENSG00000073060 | 1664 | 949 | 601040 | Q8WTV0 |
| SCCPDH | SCCPDH | ENSG00000143653 | 24275 | 51097 | NA | Q8NBX0 |
| SDF2 | SDF2 | ENSG00000132581 | 10675 | 6388 | 602934 | Q99470 |
| RRP9 | RRP9 | ENSG00000114767 | 16829 | 9136 | NA | O43818 |
| SBNO1 | SBNO1 | ENSG00000139697 | 22973 | 55206 | 614274 | A3KN83 |
| RTN4 | RTN4 | ENSG00000115310 | 14085 | 57142 | 604475 | Q9NQC3 |
| EGFR | EGFR | ENSG00000146648 | 3236 | 1956 | 131550 | P00533 |
| RPL36 | RPL36 | ENSG00000130255 | 13631 | 25873 | 617893 | Q9Y3U8 |
| ENPEP | ENPEP | ENSG00000138792 | 3355 | 2028 | 138297 | Q07075 |
| NGDN | NGDN | ENSG00000129460 | 20271 | 25983 | 610777 | Q8NEJ9 |
| NGLY1 | NGLY1 | ENSG00000151092 | 17646 | 55768 | 610661 | Q96IV0 |
| NIN | NIN | ENSG00000100503 | 14906 | 51199 | 608684 | Q8N4C6 |
| NINL | NINL | ENSG00000101004 | 29163 | 22981 | 609580 | Q9Y2I6 |
| SLC44A2 | SLC44A2 | ENSG00000129353 | 17292 | 57153 | 606106 | Q8IWA5 |
| NUP210 | NUP210 | ENSG00000132182 | 30052 | 23225 | 607703 | Q8TEM1 |
| NUP214 | NUP214 | ENSG00000126883 | 8064 | 8021 | 114350 | P35658 |
| SLC6A19 | SLC6A19 | ENSG00000174358 | 27960 | 340024 | 608893 | Q695T7 |
| SMOC1 | SMOC1 | ENSG00000198732 | 20318 | 64093 | 608488 | Q9H4F8 |
| PKP2 | PKP2 | ENSG00000057294 | 9024 | 5318 | 602861 | Q99959 |
| SLU7 | SLU7 | ENSG00000164609 | 16939 | 10569 | 605974 | O95391 |
| PITRM1 | PITRM1 | ENSG00000107959 | 17663 | 10531 | 618211 | Q5JRX3 |
| HSBP1 | HSBP1 | ENSG00000230989 | 5203 | 3281 | 604553 | O75506 |
| OS9 | OS9 | ENSG00000135506 | 16994 | 10956 | 609677 | Q13438 |
| POFUT1 | POFUT1 | ENSG00000101346 | 14988 | 23509 | 607491 | Q9H488 |
| POGLUT2 | POGLUT2 | ENSG00000134901 | 19350 | 79070 | 611613 | Q6UW63 |
| POGLUT3 | POGLUT3 | ENSG00000178202 | 28496 | 143888 | 618503 | Q7Z4H8 |
| POLA1 | POLA1 | ENSG00000101868 | 9173 | 5422 | 312040 | P09884 |
| POLA2 | POLA2 | ENSG00000014138 | 30073 | 23649 | NA | Q14181 |
| PLOD2 | PLOD2 | ENSG00000152952 | 9082 | 5352 | 601865 | O00469 |
| SNIP1 | SNIP1 | ENSG00000163877 | 30587 | 79753 | 608241 | Q8TAD8 |
| PLD3 | PLD3 | ENSG00000105223 | 17158 | 23646 | 615698 | Q8IV08 |
| PLEKHA5 | PLEKHA5 | ENSG00000052126 | 30036 | 54477 | 607770 | Q9HAU0 |
| PLEKHF2 | PLEKHF2 | ENSG00000175895 | 20757 | 79666 | 615208 | Q9H8W4 |
| SIRT5 | SIRT5 | ENSG00000124523 | 14933 | 23408 | 604483 | Q9NXA8 |
| SLC27A2 | SLC27A2 | ENSG00000140284 | 10996 | 11001 | 603247 | O14975 |
| SLC30A7 | SLC30A7 | ENSG00000162695 | 19306 | 148867 | 611149 | Q8NEW0 |
| SLC30A9 | SLC30A9 | ENSG00000014824 | 1329 | 10463 | 604604 | Q6PML9 |
| NUP54 | NUP54 | ENSG00000138750 | 17359 | 53371 | 607607 | Q7Z3B4 |
| NUP58 | NUP58 | ENSG00000139496 | 20261 | 9818 | 607615 | Q9BVL2 |
| NUP62 | NUP62 | ENSG00000213024 | 8066 | 23636 | 605815 | P37198 |
| NUP88 | NUP88 | ENSG00000108559 | 8067 | 4927 | 602552 | Q99567 |
| NUP98 | NUP98 | ENSG00000110713 | 8068 | 4928 | 601021 | P52948 |
| NUTF2 | NUTF2 | ENSG00000102898 | 13722 | 10204 | 605813 | P61970 |
| SLC25A21 | SLC25A21 | ENSG00000183032 | 14411 | 89874 | 607571 | Q9BQT8 |
| NSD2 | NSD2 | ENSG00000109685 | 12766 | 7468 | 602952 | O96028 |
| SEPSECS | SEPSECS | ENSG00000109618 | 30605 | 51091 | 613009 | Q9HD40 |
| SIGMAR1 | SIGMAR1 | ENSG00000147955 | 8157 | 10280 | 601978 | Q99720 |
| SIL1 | SIL1 | ENSG00000120725 | 24624 | 64374 | 608005 | Q9H173 |
| SRP19 | SRP19 | ENSG00000153037 | 11300 | 6728 | 182175 | P09132 |
| SRP54 | SRP54 | ENSG00000100883 | 11301 | 6729 | 604857 | P61011 |
| INS | INS | ENSG00000254647 | 6081 | 3630 | 176730 | P01308 |
| SRP72 | SRP72 | ENSG00000174780 | 11303 | 6731 | 602122 | O76094 |
| IMPDH2 | IMPDH2 | ENSG00000178035 | 6053 | 3615 | 146691 | P12268 |
| PTGES2 | PTGES2 | ENSG00000148334 | 17822 | 80142 | 608152 | Q9H7Z7 |
| INHBE | INHBE | ENSG00000139269 | 24029 | 83729 | 612031 | P58166 |
| PSMD8 | PSMD8 | ENSG00000099341 | 9566 | 5714 | 617844 | P48556 |
| INTS4 | INTS4 | ENSG00000149262 | 25048 | 92105 | 611348 | Q96HW7 |
| TOR1A | TOR1A | ENSG00000136827 | 3098 | 1861 | 605204 | O14656 |
| TOR1AIP1 | TOR1AIP1 | ENSG00000143337 | 29456 | 26092 | 614512 | Q5JTV8 |
| LARP1 | LARP1 | ENSG00000155506 | 29531 | 23367 | 612059 | Q6PKG0 |
| LARP4B | LARP4B | ENSG00000107929 | 28987 | 23185 | 616513 | Q92615 |
| LARP7 | LARP7 | ENSG00000174720 | 24912 | 51574 | 612026 | Q4G0J3 |
| TUBGCP2 | TUBGCP2 | ENSG00000130640 | 18599 | 10844 | 617817 | Q9BSJ2 |
| TUBGCP3 | TUBGCP3 | ENSG00000126216 | 18598 | 10426 | 617818 | Q96CW5 |
| UGGT2 | UGGT2 | ENSG00000102595 | 15664 | 55757 | 605898 | Q9NYU1 |
| TRIM59 | TRIM59 | ENSG00000213186 | 30834 | 286827 | 616148 | Q8IWR1 |
| UBAP2 | UBAP2 | ENSG00000137073 | 14185 | 55833 | NA | Q5T6F2 |
| TYSND1 | TYSND1 | ENSG00000156521 | 28531 | 219743 | 611017 | Q2T9J0 |
| UBAP2L | UBAP2L | ENSG00000143569 | 29877 | 9898 | 616472 | Q14157 |
| UPF1 | UPF1 | ENSG00000005007 | 9962 | 5976 | 601430 | Q92900 |
| ITGB1 | ITGB1 | ENSG00000150093 | 6153 | 3688 | 135630 | P05556 |
| PUSL1 | PUSL1 | ENSG00000169972 | 26914 | 126789 | NA | Q8N0Z8 |
| PVR | PVR | ENSG00000073008 | 9705 | 5817 | 173850 | P15151 |
| RAB2A | RAB2A | ENSG00000104388 | 9763 | 5862 | 179509 | P61019 |
| RAP1GDS1 | RAP1GDS1 | ENSG00000138698 | 9859 | 5910 | 179502 | P52306 |
| KNG1 | KNG1 | ENSG00000113889 | 6383 | 3827 | 612358 | P01042 |
| TCF12 | TCF12 | ENSG00000140262 | 11623 | 6938 | 600480 | Q99081 |
| TARS2 | TARS2 | ENSG00000143374 | 30740 | 80222 | 612805 | Q9BW92 |
| REEP5 | REEP5 | ENSG00000129625 | 30077 | 7905 | 125265 | Q00765 |
| REEP6 | REEP6 | ENSG00000115255 | 30078 | 92840 | 609346 | Q96HR9 |
| TBK1 | TBK1 | ENSG00000183735 | 11584 | 29110 | 604834 | Q9UHD2 |
| TBKBP1 | TBKBP1 | ENSG00000198933 | 30140 | 9755 | 608476 | A7MCY6 |
| RBX1 | RBX1 | ENSG00000100387 | 9928 | 9978 | 603814 | P62877 |
| RBM41 | RBM41 | ENSG00000089682 | 25617 | 55285 | NA | Q96IZ5 |
| RDX | RDX | ENSG00000137710 | 9944 | 5962 | 179410 | P35241 |
| RAB10 | RAB10 | ENSG00000084733 | 9759 | 10890 | 612672 | P61026 |
| JAKMIP1 | JAKMIP1 | ENSG00000152969 | 26460 | 152789 | 611195 | Q96N16 |
| STOM | STOM | ENSG00000148175 | 3383 | 2040 | 133090 | P27105 |
| RAB14 | RAB14 | ENSG00000119396 | 16524 | 51552 | 612673 | P61106 |
| RAB18 | RAB18 | ENSG00000099246 | 14244 | 22931 | 602207 | Q9NP72 |
| RAB1A | RAB1A | ENSG00000138069 | 9758 | 5861 | 179508 | P62820 |
| STC2 | STC2 | ENSG00000113739 | 11374 | 8614 | 603665 | O76061 |
| RALA | RALA | ENSG00000006451 | 9839 | 5898 | 179550 | P11233 |
| RAB5C | RAB5C | ENSG00000108774 | 9785 | 5878 | 604037 | P51148 |
| RAB7A | RAB7A | ENSG00000075785 | 9788 | 7879 | 602298 | P51149 |
| RAB8A | RAB8A | ENSG00000167461 | 7007 | 4218 | 165040 | P61006 |
| QSOX2 | QSOX2 | ENSG00000165661 | 30249 | 169714 | 612860 | Q6ZRP7 |
| RAE1 | RAE1 | ENSG00000101146 | 9828 | 8480 | 603343 | P78406 |
| SUN2 | SUN2 | ENSG00000100242 | 14210 | 25777 | 613569 | Q9UH99 |
| TLE1 | TLE1 | ENSG00000196781 | 11837 | 7088 | 600189 | Q04724 |
| TLE3 | TLE3 | ENSG00000140332 | 11839 | 7090 | 600190 | Q04726 |
| TLE5 | TLE5 | ENSG00000104964 | 307 | 166 | 600188 | Q08117 |
| TM2D3 | TM2D3 | ENSG00000184277 | 24128 | 80213 | 610014 | Q9BRN9 |
| TMPRSS2 | TMPRSS2 | ENSG00000184012 | 11876 | 7113 | 602060 | O15393 |
| TMPRSS4 | TMPRSS4 | ENSG00000137648 | 11878 | 56649 | 606565 | Q9NRS4 |
| IL6R | IL6R | ENSG00000160712 | 6019 | 3570 | 147880 | P08887 |
| PRKAR2B | PRKAR2B | ENSG00000005249 | 9392 | 5577 | 176912 | P31323 |
| PRIM1 | PRIM1 | ENSG00000198056 | 9369 | 5557 | 176635 | P49642 |
| PRIM2 | PRIM2 | ENSG00000146143 | 9370 | 5558 | 176636 | P49643 |
| PPT1 | PPT1 | ENSG00000131238 | 9325 | 5538 | 600722 | P50897 |
| PPIL3 | PPIL3 | ENSG00000240344 | 9262 | 53938 | 615811 | Q9H2H8 |
| TRMT1 | TRMT1 | ENSG00000104907 | 25980 | 55621 | 611669 | Q9NXH9 |
| MYCBP2 | MYCBP2 | ENSG00000005810 | 23386 | 23077 | 610392 | O75592 |
| NARS2 | NARS2 | ENSG00000137513 | 26274 | 79731 | 612803 | Q96I59 |
| CNTRL | CNTRL | ENSG00000119397 | 1858 | 11064 | 605496 | Q7Z7A1 |
| NAT14 | NAT14 | ENSG00000090971 | 28918 | 57106 | NA | Q8WUY8 |
| NOL10 | NOL10 | ENSG00000115761 | 25862 | 79954 | 616197 | Q9BSC4 |
| CUL2 | CUL2 | ENSG00000108094 | 2552 | 8453 | 603135 | Q13617 |
| MOGS | MOGS | ENSG00000115275 | 24862 | 7841 | 601336 | Q13724 |
| PLAT | PLAT | ENSG00000104368 | 9051 | 5327 | 173370 | P00750 |
| PLAUR | PLAUR | ENSG00000011422 | 9053 | 5329 | 173391 | Q03405 |
| POR | POR | ENSG00000127948 | 9208 | 5447 | 124015 | P16435 |
| HYOU1 | HYOU1 | ENSG00000149428 | 16931 | 10525 | 601746 | Q9Y4L1 |
| PMPCA | PMPCA | ENSG00000165688 | 18667 | 23203 | 613036 | Q10713 |
| PMPCB | PMPCB | ENSG00000105819 | 9119 | 9512 | 603131 | O75439 |
| IL6 | IL6 | ENSG00000136244 | 6018 | 3569 | 147620 | P05231 |
| RBM28 | RBM28 | ENSG00000106344 | 21863 | 55131 | 612074 | Q9NW13 |
| THTPA | THTPA | ENSG00000259431 | 18987 | 79178 | 611612 | Q9BU02 |
| TIMM10 | TIMM10 | ENSG00000134809 | 11814 | 26519 | 602251 | P62072 |
| TIMM10B | TIMM10B | ENSG00000132286 | 4022 | 26515 | 607388 | Q9Y5J6 |
| TIMM29 | TIMM29 | ENSG00000142444 | 25152 | 90580 | 617380 | Q9BSF4 |
| TIMM8B | TIMM8B | ENSG00000150779 | 11818 | 26521 | 606659 | Q9Y5J9 |
| TIMM9 | TIMM9 | ENSG00000100575 | 11819 | 26520 | 607384 | Q9Y5J7 |
| USP54 | USP54 | ENSG00000166348 | 23513 | 159195 | NA | Q70EL1 |
| ZC3H7A | ZC3H7A | ENSG00000122299 | 30959 | 29066 | NA | Q8IWR0 |
| ZDHHC5 | ZDHHC5 | ENSG00000156599 | 18472 | 25921 | 614586 | Q9C0B5 |
| ZC3H18 | ZC3H18 | ENSG00000158545 | 25091 | 124245 | NA | Q86VM9 |
| ZNF503 | ZNF503 | ENSG00000165655 | 23589 | 84858 | 613902 | Q96F45 |
| ZNF318 | ZNF318 | ENSG00000171467 | 13578 | 24149 | 617512 | Q5VUA4 |
| ZYG11B | ZYG11B | ENSG00000162378 | 25820 | 79699 | 618673 | Q9C0D3 |
| VPS11 | VPS11 | ENSG00000160695 | 14583 | 55823 | 608549 | Q9H270 |
| VPS39 | VPS39 | ENSG00000166887 | 20593 | 23339 | 612188 | Q96JC1 |
| WASHC4 | WASHC4 | ENSG00000136051 | 29174 | 23325 | 615748 | Q2M389 |
| USP13 | USP13 | ENSG00000058056 | 12611 | 8975 | 603591 | Q92995 |
| VEGFA | VEGFA | ENSG00000112715 | 12680 | 7422 | 192240 | P15692 |
| YIF1A | YIF1A | ENSG00000174851 | 16688 | 10897 | 611484 | O95070 |
| LMAN2 | LMAN2 | ENSG00000169223 | 16986 | 10960 | 609551 | Q12907 |
3.2. Siddha formulation as the supplement
The list of plants involved in the formulation of Kabasura kudineer, Nilavembu kudineer, and JACOM were identified from the literature source. A total of 25 plants were identified in the formulation (Table 2 ). Further, a list of phytochemical compounds from the identified 25 plants was obtained from the ChEBI database. Three hundred and fourteen phytochemical compounds were identified in these 25 plants overall (Table 3 ).
Table 2.
The list of plant involved in the formulation of Nilavembu kudineer, Kabasura kudineer, and JACOM.
| S.·no | Plants name |
|---|---|
| 1 | Zingiber officinale |
| 2 | Piper longum |
| 3 | Syzygium aromaticum |
| 4 | Tragia involucrata |
| 5 | Anacyclus pyrethrum |
| 6 | Andrographis paniculata |
| 7 | Hygrophila auriculata |
| 8 | Terminalia chebula |
| 9 | Justicia adhatoda |
| 10 | Plectranthus amboinicus |
| 11 | Saussurea lappa |
| 12 | Tinospora cordifolia |
| 13 | Rotheca serrata |
| 14 | Cypreus rotundus |
| 15 | Sida acuta Burm.f.L |
| 16 | Adeloda serrata raf |
| 17 | Carica Papaya |
| 18 | A. paniculata |
| 19 | Ocimum tenuiflorum |
| 20 | Vetiveria zizanioides |
| 21 | Santalum album |
| 22 | Piper nigrum |
| 23 | Hedyotis corymbosa |
| 24 | Plectranthus vettiveroides |
| 25 | Trichosanthes cucumerina |
Table 3.
The list of phytochemical compounds found in the plant involved in the formulation of Nilavembu kudineer, Kabasura kudineer, and JACOM with ChEBI ID.
| S.·no | Name of the plant | ChEBI ID | Name of the compound |
|---|---|---|---|
| 1 | Zingiber officinale | CHEBI:142262 | Gingerenone B |
| 2 | CHEBI:64361 | Beta-sesquiphellandrene | |
| 3 | CHEBI:69294 | 3-(3,4-Dimethoxyphenyl)-4-[(Z)-3,4-dimethoxystyryl]cyclohex-1-ene | |
| 4 | CHEBI:69295 | 3-(3,4-Dimethoxyphenyl)-4-[(E)-3,4-dimethoxystyryl]cyclohex-1-ene | |
| 5 | CHEBI:26137 | Pinocarveol | |
| 6 | CHEBI:66502 | Zerumboneoxide | |
| 7 | CHEBI:68065 | Ramonanin A, (rel)- | |
| 8 | CHEBI:68066 | Ramonanin B, (rel)- | |
| 9 | CHEBI:68067 | Ramonanin C, (rel)- | |
| 10 | CHEBI:68068 | Ramonanin D, (rel)- | |
| 11 | CHEBI:63892 | Zerumbone | |
| 12 | CHEBI:66050 | 5-Hydroxyzerumbone | |
| 13 | CHEBI:138043 | (2E,6E)-hedycaryol | |
| 14 | CHEBI:28817 | Dodecane | |
| 15 | CHEBI:146145 | 7,4′-Dimethylkaempferol | |
| 16 | CHEBI:10115 | Zingiberene | |
| 17 | CHEBI:5414 | Glucoputranjivin(1-) | |
| 18 | CHEBI:79331 | Glucoputranjivin | |
| 19 | CHEBI:32389 | All-cis-octadeca-6,9,12,15-tetraenoic acid | |
| 20 | Piper longum | CHEBI:132658 | Pipataline |
| 21 | CHEBI:66757 | Pipercyclobutanamide A(rel) | |
| 22 | CHEBI:69686 | Pellitorine | |
| 23 | CHEBI:67582 | Gaudichaudianic acid, (− rac) | |
| 24 | CHEBI:69675 | Sarmentosumin A | |
| 25 | CHEBI:69676 | Sarmentosumin B | |
| 26 | CHEBI:69677 | Sarmentosumin C | |
| 27 | CHEBI:69678 | Sarmentosumin D | |
| 28 | CHEBI:69679 | Isochamanetin | |
| 29 | CHEBI:69680 | 7-Methoxychamanetin | |
| 30 | CHEBI:69681 | Dichamanetin | |
| 31 | CHEBI:69682 | 7-Methoxydichamanetin | |
| 32 | CHEBI:69683 | 5″-(2⁗-Hydroxybenzyl)uvarinol | |
| 33 | CHEBI:69687 | 2,4-Dodecadienamide | |
| 34 | CHEBI:69689 | 7-Methoxyisochamanetin | |
| 35 | CHEBI:70083 | (2E,4E)-N-[2-(methylsulfinyl)ethyl]-2,4-decadienamide | |
| 36 | CHEBI:70084 | (2E,4E)-N-[(4-hydroxy-3-methoxyphenyl)ethyl]-2,4-decadienamide | |
| 37 | CHEBI:70085 | 3-(4-Hydroxy-3,5-dimethoxyphenyl)propanoylpyrrole | |
| 38 | CHEBI:70086 | 3-(3,4,5-Timethoxyphenyl)propanoylpyrrole | |
| 39 | CHEBI:70087 | 1-[(2E,4E,6E)-2,4,6-dodecatrienoyl]pyrrolidine | |
| 40 | CHEBI:70088 | 1-[(2E,4Z,8E)-9-(3,4-methylenedioxyphenyl)-2,4,8-nonatrienoyl]pyrrolidine | |
| 41 | CHEBI:70089 | 1-[(2E,4E,10E)-10-(3,4-methylenedioxyphenyl)-2,4,10-undecatrienoyl]pyrrolidine | |
| 42 | CHEBI:70090 | 1-[(4E,10E)-11-(3,4-methylenedioxyphenyl)-4,10-undecadienoyl]pyrrolidine | |
| 43 | CHEBI:70091 | 1-[(9E)-10-(3,4-methylenedioxyphenyl)-9-decenoyl]pyrrolidine | |
| 44 | CHEBI:70092 | 1-[(2E,4E)-2,4-decadienoyl]pyrrolidine | |
| 45 | CHEBI:70093 | 1-[(2E,4E)-2,4-dodecadienoyl]pyrrolidine | |
| 46 | CHEBI:70094 | 1-[(2E)-7-(3,4-methylenedioxyphenyl)-2-heptenoyl]pyrrolidine | |
| 47 | CHEBI:70095 | 1-[(2E,4E)-7-(3,4-methylenedioxyphenyl)-2,4-heptadienoyl]pyrrolidine | |
| 48 | CHEBI:70096 | 1-[(2E,8E)-9-(3,4-methylenedioxyphenyl)-2,8-nonadienoyl]pyrrolidine | |
| 49 | CHEBI:70097 | 1-[(8E)-9-(3,4-methylenedioxyphenyl)-8-nonenoyl]pyrrolidine | |
| 50 | CHEBI:70098 | 1-[(2E,4E,8E)-9-(3,4-methylenedioxyphenyl)-2,4,8-nonatrienoyl]pyrrolidine | |
| 51 | CHEBI:70099 | 1-[(2E,4E)-11-(3,4-methylenedioxyphenyl)-2,4-undecadienoyl]pyrrolidine | |
| 52 | CHEBI:70100 | 1-[(2E,10E)-11-(3,4-methylenedioxyphenyl)-2,10-undecadienoyl]pyrrolidine | |
| 53 | CHEBI:70101 | (2E,4E)-N-isobutyl-2,4-dodecadienamide | |
| 54 | CHEBI:70102 | (2E,4E)-N-isobutyl-7-(3,4-methylenedioxyphenyl)-hepta-2,4-dienamide | |
| 55 | CHEBI:70103 | (8E)-N-isobutyl-9-(3,4-methylenedioxyphenyl)nona-8-enamide | |
| 56 | CHEBI:70104 | (2E,4E,8E)-N-isobutyl-11-(3,4-methylenedioxyphenyl)undeca-2,4,8-trienamide | |
| 57 | CHEBI:70105 | N-trans-sinapoyltyramine | |
| 58 | CHEBI:70490 | Dihydrocubebin, rel- | |
| 59 | CHEBI:70491 | Justiflorinol | |
| 60 | CHEBI:70485 | (−)-Sanguinolignan A | |
| 61 | CHEBI:70486 | (−)-Sanguinolignan B | |
| 62 | CHEBI:70487 | (−)-Sanguinolignan C | |
| 63 | CHEBI:70488 | (−)-Sanguinolignan D | |
| 64 | CHEBI:70489 | (7′S)-parabenzlactone | |
| 65 | CHEBI:65899 | Flavokawain B | |
| 66 | CHEBI:66709 | Methyl 3,4-dihydroxy-5-(3′-methyl-2′-butenyl)benzoate | |
| 67 | CHEBI:69688 | Pipercallosidine | |
| 68 | CHEBI:132651 | Kadsurenin C | |
| 69 | CHEBI:132652 | Kadsurenin K | |
| 70 | CHEBI:132653 | Kadsurenin L | |
| 71 | CHEBI:69685 | Pipercallosine | |
| 72 | CHEBI:70483 | (S)-1′-methylhexyl caffeate | |
| 73 | CHEBI:132647 | Futoenone | |
| 74 | CHEBI:65684 | (−)-Cubebin | |
| 75 | CHEBI:65685 | (−)-3,4-Dimethoxy-3,4-desmethylenedioxycubebin | |
| 76 | CHEBI:70482 | (S)-1′-methylbutyl caffeate | |
| 77 | CHEBI:70484 | (S)-1′-methyloctyl caffeate | |
| 78 | CHEBI:65937 | Futokadsurin B | |
| 79 | CHEBI:65938 | Futokadsurin C | |
| 80 | CHEBI:28821 | Piperine | |
| 81 | CHEBI:65936 | Futokadsurin A | |
| 82 | CHEBI:132650 | Burchellin | |
| 83 | CHEBI:35697 | Trans-cinnamic acid | |
| 84 | CHEBI:80484 | Pinocembrin chalcone | |
| 85 | CHEBI:132657 | Piperlactam S | |
| 86 | CHEBI:2871 | Asebogenin | |
| 87 | CHEBI:66470 | (+)-Sesamin | |
| 88 | CHEBI:30746 | Benzoic acid | |
| 89 | CHEBI:17818 | N-feruloyltyramine | |
| 90 | CHEBI:132654 | Kadsurenin M | |
| 91 | CHEBI:156227 | (−)-Epicubenol | |
| 92 | CHEBI:70626 | Acacetin-8-C-neohesperidoside | |
| 93 | CHEBI:70148 | Monocerin | |
| 94 | CHEBI:70149 | (2S,3aR,9bR)-6,7-dihydroxy-8-methoxy-2-propyl-3,3a-dihydro-2H-furo[3,2-c]isochromen-5(9bH)-one | |
| 95 | CHEBI:70150 | Fusarentin 6,7-dimethyl ether | |
| 96 | CHEBI:70151 | Fusarentin 6-methyl ether | |
| 97 | CHEBI:70152 | (3R,4R)-4,8-dihydroxy-3-((R)-2-hydroxypentyl)-6,7-dimethoxyisochroman-1-one | |
| 98 | CHEBI:70153 | Colletotrialide, (+)- | |
| 99 | CHEBI:132648 | Galgravin | |
| 100 | CHEBI:8240 | Piperonal | |
| 101 | CHEBI:28157 | Pinocembrin | |
| 102 | CHEBI:80788 | 1,4-Cineole | |
| 103 | CHEBI:113455 | Sodium benzoate | |
| 104 | CHEBI:156224 | (−)-Cubenol | |
| 105 | CHEBI:37316 | (E,E)-piperic acid | |
| 106 | CHEBI:6116 | Kavapyrone | |
| 107 | CHEBI:10224 | Alpha-cubebene | |
| 108 | CHEBI:132649 | Acuminatin | |
| 109 | Syzygium aromaticum | CHEBI:65486 | Betulin di(3-carboxybutanoate) |
| 110 | CHEBI:65485 | Dihydrobetulinic acid | |
| 111 | CHEBI:65484 | Bevirimat | |
| 112 | CHEBI:65487 | Platanic acid | |
| 113 | CHEBI:132345 | Canophyllal | |
| 114 | CHEBI:27386 | Cinnamic acid | |
| 115 | CHEBI:69305 | Cinnamtannin D-1 | |
| 116 | CHEBI:69307 | Cassiatannin A | |
| 117 | CHEBI:69304 | Cinnamtannin B-1 | |
| 118 | CHEBI:69306 | Parameritannin A-1 | |
| 119 | CHEBI:133634 | Methyl linolenate | |
| 120 | CHEBI:63892 | Zerumbone | |
| 121 | CHEBI:89729 | (Z)-3-phenyl-2-propenal | |
| 122 | CHEBI:136676 | (E)-2-methoxycinnamic acid | |
| 123 | CHEBI:3087 | Betulinic acid | |
| 124 | Tragia involucrata | CHEBI:133981 | Heptacosan-1-ol |
| 125 | Anacyclus pyrethrum | CHEBI:27815 | Pyrethrin I |
| 126 | CHEBI:27474 | Pyrethrin II | |
| 127 | Andrographis paniculata | CHEBI:69808 | 14-Deoxy-11,12-didehydroandrographolide |
| 128 | CHEBI:65408 | Andrographolide | |
| 129 | CHEBI:86612 | Dihydroferulic acid | |
| 130 | CHEBI:132373 | Mesembryanthemoidigenic acid | |
| 131 | CHEBI:142267 | Methyl N-methylanthranilate | |
| 132 | CHEBI:132830 | Delta-elemene | |
| 133 | CHEBI:65732 | Decussatin | |
| 134 | Hygrophila auriculata | CHEBI:3367 | Capensinidin |
| 135 | Terminalia chebula | CHEBI:66202 | Termilignan B |
| 136 | CHEBI:69692 | (Z)-9-hydroxybenzo[c]oxepin-3(1H)-one | |
| 137 | CHEBI:69693 | Cyclosordariolone, (rac)- | |
| 138 | CHEBI:69694 | (R)-3-Hydroxy-1-[(R)-4-hydroxy-1,3-dihydroisobenzofuran-1-yl]butan-2-one | |
| 139 | CHEBI:69695 | (R)-3-Hydroxy-1-[(S)-4-hydroxy-1,3-dihydroisobenzofuran-1-yl]butan-2-one | |
| 140 | CHEBI:69696 | (E)-2-(Hydroxymethyl)-3-(4-hydroxypent-1-enyl)phenol | |
| 141 | CHEBI:69697 | 1-(3,9-Dihydroxy-1,3-dihydrobenzo[c]oxepin-3-yl)ethanone, (rac)- | |
| 142 | CHEBI:69698 | Pestalospirane A | |
| 143 | CHEBI:69699 | Pestalospirane B | |
| 144 | CHEBI:145828 | Methyl 3,4,5-trihydroxybenzoate | |
| 145 | CHEBI:9908 | Ursolic acid | |
| 146 | Justicia adhatoda | CHEBI:2814 | Arecoline |
| 147 | CHEBI:156072 | Propyl benzoate | |
| 148 | Plectranthus amboinicus | CHEBI:66763 | Plectranthol A |
| 149 | CHEBI:66764 | Plectranthol B | |
| 150 | CHEBI:138963 | 11,20-Dihydroxysugiol | |
| 151 | CHEBI:138962 | 11-Hydroxysugiol | |
| 152 | CHEBI:86062 | Abietatriene | |
| 153 | Saussurea lappa | CHEBI:66024 | 1beta-hydroxy arbusculin A |
| 154 | CHEBI:2540 | Alantolactone | |
| 155 | CHEBI:244418 | Dehydrocostus lactone | |
| 156 | CHEBI:3900 | Costunolide | |
| 157 | CHEBI:132820 | Matairesinoside | |
| 158 | CHEBI:138251 | 3-Hydroxyhexane-2,5-dione | |
| 159 | CHEBI:133981 | Heptacosan-1-ol | |
| 160 | Tinospora cordifolia | CHEBI:66046 | 6-Hydroxyluteolin 7-O-laminaribioside |
| 161 | CHEBI:89715 | Cyclotetradecane | |
| 162 | CHEBI:142915 | Cycloeucalenone | |
| 163 | CHEBI:141063 | Rubimaillin | |
| 164 | CHEBI:132345 | Canophyllal | |
| 165 | CHEBI:132718 | Stepharanine | |
| 166 | CHEBI:134479 | (Z)-icos-13-enoic acid | |
| 167 | CHEBI:145828 | Methyl 3,4,5-trihydroxybenzoate | |
| 168 | Rotheca serrata | CHEBI:156193 | Serratol |
| 169 | CHEBI:78330 | Huperzine A | |
| 170 | CHEBI:6701 | Maytansine | |
| 171 | Cypreus rotundus | CHEBI:66416 | Mustakone |
| 172 | CHEBI:81377 | (+)-Nootkatone | |
| 173 | Sida acuta Burm.f.L | CHEBI:142397 | Pectenotoxin-11 |
| 174 | CHEBI:145981 | Dinophysistoxin 2 | |
| 175 | CHEBI:142495 | 1-Icosanoylglycerol | |
| 176 | CHEBI:131838 | Swertisin | |
| 177 | CHEBI:156193 | Serratol | |
| 178 | CHEBI:16634 | Raffinose | |
| 179 | CHEBI:90295 | PLX-4720 | |
| 180 | CHEBI:78330 | Huperzine A | |
| 181 | CHEBI:6701 | Maytansine | |
| 182 | CHEBI:143119 | N-(2-methoxyethyl)-4-{[6-(pyridin-4-yl)quinazolin-2-yl]amino}benzamide | |
| 183 | Carica Papaya | CHEBI:17127 | Glucotropeolin |
| 184 | CHEBI:58021 | Glucotropeolin(1-) | |
| 185 | CHEBI:142658 | Methyl 12-methyltetradecanoate | |
| 186 | CHEBI:91143 | (3S,5R,6S)-beta-cryptoxanthin 5,6-epoxide | |
| 187 | CHEBI:3433 | Carpaine | |
| 188 | CHEBI:10362 | Beta-cryptoxanthin | |
| 189 | CHEBI:1307 | 24-Methylenecycloartanol | |
| 190 | CHEBI:141360 | Helvolic acid methyl ester | |
| 191 | CHEBI:4316 | Danielone | |
| 192 | CHEBI:133683 | 2-Isobutylthiazole | |
| 193 | A. paniculata | CHEBI:69808 | 14-Deoxy-11,12-didehydroandrographolide |
| 194 | CHEBI:65408 | Andrographolide | |
| 195 | CHEBI:86612 | Dihydroferulic acid | |
| 196 | CHEBI:132373 | Mesembryanthemoidigenic acid | |
| 197 | CHEBI:142267 | Methyl N-methylanthranilate | |
| 198 | CHEBI:132830 | Delta-elemene | |
| 199 | CHEBI:65732 | Decussatin | |
| 200 | Ocimum tenuiflorum | CHEBI:17580 | Linalool |
| 201 | CHEBI:7545 | Nevadensin | |
| 202 | CHEBI:63710 | 7-Epi-sesquithujene | |
| 203 | Vetiveria zizanioides | CHEBI:138051 | Selina-4(15),7(11)-diene |
| 204 | Santalum album | CHEBI:16714 | Codeine |
| 205 | CHEBI:45373 | Sulfanilamide | |
| 206 | CHEBI:65460 | Avicularin | |
| 207 | Piper nigrum | CHEBI:66757 | Pipercyclobutanamide A(rel) |
| 208 | CHEBI:65684 | (−)-cubebin | |
| 209 | CHEBI:65685 | (−)-3,4-dimethoxy-3,4-desmethylenedioxycubebin | |
| 210 | CHEBI:28821 | Piperine | |
| 211 | CHEBI:69686 | Pellitorine | |
| 212 | CHEBI:67582 | Gaudichaudianic acid, (− rac) | |
| 213 | CHEBI:69675 | Sarmentosumin A | |
| 214 | CHEBI:69676 | Sarmentosumin B | |
| 215 | CHEBI:69677 | Sarmentosumin C | |
| 216 | CHEBI:69678 | Sarmentosumin D | |
| 217 | CHEBI:69679 | Isochamanetin | |
| 218 | CHEBI:69680 | 7-Methoxychamanetin | |
| 219 | CHEBI:69681 | Dichamanetin | |
| 220 | CHEBI:69682 | 7-Methoxydichamanetin | |
| 221 | CHEBI:69683 | 5″-(2⁗-Hydroxybenzyl)uvarinol | |
| 222 | CHEBI:69687 | 2,4-Dodecadienamide | |
| 223 | CHEBI:69689 | 7-Methoxyisochamanetin | |
| 224 | CHEBI:70083 | (2E,4E)-N-[2-(methylsulfinyl)ethyl]-2,4-decadienamide | |
| 225 | CHEBI:70084 | (2E,4E)-N-[(4-hydroxy-3-methoxyphenyl)ethyl]-2,4-decadienamide | |
| 226 | CHEBI:70085 | 3-(4-hydroxy-3,5-dimethoxyphenyl)propanoylpyrrole | |
| 227 | CHEBI:70086 | 3-(3,4,5-timethoxyphenyl)propanoylpyrrole | |
| 228 | CHEBI:70087 | 1-[(2E,4E,6E)-2,4,6-dodecatrienoyl]pyrrolidine | |
| 229 | CHEBI:70088 | 1-[(2E,4Z,8E)-9-(3,4-methylenedioxyphenyl)-2,4,8-nonatrienoyl]pyrrolidine | |
| 230 | CHEBI:70089 | 1-[(2E,4E,10E)-10-(3,4-methylenedioxyphenyl)-2,4,10-undecatrienoyl]pyrrolidine | |
| 231 | CHEBI:70090 | 1-[(4E,10E)-11-(3,4-methylenedioxyphenyl)-4,10-undecadienoyl]pyrrolidine | |
| 232 | CHEBI:70091 | 1-[(9E)-10-(3,4-methylenedioxyphenyl)-9-decenoyl]pyrrolidine | |
| 233 | CHEBI:70092 | 1-[(2E,4E)-2,4-decadienoyl]pyrrolidine | |
| 234 | CHEBI:70093 | 1-[(2E,4E)-2,4-dodecadienoyl]pyrrolidine | |
| 235 | CHEBI:70094 | 1-[(2E)-7-(3,4-methylenedioxyphenyl)-2-heptenoyl]pyrrolidine | |
| 236 | CHEBI:70095 | 1-[(2E,4E)-7-(3,4-methylenedioxyphenyl)-2,4-heptadienoyl]pyrrolidine | |
| 237 | CHEBI:70096 | 1-[(2E,8E)-9-(3,4-methylenedioxyphenyl)-2,8-nonadienoyl]pyrrolidine | |
| 238 | CHEBI:70097 | 1-[(8E)-9-(3,4-methylenedioxyphenyl)-8-nonenoyl]pyrrolidine | |
| 239 | CHEBI:70098 | 1-[(2E,4E,8E)-9-(3,4-methylenedioxyphenyl)-2,4,8-nonatrienoyl]pyrrolidine | |
| 240 | CHEBI:70099 | 1-[(2E,4E)-11-(3,4-methylenedioxyphenyl)-2,4-undecadienoyl]pyrrolidine | |
| 241 | CHEBI:70100 | 1-[(2E,10E)-11-(3,4-methylenedioxyphenyl)-2,10-undecadienoyl]pyrrolidine | |
| 242 | CHEBI:70101 | (2E,4E)-N-isobutyl-2,4-dodecadienamide | |
| 243 | CHEBI:70102 | (2E,4E)-N-isobutyl-7-(3,4-methylenedioxyphenyl)-hepta-2,4-dienamide | |
| 244 | CHEBI:70103 | (8E)-N-isobutyl-9-(3,4-methylenedioxyphenyl)nona-8-enamide | |
| 245 | CHEBI:70104 | (2E,4E,8E)-N-isobutyl-11-(3,4-methylenedioxyphenyl)undeca-2,4,8-trienamide | |
| 246 | CHEBI:70105 | N-trans-sinapoyltyramine | |
| 247 | CHEBI:70490 | Dihydrocubebin, rel- | |
| 248 | CHEBI:70491 | Justiflorinol | |
| 249 | CHEBI:70485 | (−)-Sanguinolignan A | |
| 250 | CHEBI:70486 | (−)-Sanguinolignan B | |
| 251 | CHEBI:70487 | (−)-Sanguinolignan C | |
| 252 | CHEBI:70488 | (−)-Sanguinolignan D | |
| 253 | CHEBI:70489 | (7′S)-parabenzlactone | |
| 254 | CHEBI:65899 | Flavokawain B | |
| 255 | CHEBI:66709 | Methyl 3,4-dihydroxy-5-(3′-methyl-2′-butenyl)benzoate | |
| 256 | CHEBI:69688 | Pipercallosidine | |
| 257 | CHEBI:132651 | Kadsurenin C | |
| 258 | CHEBI:132652 | Kadsurenin K | |
| 259 | CHEBI:132653 | Kadsurenin L | |
| 260 | CHEBI:69685 | Pipercallosine | |
| 261 | CHEBI:70483 | (S)-1′-methylhexyl caffeate | |
| 262 | CHEBI:132647 | Futoenone | |
| 263 | CHEBI:70482 | (S)-1′-methylbutyl caffeate | |
| 264 | CHEBI:70484 | (S)-1′-methyloctyl caffeate | |
| 265 | CHEBI:65937 | Futokadsurin B | |
| 266 | CHEBI:65938 | Futokadsurin C | |
| 267 | CHEBI:65936 | Futokadsurin A | |
| 268 | CHEBI:132650 | Burchellin | |
| 269 | CHEBI:8240 | Piperonal | |
| 270 | CHEBI:156224 | (−)-Cubenol | |
| 271 | CHEBI:37316 | (E,E)-piperic acid | |
| 272 | CHEBI:35697 | Trans-cinnamic acid | |
| 273 | CHEBI:80484 | Pinocembrin chalcone | |
| 274 | CHEBI:132657 | Piperlactam S | |
| 275 | CHEBI:2871 | Asebogenin | |
| 276 | CHEBI:51226 | Epicocconone | |
| 277 | CHEBI:66470 | (+)-Sesamin | |
| 278 | CHEBI:132658 | Pipataline | |
| 279 | CHEBI:143911 | (−)-Antofine | |
| 280 | CHEBI:30746 | Benzoic acid | |
| 281 | CHEBI:88764 | Ethyl butyrate | |
| 282 | CHEBI:17818 | N-feruloyltyramine | |
| 283 | CHEBI:132654 | Kadsurenin M | |
| 284 | CHEBI:156227 | (−)-Epicubenol | |
| 285 | CHEBI:70626 | Acacetin-8-C-neohesperidoside | |
| 286 | CHEBI:70148 | Monocerin | |
| 287 | CHEBI:70149 | (2S,3aR,9bR)-6,7-dihydroxy-8-methoxy-2-propyl-3,3a-dihydro-2H-furo[3,2-c]isochromen-5(9bH)-one | |
| 288 | CHEBI:70150 | Fusarentin 6,7-dimethyl ether | |
| 289 | CHEBI:70151 | Fusarentin 6-methyl ether | |
| 290 | CHEBI:70152 | (3R,4R)-4,8-dihydroxy-3-((R)-2-hydroxypentyl)-6,7-dimethoxyisochroman-1-one | |
| 291 | CHEBI:70153 | Colletotrialide, (+)- | |
| 292 | CHEBI:132648 | Galgravin | |
| 293 | CHEBI:133381 | 9,10-Epoxy-18-hydroxyoctadecanoic acid | |
| 294 | CHEBI:28157 | Pinocembrin | |
| 295 | CHEBI:80788 | 1,4-Cineole | |
| 296 | CHEBI:113455 | Sodium benzoate | |
| 297 | CHEBI:133325 | 9,10,18-Trihydroxyoctadecanoic acid | |
| 298 | CHEBI:6116 | Kavapyrone | |
| 299 | CHEBI:10224 | Alpha-cubebene | |
| 300 | CHEBI:132649 | Acuminatin | |
| 301 | Hedyotis corymbosa | CHEBI:66123 | Jerantinine E |
| 302 | CHEBI:66124 | Jerantinine F | |
| 303 | CHEBI:66121 | Jerantinine C | |
| 304 | CHEBI:66120 | Jerantinine B | |
| 305 | CHEBI:66122 | Jerantinine D | |
| 306 | CHEBI:66119 | Jerantinine A | |
| 307 | CHEBI:142075 | Tabernaemontanine | |
| 308 | CHEBI:6682 | Mangiferin | |
| 309 | Plectranthus vettiveroides | CHEBI:66763 | Plectranthol A |
| 310 | CHEBI:66764 | Plectranthol B | |
| 311 | CHEBI:138963 | 11,20-Dihydroxysugiol | |
| 312 | CHEBI:138962 | 11-Hydroxysugiol | |
| 313 | CHEBI:86062 | Abietatriene | |
| 314 | Trichosanthes cucumerina | CHEBI:66838 | 7-Oxo-10α-cucurbitadienol |
3.3. Drug likeliness of phytochemical compounds
For a compound to persuade as a drug, the compound must satisfy a set of parameters that will prove its compatibility, efficiency, and toxicity. Lipinski's “rule of five” highlights possible bioavailability problems if two or more properties were violated. The SwissADME server was used to identify the Lipinski rule of 5 violations. The overall results from the SwissADME are tabulated in Table 4 . The list of 285 compounds that satisfy the Lipinski rule of 5 is tabulated in Table 5 .
Table 4.
The Lipinski rule calculation for the phytochemical compounds of the plant involved in the formulation of Nilavembu kudineer, Kabasura kudineer, and JACOM.
| S.·no | Compounds | MW | #H-bond acceptors | #H-bond donors | iLOGP | Lipinski #violations |
|---|---|---|---|---|---|---|
| 1 | Gingerenone B | 386.44 | 6 | 2 | 3.7 | 0 |
| 2 | Beta-sesquiphellandrene | 204.35 | 0 | 0 | 3.65 | 1 |
| 3 | 3-(3,4-Dimethoxyphenyl)-4-[(Z)-3,4-dimethoxystyryl]cyclohex-1-ene | 380.48 | 4 | 0 | 4.43 | 0 |
| 4 | 3-(3,4-Dimethoxyphenyl)-4-[(E)-3,4-dimethoxystyryl]cyclohex-1-ene | 380.48 | 4 | 0 | 4.43 | 0 |
| 5 | Pinocarveol | 152.23 | 1 | 1 | 2.12 | 0 |
| 6 | Zerumboneoxide | 234.33 | 2 | 0 | 2.71 | 0 |
| 7 | Ramonanin A, (rel)- | 680.74 | 10 | 4 | 4.14 | 1 |
| 8 | Ramonanin B, (rel)- | 680.74 | 10 | 4 | 4.51 | 1 |
| 9 | Ramonanin C, (rel)- | 680.74 | 10 | 4 | 4.5 | 1 |
| 10 | Ramonanin D, (rel)- | 680.74 | 10 | 4 | 5.29 | 1 |
| 11 | Zerumbone | 218.33 | 1 | 0 | 2.72 | 0 |
| 12 | 5-Hydroxyzerumbone | 234.33 | 2 | 1 | 2.14 | 0 |
| 13 | (2E,6E)-hedycaryol | 222.37 | 1 | 1 | 3.05 | 0 |
| 14 | Dodecane | 170.33 | 0 | 0 | 3.82 | 1 |
| 15 | 7,4′-Dimethylkaempferol | 314.29 | 6 | 2 | 2.94 | 0 |
| 16 | Zingiberene | 204.35 | 0 | 0 | 3.63 | 1 |
| 17 | Glucoputranjivin(1-) | 360.38 | 10 | 4 | 0.86 | 0 |
| 18 | Glucoputranjivin | 361.39 | 10 | 5 | 0.14 | 0 |
| 19 | All-cis-octadeca-6,9,12,15-tetraenoic acid | 276.41 | 2 | 1 | 3.29 | 1 |
| 20 | Pipataline | 288.42 | 2 | 0 | 4.61 | 1 |
| 21 | Pipercyclobutanamide A(rel) | 570.68 | 6 | 0 | 5.26 | 1 |
| 22 | pellitorine | 223.35 | 1 | 1 | 3.61 | 0 |
| 23 | Gaudichaudianic acid, (− rac) | 340.46 | 3 | 1 | 3.85 | 1 |
| 24 | Sarmentosumin A | 680.74 | 8 | 6 | 3.7 | 2 |
| 25 | Sarmentosumin B | 680.74 | 8 | 6 | 3.97 | 2 |
| 26 | Sarmentosumin C | 786.86 | 9 | 7 | 4.13 | 3 |
| 27 | Sarmentosumin D | 786.86 | 9 | 7 | 4.06 | 3 |
| 28 | Isochamanetin | 362.38 | 5 | 3 | 2.68 | 0 |
| 29 | 7-Methoxychamanetin | 376.4 | 5 | 2 | 3.2 | 0 |
| 30 | Dichamanetin | 468.5 | 6 | 4 | 3.01 | 0 |
| 31 | 7-Methoxydichamanetin | 482.52 | 6 | 3 | 3.58 | 0 |
| 32 | 5″-(2⁗-Hydroxybenzyl)uvarinol | 694.77 | 8 | 5 | 4.16 | 1 |
| 33 | 2,4-Dodecadienamide | 195.3 | 1 | 1 | 2.87 | 0 |
| 34 | 7-Methoxyisochamanetin | 376.4 | 5 | 2 | 3.5 | 0 |
| 35 | (2E,4E)-N-[2-(methylsulfinyl)ethyl]-2,4-decadienamide | 257.39 | 2 | 1 | 3.05 | 0 |
| 36 | (2E,4E)-N-[(4-hydroxy-3-methoxyphenyl)ethyl]-2,4-decadienamide | 317.42 | 3 | 2 | 3.76 | 0 |
| 37 | 3-(4-Hydroxy-3,5-dimethoxyphenyl)propanoylpyrrole | 275.3 | 4 | 1 | 2.63 | 0 |
| 38 | 3-(3,4,5-Timethoxyphenyl)propanoylpyrrole | 289.33 | 4 | 0 | 3.21 | 0 |
| 39 | 1-[(2E,4E,6E)-2,4,6-dodecatrienoyl]pyrrolidine | 247.38 | 1 | 0 | 3.86 | 0 |
| 40 | 1-[(2E,4Z,8E)-9-(3,4-methylenedioxyphenyl)-2,4,8-nonatrienoyl]pyrrolidine | 325.4 | 3 | 0 | 3.88 | 0 |
| 41 | 1-[(2E,4E,10E)-10-(3,4-methylenedioxyphenyl)-2,4,10-undecatrienoyl]pyrrolidine | 353.45 | 3 | 0 | 4.53 | 0 |
| 42 | 1-[(4E,10E)-11-(3,4-methylenedioxyphenyl)-4,10-undecadienoyl]pyrrolidine | 355.47 | 3 | 0 | 4.6 | 0 |
| 43 | 1-[(9E)-10-(3,4-methylenedioxyphenyl)-9-decenoyl]pyrrolidine | 343.46 | 3 | 0 | 4.43 | 0 |
| 44 | 1-[(2E,4E)-2,4-decadienoyl]pyrrolidine | 221.34 | 1 | 0 | 3.45 | 0 |
| 45 | 1-[(2E,4E)-2,4-dodecadienoyl]pyrrolidine | 249.39 | 1 | 0 | 3.95 | 0 |
| 46 | 1-[(2E)-7-(3,4-methylenedioxyphenyl)-2-heptenoyl]pyrrolidine | 301.38 | 3 | 0 | 3.64 | 0 |
| 47 | 1-[(2E,4E)-7-(3,4-methylenedioxyphenyl)-2,4-heptadienoyl]pyrrolidine | 299.36 | 3 | 0 | 3.65 | 0 |
| 48 | 1-[(2E,8E)-9-(3,4-methylenedioxyphenyl)-2,8-nonadienoyl]pyrrolidine | 327.42 | 3 | 0 | 4.04 | 0 |
| 49 | 1-[(8E)-9-(3,4-methylenedioxyphenyl)-8-nonenoyl]pyrrolidine | 329.43 | 3 | 0 | 4.16 | 0 |
| 50 | 1-[(2E,4E,8E)-9-(3,4-methylenedioxyphenyl)-2,4,8-nonatrienoyl]pyrrolidine | 325.4 | 3 | 0 | 3.88 | 0 |
| 51 | 1-[(2E,4E)-11-(3,4-methylenedioxyphenyl)-2,4-undecadienoyl]pyrrolidine | 355.47 | 3 | 0 | 4.51 | 0 |
| 52 | 1-[(2E,10E)-11-(3,4-methylenedioxyphenyl)-2,10-undecadienoyl]pyrrolidine | 355.47 | 3 | 0 | 4.16 | 0 |
| 53 | (2E,4E)-N-isobutyl-2,4-dodecadienamide | 251.41 | 1 | 1 | 4.06 | 0 |
| 54 | (2E,4E)-N-isobutyl-7-(3,4-methylenedioxyphenyl)-hepta-2,4-dienamide | 301.38 | 3 | 1 | 3.67 | 0 |
| 55 | (8E)-N-isobutyl-9-(3,4-methylenedioxyphenyl)nona-8-enamide | 331.45 | 3 | 1 | 4.18 | 0 |
| 56 | (2E,4E,8E)-N-isobutyl-11-(3,4-methylenedioxyphenyl)undeca-2,4,8-trienamide | 355.47 | 3 | 1 | 4.42 | 0 |
| 57 | N-trans-sinapoyltyramine | 343.37 | 5 | 3 | 2.71 | 0 |
| 58 | Dihydrocubebin, rel- | 358.39 | 6 | 2 | 3.19 | 0 |
| 59 | Justiflorinol | 356.33 | 7 | 1 | 2.76 | 0 |
| 60 | (−)-Sanguinolignan A | 384.34 | 8 | 1 | 2.82 | 0 |
| 61 | (−)-Sanguinolignan B | 384.34 | 8 | 1 | 2.79 | 0 |
| 62 | (−)-Sanguinolignan C | 442.42 | 9 | 0 | 3.36 | 0 |
| 63 | (−)-Sanguinolignan D | 426.37 | 9 | 0 | 2.74 | 0 |
| 64 | (7′S)-parabenzlactone | 370.35 | 7 | 1 | 3.12 | 0 |
| 65 | Flavokawain B | 284.31 | 4 | 1 | 2.63 | 0 |
| 66 | Methyl 3,4-dihydroxy-5-(3′-methyl-2′-butenyl)benzoate | 236.26 | 4 | 2 | 2.62 | 0 |
| 67 | Pipercallosidine | 303.4 | 3 | 1 | 3.76 | 0 |
| 68 | Kadsurenin C | 358.43 | 5 | 1 | 3.37 | 0 |
| 69 | Kadsurenin K | 342.39 | 5 | 1 | 3.02 | 0 |
| 70 | Kadsurenin L | 400.46 | 6 | 0 | 3.42 | 0 |
| 71 | Pipercallosine | 329.43 | 3 | 1 | 4.04 | 0 |
| 72 | (S)-1′-methylhexyl caffeate | 278.34 | 4 | 2 | 3.41 | 0 |
| 73 | Futoenone | 340.37 | 5 | 0 | 3.15 | 0 |
| 74 | (−)-Cubebin | 356.37 | 6 | 1 | 3.18 | 0 |
| 75 | (−)-3,4-Dimethoxy-3,4-desmethylenedioxycubebin | 372.41 | 6 | 1 | 3.06 | 0 |
| 76 | (S)-1′-methylbutyl caffeate | 250.29 | 4 | 2 | 2.81 | 0 |
| 77 | (S)-1′-methyloctyl caffeate | 306.4 | 4 | 2 | 3.66 | 0 |
| 78 | Futokadsurin B | 356.41 | 5 | 0 | 3.89 | 0 |
| 79 | Futokadsurin C | 356.41 | 5 | 0 | 3.77 | 0 |
| 80 | Piperine | 285.34 | 3 | 0 | 3.42 | 0 |
| 81 | Futokadsurin A | 358.43 | 5 | 1 | 3.46 | 0 |
| 82 | Burchellin | 340.37 | 5 | 0 | 3.26 | 0 |
| 83 | Trans-cinnamic acid | 148.16 | 2 | 1 | 1.55 | 0 |
| 84 | Pinocembrin chalcone | 256.25 | 4 | 3 | 1.09 | 0 |
| 85 | Piperlactam S | 295.29 | 4 | 1 | 2.54 | 0 |
| 86 | Asebogenin | 288.3 | 5 | 3 | 1.97 | 0 |
| 87 | (+)-Sesamin | 354.35 | 6 | 0 | 3.46 | 0 |
| 88 | Benzoic acid | 122.12 | 2 | 1 | 1.11 | 0 |
| 89 | N-feruloyltyramine | 313.35 | 4 | 3 | 2.58 | 0 |
| 90 | Kadsurenin M | 328.36 | 5 | 0 | 3.15 | 0 |
| 91 | (−)-Epicubenol | 222.37 | 1 | 1 | 3.11 | 0 |
| 92 | Acacetin-8-C-neohesperidoside | 592.55 | 14 | 8 | 1.49 | 3 |
| 93 | Monocerin | 308.33 | 6 | 1 | 2.99 | 0 |
| 94 | (2S,3aR,9bR)-6,7-dihydroxy-8-methoxy-2-propyl-3,3a-dihydro-2H-furo[3,2-c]isochromen-5(9bH)-one | 294.3 | 6 | 2 | 2.51 | 0 |
| 95 | Fusarentin 6,7-dimethyl ether | 310.34 | 6 | 2 | 2.85 | 0 |
| 96 | Fusarentin 6-methyl ether | 296.32 | 6 | 3 | 2.27 | 0 |
| 97 | (3R,4R)-4,8-dihydroxy-3-((R)-2-hydroxypentyl)-6,7-dimethoxyisochroman-1-one | 326.34 | 7 | 3 | 2.59 | 0 |
| 98 | Colletotrialide, (+)- | 308.33 | 6 | 1 | 2.49 | 0 |
| 99 | Galgravin | 372.45 | 5 | 0 | 3.97 | 0 |
| 100 | Piperonal | 150.13 | 3 | 0 | 1.6 | 0 |
| 101 | Pinocembrin | 256.25 | 4 | 2 | 2.11 | 0 |
| 102 | 1,4-Cineole | 154.25 | 1 | 0 | 2.68 | 0 |
| 103 | Sodium benzoate | 144.1 | 2 | 0 | − 11.15 | 0 |
| 104 | (−)-Cubenol | 222.37 | 1 | 1 | 3.24 | 0 |
| 105 | (E,E)-piperic acid | 218.21 | 4 | 1 | 2.2 | 0 |
| 106 | Kavapyrone | 244.24 | 4 | 0 | 2.6 | 0 |
| 107 | Alpha-cubebene | 204.35 | 0 | 0 | 3.4 | 1 |
| 108 | Acuminatin | 340.41 | 4 | 0 | 4.05 | 0 |
| 109 | Betulin di(3-carboxybutanoate) | 670.92 | 8 | 2 | 4.69 | 2 |
| 110 | Dihydrobetulinic acid | 458.72 | 3 | 2 | 3.91 | 1 |
| 111 | Bevirimat | 584.83 | 6 | 2 | 4.25 | 2 |
| 112 | Platanic acid | 458.67 | 4 | 2 | 3.52 | 1 |
| 113 | Canophyllal | 440.7 | 2 | 0 | 4.07 | 1 |
| 114 | Cinnamic acid | 148.16 | 2 | 1 | 1.55 | 0 |
| 115 | Cinnamtannin D-1 | 864.76 | 18 | 14 | 2.04 | 3 |
| 116 | Cassiatannin A | 1153.01 | 24 | 19 | 2.75 | 3 |
| 117 | Cinnamtannin B-1 | 864.76 | 18 | 14 | 2.25 | 3 |
| 118 | Parameritannin A-1 | 1153.01 | 24 | 19 | 2.6 | 3 |
| 119 | Methyl linolenate | 292.46 | 2 | 0 | 4.94 | 1 |
| 120 | Zerumbone | 218.33 | 1 | 0 | 2.72 | 0 |
| 121 | (Z)-3-phenyl-2-propenal | 132.16 | 1 | 0 | 1.65 | 0 |
| 122 | (E)-2-methoxycinnamic acid | 178.18 | 3 | 1 | 1.76 | 0 |
| 123 | Betulinic acid | 456.7 | 3 | 2 | 3.81 | 1 |
| 124 | Heptacosan-1-ol | 396.73 | 1 | 1 | 6.93 | 1 |
| 125 | Pyrethrin I | 328.45 | 3 | 0 | 4.22 | 0 |
| 126 | Pyrethrin II | 372.45 | 5 | 0 | 4.2 | 0 |
| 127 | 14-Deoxy-11,12-didehydroandrographolide | 332.43 | 4 | 2 | 3.07 | 0 |
| 128 | Andrographolide | 350.45 | 5 | 3 | 2.61 | 0 |
| 129 | Dihydroferulic acid | 196.2 | 4 | 2 | 1.62 | 0 |
| 130 | Mesembryanthemoidigenic acid | 472.7 | 4 | 3 | 3.56 | 1 |
| 131 | Methyl N-methylanthranilate | 165.19 | 2 | 1 | 2.02 | 0 |
| 132 | Delta-elemene | 204.35 | 0 | 0 | 3.43 | 1 |
| 133 | Decussatin | 302.28 | 6 | 1 | 2.96 | 0 |
| 134 | Capensinidin | 345.32 | 7 | 3 | − 0.95 | 0 |
| 135 | Termilignan B | 294.34 | 3 | 1 | 2.74 | 0 |
| 136 | (Z)-9-hydroxybenzo[c]oxepin-3(1H)-one | 176.17 | 3 | 1 | 1.37 | 0 |
| 137 | Cyclosordariolone, (rac)- | 220.22 | 4 | 3 | 1.44 | 0 |
| 138 | (R)-3-Hydroxy-1-[(R)-4-hydroxy-1,3-dihydroisobenzofuran-1-yl]butan-2-one | 222.24 | 4 | 2 | 1.7 | 0 |
| 139 | (R)-3-Hydroxy-1-[(S)-4-hydroxy-1,3-dihydroisobenzofuran-1-yl]butan-2-one | 222.24 | 4 | 2 | 1.51 | 0 |
| 140 | (E)-2-(Hydroxymethyl)-3-(4-hydroxypent-1-enyl)phenol | 208.25 | 3 | 3 | 1.99 | 0 |
| 141 | 1-(3,9-Dihydroxy-1,3-dihydrobenzo[c]oxepin-3-yl)ethanone, (rac)- | 220.22 | 4 | 2 | 1.53 | 0 |
| 142 | Pestalospirane A | 408.44 | 6 | 2 | 2.89 | 0 |
| 143 | Pestalospirane B | 408.44 | 6 | 2 | 3.02 | 0 |
| 144 | Methyl 3,4,5-trihydroxybenzoate | 184.15 | 5 | 3 | 0.96 | 0 |
| 145 | Ursolic acid | 456.7 | 3 | 2 | 3.95 | 1 |
| 146 | Arecoline | 155.19 | 3 | 0 | 2.26 | 0 |
| 147 | Propyl benzoate | 164.2 | 2 | 0 | 2.46 | 0 |
| 148 | Plectranthol A | 450.52 | 6 | 4 | 3.49 | 0 |
| 149 | Plectranthol B | 536.66 | 7 | 3 | 4.59 | 2 |
| 150 | 11,20-Dihydroxysugiol | 332.43 | 4 | 3 | 2.87 | 0 |
| 151 | 11-Hydroxysugiol | 316.43 | 3 | 2 | 3.15 | 0 |
| 152 | Abietatriene | 270.45 | 0 | 0 | 3.86 | 1 |
| 153 | 1beta-hydroxy arbusculin A | 266.33 | 4 | 2 | 2.23 | 0 |
| 154 | Alantolactone | 232.32 | 2 | 0 | 2.71 | 0 |
| 155 | Dehydrocostus lactone | 230.3 | 2 | 0 | 2.67 | 0 |
| 156 | Costunolide | 232.32 | 2 | 0 | 2.72 | 0 |
| 157 | Matairesinoside | 520.53 | 11 | 5 | 2.37 | 2 |
| 158 | 3-Hydroxyhexane-2,5-dione | 130.14 | 3 | 1 | 0.6 | 0 |
| 159 | Heptacosan-1-ol | 396.73 | 1 | 1 | 6.93 | 1 |
| 160 | 6-Hydroxyluteolin 7-O-laminaribioside | 626.52 | 17 | 11 | 1.44 | 3 |
| 161 | Cyclotetradecane | 196.37 | 0 | 0 | 3.35 | 1 |
| 162 | Cycloeucalenone | 424.7 | 1 | 0 | 4.98 | 1 |
| 163 | Rubimaillin | 284.31 | 4 | 1 | 2.87 | 0 |
| 164 | Canophyllal | 440.7 | 2 | 0 | 4.07 | 1 |
| 165 | Stepharanine | 324.35 | 4 | 2 | − 0.53 | 0 |
| 166 | (Z)-icos-13-enoic acid | 310.51 | 2 | 1 | 4.26 | 1 |
| 167 | Methyl 3,4,5-trihydroxybenzoate | 184.15 | 5 | 3 | 0.96 | 0 |
| 168 | Serratol | 290.48 | 1 | 1 | 3.82 | 1 |
| 169 | Huperzine A | 242.32 | 2 | 2 | 2.42 | 0 |
| 170 | Maytansine | 692.2 | 10 | 2 | 4.27 | 2 |
| 171 | Mustakone | 218.33 | 1 | 0 | 2.95 | 0 |
| 172 | (+)-nootkatone | 218.33 | 1 | 0 | 2.83 | 0 |
| 173 | Pectenotoxin-11 | 875.05 | 15 | 4 | 4.09 | 2 |
| 174 | Dinophysistoxin 2 | 805 | 13 | 5 | 6.22 | 2 |
| 175 | 1-Icosanoylglycerol | 386.61 | 4 | 2 | 4.71 | 0 |
| 176 | Swertisin | 446.4 | 10 | 6 | 2.5 | 1 |
| 177 | Serratol | 290.48 | 1 | 1 | 3.82 | 1 |
| 178 | Raffinose | 504.44 | 16 | 11 | 1.12 | 3 |
| 179 | PLX-4720 | 413.83 | 6 | 2 | 2.42 | 0 |
| 180 | Huperzine A | 242.32 | 2 | 2 | 2.42 | 0 |
| 181 | Maytansine | 692.2 | 10 | 2 | 4.27 | 2 |
| 182 | N-(2-methoxyethyl)-4-{[6-(pyridin-4-yl)quinazolin-2-yl]amino}benzamide | 399.45 | 5 | 2 | 2.8 | 0 |
| 183 | Glucotropeolin | 409.43 | 10 | 5 | 0.57 | 0 |
| 184 | Glucotropeolin(1-) | 408.42 | 10 | 4 | 1.04 | 0 |
| 185 | Methyl 12-methyltetradecanoate | 256.42 | 2 | 0 | 4.08 | 1 |
| 186 | (3S,5R,6S)-beta-cryptoxanthin 5,6-epoxide | 568.87 | 2 | 1 | 7.8 | 2 |
| 187 | Carpaine | 478.71 | 6 | 2 | 4.4 | 0 |
| 188 | Beta-cryptoxanthin | 552.87 | 1 | 1 | 7.6 | 2 |
| 189 | 24-Methylenecycloartanol | 440.74 | 1 | 1 | 5.31 | 1 |
| 190 | Helvolic acid methyl ester | 582.72 | 8 | 0 | 4.5 | 1 |
| 191 | Danielone | 212.2 | 5 | 2 | 1.63 | 0 |
| 192 | 2-Isobutylthiazole | 141.23 | 1 | 0 | 2.37 | 0 |
| 193 | 14-Deoxy-11,12-didehydroandrographolide | 332.43 | 4 | 2 | 3.07 | 0 |
| 194 | Andrographolide | 350.45 | 5 | 3 | 2.61 | 0 |
| 195 | Dihydroferulic acid | 196.2 | 4 | 2 | 1.62 | 0 |
| 196 | Mesembryanthemoidigenic acid | 472.7 | 4 | 3 | 3.56 | 1 |
| 197 | Methyl N-methylanthranilate | 165.19 | 2 | 1 | 2.02 | 0 |
| 198 | Delta-elemene | 204.35 | 0 | 0 | 3.43 | 1 |
| 199 | Decussatin | 302.28 | 6 | 1 | 2.96 | 0 |
| 200 | Linalool | 154.25 | 1 | 1 | 2.7 | 0 |
| 201 | Nevadensin | 344.32 | 7 | 2 | 3 | 0 |
| 202 | 7-Epi-sesquithujene | 204.35 | 0 | 0 | 3.37 | 1 |
| 203 | Selina-4(15),7(11)-diene | 204.35 | 0 | 0 | 3.31 | 1 |
| 204 | Codeine | 299.36 | 4 | 1 | 2.67 | 0 |
| 205 | Sulfanilamide | 172.2 | 3 | 2 | 0.61 | 0 |
| 206 | Avicularin | 434.35 | 11 | 7 | 1.86 | 2 |
| 207 | Pipercyclobutanamide A(rel) | 570.68 | 6 | 0 | 5.26 | 1 |
| 208 | (−)-Cubebin | 356.37 | 6 | 1 | 3.18 | 0 |
| 209 | (−)-3,4-Dimethoxy-3,4-desmethylenedioxycubebin | 372.41 | 6 | 1 | 3.06 | 0 |
| 210 | Piperine | 285.34 | 3 | 0 | 3.42 | 0 |
| 211 | Pellitorine | 223.35 | 1 | 1 | 3.61 | 0 |
| 212 | Gaudichaudianic acid, (− rac) | 340.46 | 3 | 1 | 3.85 | 1 |
| 213 | Sarmentosumin A | 680.74 | 8 | 6 | 3.7 | 2 |
| 214 | Sarmentosumin B | 680.74 | 8 | 6 | 3.97 | 2 |
| 215 | Sarmentosumin C | 786.86 | 9 | 7 | 4.13 | 3 |
| 216 | Sarmentosumin D | 786.86 | 9 | 7 | 4.06 | 3 |
| 217 | Isochamanetin | 362.38 | 5 | 3 | 2.68 | 0 |
| 218 | 7-Methoxychamanetin | 376.4 | 5 | 2 | 3.2 | 0 |
| 219 | Dichamanetin | 468.5 | 6 | 4 | 3.01 | 0 |
| 220 | 7-Methoxydichamanetin | 482.52 | 6 | 3 | 3.58 | 0 |
| 221 | 5″-(2⁗-Hydroxybenzyl)uvarinol | 694.77 | 8 | 5 | 4.16 | 1 |
| 222 | 2,4-Dodecadienamide | 195.3 | 1 | 1 | 2.87 | 0 |
| 223 | 7-Methoxyisochamanetin | 376.4 | 5 | 2 | 3.5 | 0 |
| 224 | (2E,4E)-N-[2-(methylsulfinyl)ethyl]-2,4-decadienamide | 257.39 | 2 | 1 | 3.05 | 0 |
| 225 | (2E,4E)-N-[(4-hydroxy-3-methoxyphenyl)ethyl]-2,4-decadienamide | 317.42 | 3 | 2 | 3.76 | 0 |
| 226 | 3-(4-Hydroxy-3,5-dimethoxyphenyl)propanoylpyrrole | 275.3 | 4 | 1 | 2.63 | 0 |
| 227 | 3-(3,4,5-Timethoxyphenyl)propanoylpyrrole | 289.33 | 4 | 0 | 3.21 | 0 |
| 228 | 1-[(2E,4E,6E)-2,4,6-dodecatrienoyl]pyrrolidine | 247.38 | 1 | 0 | 3.86 | 0 |
| 229 | 1-[(2E,4Z,8E)-9-(3,4-methylenedioxyphenyl)-2,4,8-nonatrienoyl]pyrrolidine | 325.4 | 3 | 0 | 3.88 | 0 |
| 230 | 1-[(2E,4E,10E)-10-(3,4-methylenedioxyphenyl)-2,4,10-undecatrienoyl]pyrrolidine | 353.45 | 3 | 0 | 4.53 | 0 |
| 231 | 1-[(4E,10E)-11-(3,4-methylenedioxyphenyl)-4,10-undecadienoyl]pyrrolidine | 355.47 | 3 | 0 | 4.6 | 0 |
| 232 | 1-[(9E)-10-(3,4-methylenedioxyphenyl)-9-decenoyl]pyrrolidine | 343.46 | 3 | 0 | 4.43 | 0 |
| 233 | 1-[(2E,4E)-2,4-decadienoyl]pyrrolidine | 221.34 | 1 | 0 | 3.45 | 0 |
| 234 | 1-[(2E,4E)-2,4-dodecadienoyl]pyrrolidine | 249.39 | 1 | 0 | 3.95 | 0 |
| 235 | 1-[(2E)-7-(3,4-methylenedioxyphenyl)-2-heptenoyl]pyrrolidine | 301.38 | 3 | 0 | 3.64 | 0 |
| 236 | 1-[(2E,4E)-7-(3,4-methylenedioxyphenyl)-2,4-heptadienoyl]pyrrolidine | 299.36 | 3 | 0 | 3.65 | 0 |
| 237 | 1-[(2E,8E)-9-(3,4-methylenedioxyphenyl)-2,8-nonadienoyl]pyrrolidine | 327.42 | 3 | 0 | 4.04 | 0 |
| 238 | 1-[(8E)-9-(3,4-methylenedioxyphenyl)-8-nonenoyl]pyrrolidine | 329.43 | 3 | 0 | 4.16 | 0 |
| 239 | 1-[(2E,4E,8E)-9-(3,4-methylenedioxyphenyl)-2,4,8-nonatrienoyl]pyrrolidine | 325.4 | 3 | 0 | 3.88 | 0 |
| 240 | 1-[(2E,4E)-11-(3,4-methylenedioxyphenyl)-2,4-undecadienoyl]pyrrolidine | 355.47 | 3 | 0 | 4.51 | 0 |
| 241 | 1-[(2E,10E)-11-(3,4-methylenedioxyphenyl)-2,10-undecadienoyl]pyrrolidine | 355.47 | 3 | 0 | 4.16 | 0 |
| 242 | (2E,4E)-N-isobutyl-2,4-dodecadienamide | 251.41 | 1 | 1 | 4.06 | 0 |
| 243 | (2E,4E)-N-isobutyl-7-(3,4-methylenedioxyphenyl)-hepta-2,4-dienamide | 301.38 | 3 | 1 | 3.67 | 0 |
| 244 | (8E)-N-isobutyl-9-(3,4-methylenedioxyphenyl)nona-8-enamide | 331.45 | 3 | 1 | 4.18 | 0 |
| 245 | (2E,4E,8E)-N-isobutyl-11-(3,4-methylenedioxyphenyl)undeca-2,4,8-trienamide | 355.47 | 3 | 1 | 4.42 | 0 |
| 246 | N-trans-sinapoyltyramine | 343.37 | 5 | 3 | 2.71 | 0 |
| 247 | Dihydrocubebin, rel- | 358.39 | 6 | 2 | 3.19 | 0 |
| 248 | Justiflorinol | 356.33 | 7 | 1 | 2.76 | 0 |
| 249 | (−)-Sanguinolignan A | 384.34 | 8 | 1 | 2.82 | 0 |
| 250 | (−)-Sanguinolignan B | 384.34 | 8 | 1 | 2.79 | 0 |
| 251 | (−)-Sanguinolignan C | 442.42 | 9 | 0 | 3.36 | 0 |
| 252 | (−)-Sanguinolignan D | 426.37 | 9 | 0 | 2.74 | 0 |
| 253 | (7′S)-parabenzlactone | 370.35 | 7 | 1 | 3.12 | 0 |
| 254 | Flavokawain B | 284.31 | 4 | 1 | 2.63 | 0 |
| 255 | Methyl 3,4-dihydroxy-5-(3′-methyl-2′-butenyl)benzoate | 236.26 | 4 | 2 | 2.62 | 0 |
| 256 | Pipercallosidine | 303.4 | 3 | 1 | 3.76 | 0 |
| 257 | Kadsurenin C | 358.43 | 5 | 1 | 3.37 | 0 |
| 258 | Kadsurenin K | 342.39 | 5 | 1 | 3.02 | 0 |
| 259 | Kadsurenin L | 400.46 | 6 | 0 | 3.42 | 0 |
| 260 | Pipercallosine | 329.43 | 3 | 1 | 4.04 | 0 |
| 261 | (S)-1′-methylhexyl caffeate | 278.34 | 4 | 2 | 3.41 | 0 |
| 262 | Futoenone | 340.37 | 5 | 0 | 3.15 | 0 |
| 263 | (S)-1′-methylbutyl caffeate | 250.29 | 4 | 2 | 2.81 | 0 |
| 264 | (S)-1′-methyloctyl caffeate | 306.4 | 4 | 2 | 3.66 | 0 |
| 265 | Futokadsurin B | 356.41 | 5 | 0 | 3.89 | 0 |
| 266 | Futokadsurin C | 356.41 | 5 | 0 | 3.77 | 0 |
| 267 | Futokadsurin A | 358.43 | 5 | 1 | 3.46 | 0 |
| 268 | Burchellin | 340.37 | 5 | 0 | 3.26 | 0 |
| 269 | Piperonal | 150.13 | 3 | 0 | 1.6 | 0 |
| 270 | (−)-Cubenol | 222.37 | 1 | 1 | 3.24 | 0 |
| 271 | (E,E)-piperic acid | 218.21 | 4 | 1 | 2.2 | 0 |
| 272 | Trans-cinnamic acid | 148.16 | 2 | 1 | 1.55 | 0 |
| 273 | Pinocembrin chalcone | 256.25 | 4 | 3 | 1.09 | 0 |
| 274 | Piperlactam S | 295.29 | 4 | 1 | 2.54 | 0 |
| 275 | Asebogenin | 288.3 | 5 | 3 | 1.97 | 0 |
| 276 | Epicocconone | 410.42 | 7 | 2 | 3.17 | 0 |
| 277 | (+)-Sesamin | 354.35 | 6 | 0 | 3.46 | 0 |
| 278 | Pipataline | 288.42 | 2 | 0 | 4.61 | 1 |
| 279 | (−)-Antofine | 363.45 | 4 | 0 | 3.8 | 0 |
| 280 | Benzoic acid | 122.12 | 2 | 1 | 1.11 | 0 |
| 281 | Ethyl butyrate | 116.16 | 2 | 0 | 1.9 | 0 |
| 282 | N-feruloyltyramine | 313.35 | 4 | 3 | 2.58 | 0 |
| 283 | Kadsurenin M | 328.36 | 5 | 0 | 3.15 | 0 |
| 284 | (−)-Epicubenol | 222.37 | 1 | 1 | 3.11 | 0 |
| 285 | Acacetin-8-C-neohesperidoside | 592.55 | 14 | 8 | 1.49 | 3 |
| 286 | Monocerin | 308.33 | 6 | 1 | 2.99 | 0 |
| 287 | (2S,3aR,9bR)-6,7-dihydroxy-8-methoxy-2-propyl-3,3a-dihydro-2H-furo[3,2-c]isochromen-5(9bH)-one | 294.3 | 6 | 2 | 2.51 | 0 |
| 288 | Fusarentin 6,7-dimethyl ether | 310.34 | 6 | 2 | 2.85 | 0 |
| 289 | Fusarentin 6-methyl ether | 296.32 | 6 | 3 | 2.27 | 0 |
| 290 | (3R,4R)-4,8-dihydroxy-3-((R)-2-hydroxypentyl)-6,7-dimethoxyisochroman-1-one | 326.34 | 7 | 3 | 2.59 | 0 |
| 291 | Colletotrialide, (+)- | 308.33 | 6 | 1 | 2.49 | 0 |
| 292 | Galgravin | 372.45 | 5 | 0 | 3.97 | 0 |
| 293 | 9,10-Epoxy-18-hydroxyoctadecanoic acid | 314.46 | 4 | 2 | 3.8 | 0 |
| 294 | Pinocembrin | 256.25 | 4 | 2 | 2.11 | 0 |
| 295 | 1,4-Cineole | 154.25 | 1 | 0 | 2.68 | 0 |
| 296 | Sodium benzoate | 144.1 | 2 | 0 | − 11.15 | 0 |
| 297 | 9,10,18-Trihydroxyoctadecanoic acid | 332.48 | 5 | 4 | 3.25 | 0 |
| 298 | Kavapyrone | 244.24 | 4 | 0 | 2.6 | 0 |
| 299 | Alpha-cubebene | 204.35 | 0 | 0 | 3.4 | 1 |
| 300 | Acuminatin | 340.41 | 4 | 0 | 4.05 | 0 |
| 301 | Jerantinine E | 384.47 | 5 | 2 | 3.52 | 0 |
| 302 | Jerantinine F | 398.45 | 6 | 2 | 3.4 | 0 |
| 303 | Jerantinine C | 396.44 | 5 | 2 | 3.13 | 0 |
| 304 | Jerantinine B | 398.45 | 6 | 2 | 3.29 | 0 |
| 305 | Jerantinine D | 412.44 | 6 | 2 | 3.07 | 0 |
| 306 | Jerantinine A | 382.45 | 5 | 2 | 3.49 | 0 |
| 307 | Tabernaemontanine | 354.44 | 4 | 1 | 2.79 | 0 |
| 308 | Mangiferin | 422.34 | 11 | 8 | 0.89 | 2 |
| 309 | Plectranthol A | 450.52 | 6 | 4 | 3.49 | 0 |
| 310 | Plectranthol B | 536.66 | 7 | 3 | 4.59 | 2 |
| 311 | 11,20-Dihydroxysugiol | 332.43 | 4 | 3 | 2.87 | 0 |
| 312 | 11-Hydroxysugiol | 316.43 | 3 | 2 | 3.15 | 0 |
| 313 | Abietatriene | 270.45 | 0 | 0 | 3.86 | 1 |
| 314 | 7-oxo-10α-cucurbitadienol | 440.7 | 2 | 1 | 4.65 | 1 |
Table 5.
The list of phytochemical those satisfies the Lipinski rule of 5 and their structures in SMILES format.
| S.·no | Compound | SMILES (chEBI) |
|---|---|---|
| 1 | Gingerenone B | C = 1(C(= CC = C(C1)CCC(/C = C/CCC = 2C = C(C(= C(C2)OC)O)OC)=O)O)OC |
| 2 | Beta-sesquiphellandrene | [H][C@@]1(CCC(= C)C = C1)[C@@H](C)CCC = C(C)C |
| 3 | 3-(3,4-Dimethoxyphenyl)-4-[(Z)-3,4-dimethoxystyryl]cyclohex-1-ene | COc1ccc(\C = C/C2CCC = CC2c2ccc(OC)c(OC)c2)cc1OC |
| 4 | 3-(3,4-Dimethoxyphenyl)-4-[(E)-3,4-dimethoxystyryl]cyclohex-1-ene | COc1ccc(\C = C\C2CCC = CC2c2ccc(OC)c(OC)c2)cc1OC |
| 5 | Pinocarveol | CC1(C)C2CC(O)C(= C)C1C2 |
| 6 | Zerumboneoxide | [H][C@]12CC(C)(C)\C = C\C(= O)\C(C)=C\CC[C@@]1(C)O2 |
| 7 | Ramonanin A, (rel)- | COc1cc(ccc1O)[C@H]1O[C@@H](c2ccc(O)c(OC)c2)[C@@]2(CCC3 = C(C2)[C@H](O[C@H]3c2ccc(O)c(OC)c2)c2ccc(O)c(OC)c2)C1 = C |
| 8 | Ramonanin B, (rel)- | COc1cc(ccc1O)[C@@H]1O[C@H](c2ccc(O)c(OC)c2)[C@@]2(CCC3 = C(C2)[C@H](O[C@H]3c2ccc(O)c(OC)c2)c2ccc(O)c(OC)c2)C1 = C |
| 9 | Ramonanin C, (rel)- | COc1cc(ccc1O)[C@H]1O[C@@H](c2ccc(O)c(OC)c2)[C@@]2(CCC3 = C(C2)[C@@H](O[C@@H]3c2ccc(O)c(OC)c2)c2ccc(O)c(OC)c2)C1 = C |
| 10 | Ramonanin D, (rel)- | COc1cc(ccc1O)[C@H]1O[C@@H](c2ccc(O)c(OC)c2)[C@@]2(CCC3 = C(C2)[C@H](O[C@@H]3c2ccc(O)c(OC)c2)c2ccc(O)c(OC)c2)C1 = C |
| 11 | Zerumbone | C\C1 = C/CC(C)(C)\C = C\C(= O)\C(C)=C\CC1 |
| 12 | 5-Hydroxyzerumbone | C\C1 = C/CC(C)(C)\C = C\C(= O)\C(C)=C\C(O)C1 |
| 13 | (2E,6E)-hedycaryol | C\C1 = C/CC\C(C)=C\C[C@@H](CC1)C(C)(C)O |
| 14 | Dodecane | CCCCCCCCCCCC |
| 15 | 7,4′-Dimethylkaempferol | C12 = C(OC(C3 = CC = C(OC)C = C3)=C(C1 = O)O)C = C(OC)C = C2O |
| 16 | Zingiberene | [H][C@@]1(CC = C(C)C = C1)[C@@H](C)CCC = C(C)C |
| 17 | Glucoputranjivin(1-) | [C@H]1(O[C@@H]([C@@H](O)[C@@H]([C@H]1O)O)CO)S/C(= N\OS([O-])(= O)=O)/C(C)C |
| 18 | Glucoputranjivin | [C@H]1(O[C@@H]([C@@H](O)[C@@H]([C@H]1O)O)CO)S/C(= N\OS(O)(= O)=O)/C(C)C |
| 19 | All-cis-octadeca-6,9,12,15-tetraenoic acid | CC\C = C/C\C = C/C\C = C/C\C = C/CCCCC(O)=O |
| 20 | Pipataline | O1C = 2C = C(\C = C\CCCCCCCCCC)C = CC2OC1 |
| 21 | Pipercyclobutanamide A(rel) | O = C(\C = C/[C@@H]1[C@@H](\C = C\c2ccc3OCOc3c2)[C@@H]([C@H]1c1ccc2OCOc2c1)C(= O)N1CCCCC1)N1CCCCC1 |
| 22 | Pellitorine | CCCCC\C = C\C = C\C(= O)NCC(C)C |
| 23 | Gaudichaudianic acid, (− rac) | CC(C)=CCCC1(C)Oc2c(CC = C(C)C)cc(cc2C = C1)C(O)=O |
| 24 | Isochamanetin | Oc1ccccc1Cc1c(O)cc2O[C@@H](CC(= O)c2c1O)c1ccccc1 |
| 25 | 7-Methoxychamanetin | COc1cc(O)c2C(= O)C[C@H](Oc2c1Cc1ccccc1O)c1ccccc1 |
| 26 | Dichamanetin | Oc1ccccc1Cc1c(O)c(Cc2ccccc2O)c2O[C@@H](CC(= O)c2c1O)c1ccccc1 |
| 27 | 7-Methoxydichamanetin | COc1c(Cc2ccccc2O)c(O)c2C(= O)C[C@H](Oc2c1Cc1ccccc1O)c1ccccc1 |
| 28 | 5″-(2⁗-Hydroxybenzyl)uvarinol | COc1c(Cc2cc(Cc3ccccc3O)ccc2O)c(O)c2C(= O)C[C@H](Oc2c1Cc1cc(Cc2ccccc2O)ccc1O)c1ccccc1 |
| 29 | 2,4-Dodecadienamide | CCCCCCC\C = C\C = C\C(N)=O |
| 30 | 7-Methoxyisochamanetin | COc1cc2O[C@@H](CC(= O)c2c(O)c1Cc1ccccc1O)c1ccccc1 |
| 31 | (2E,4E)-N-[2-(methylsulfinyl)ethyl]-2,4-decadienamide | CCCCC\C = C\C = C\C(= O)NCCS(C)=O |
| 32 | (2E,4E)-N-[(4-hydroxy-3-methoxyphenyl)ethyl]-2,4-decadienamide | CCCCC\C = C\C = C\C(= O)NCCc1ccc(O)c(OC)c1 |
| 33 | 3-(4-Hydroxy-3,5-dimethoxyphenyl)propanoylpyrrole | COc1cc(CCC(= O)n2cccc2)cc(OC)c1O |
| 34 | 3-(3,4,5-Timethoxyphenyl)propanoylpyrrole | COc1cc(CCC(= O)n2cccc2)cc(OC)c1OC |
| 35 | 1-[(2E,4E,6E)-2,4,6-dodecatrienoyl]pyrrolidine | CCCCC\C = C\C = C\C = C\C(= O)N1CCCC1 |
| 36 | 1-[(2E,4Z,8E)-9-(3,4-methylenedioxyphenyl)-2,4,8-nonatrienoyl]pyrrolidine | O = C(\C = C\C = C/CC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 37 | 1-[(2E,4E,10E)-10-(3,4-methylenedioxyphenyl)-2,4,10-undecatrienoyl]pyrrolidine | O = C(\C = C\C = C\CCCC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 38 | 1-[(4E,10E)-11-(3,4-methylenedioxyphenyl)-4,10-undecadienoyl]pyrrolidine | O = C(CC\C = C\CCCC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 39 | 1-[(9E)-10-(3,4-methylenedioxyphenyl)-9-decenoyl]pyrrolidine | O = C(CCCCCCC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 40 | 1-[(2E,4E)-2,4-decadienoyl]pyrrolidine | CCCCC\C = C\C = C\C(= O)N1CCCC1 |
| 41 | 1-[(2E,4E)-2,4-dodecadienoyl]pyrrolidine | CCCCCCC\C = C\C = C\C(= O)N1CCCC1 |
| 42 | 1-[(2E)-7-(3,4-methylenedioxyphenyl)-2-heptenoyl]pyrrolidine | O = C(\C = C\CCCCc1ccc2OCOc2c1)N1CCCC1 |
| 43 | 1-[(2E,4E)-7-(3,4-methylenedioxyphenyl)-2,4-heptadienoyl]pyrrolidine | O = C(\C = C\C = C\CCc1ccc2OCOc2c1)N1CCCC1 |
| 44 | 1-[(2E,8E)-9-(3,4-methylenedioxyphenyl)-2,8-nonadienoyl]pyrrolidine | O = C(\C = C\CCCC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 45 | 1-[(8E)-9-(3,4-methylenedioxyphenyl)-8-nonenoyl]pyrrolidine | O = C(CCCCCC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 46 | 1-[(2E,4E,8E)-9-(3,4-methylenedioxyphenyl)-2,4,8-nonatrienoyl]pyrrolidine | O = C(\C = C\C = C\CC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 47 | 1-[(2E,4E)-11-(3,4-methylenedioxyphenyl)-2,4-undecadienoyl]pyrrolidine | O = C(\C = C\C = C\CCCCCCc1ccc2OCOc2c1)N1CCCC1 |
| 48 | 1-[(2E,10E)-11-(3,4-methylenedioxyphenyl)-2,10-undecadienoyl]pyrrolidine | O = C(\C = C\CCCCCC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 49 | (2E,4E)-N-isobutyl-2,4-dodecadienamide | CCCCCCC\C = C\C = C\C(= O)NCC(C)C |
| 50 | (2E,4E)-N-isobutyl-7-(3,4-methylenedioxyphenyl)-hepta-2,4-dienamide | CC(C)CNC(= O)\C = C\C = C\CCc1ccc2OCOc2c1 |
| 51 | (8E)-N-isobutyl-9-(3,4-methylenedioxyphenyl)nona-8-enamide | CC(C)CNC(= O)CCCCCC\C = C\c1ccc2OCOc2c1 |
| 52 | (2E,4E,8E)-N-isobutyl-11-(3,4-methylenedioxyphenyl)undeca-2,4,8-trienamide | CC(C)CNC(= O)\C = C\C = C\CC\C = C\CCc1ccc2OCOc2c1 |
| 53 | N-trans-sinapoyltyramine | COc1cc(cc(OC)c1O)\C = C\C(= O)NCCc1ccc(O)cc1 |
| 54 | Dihydrocubebin, rel- | OC[C@@H](Cc1ccc2OCOc2c1)[C@@H](CO)Cc1ccc2OCOc2c1 |
| 55 | Justiflorinol | OCC(CC(= O)c1ccc2OCOc2c1)C(= O)c1ccc2OCOc2c1 |
| 56 | (−)-Sanguinolignan A | O[C@@H]([C@H]1COC(= O)[C@@H]1C(= O)c1ccc2OCOc2c1)c1ccc2OCOc2c1 |
| 57 | (−)-Sanguinolignan B | O[C@@H]([C@@H]1COC(= O)[C@@H]1C(= O)c1ccc2OCOc2c1)c1ccc2OCOc2c1 |
| 58 | (−)-Sanguinolignan C | COc1ccc(cc1OC)[C@@H](OC(C)=O)[C@H]1COC(= O)[C@@H]1C(= O)c1ccc2OCOc2c1 |
| 59 | (−)-Sanguinolignan D | CC(= O)O[C@@H]([C@@H]1[C@H](COC1 = O)C(= O)c1ccc2OCOc2c1)c1ccc2OCOc2c1 |
| 60 | (7′S)-parabenzlactone | O[C@@H]([C@H]1COC(= O)[C@@H]1Cc1ccc2OCOc2c1)c1ccc2OCOc2c1 |
| 61 | Flavokawain B | COc1cc(O)c(C(= O)\C = C\c2ccccc2)c(OC)c1 |
| 62 | Methyl 3,4-dihydroxy-5-(3′-methyl-2′-butenyl)benzoate | COC(= O)c1cc(O)c(O)c(CC = C(C)C)c1 |
| 63 | Pipercallosidine | C(/C = C/CCCCC = 1C = C2C(= CC1)OCO2)(NCC(C)C)=O |
| 64 | Kadsurenin C | O([C@@]12[C@@H]([C@H]([C@@]([C@H]1O)(C(= O)C(= C2)CC = C)[H])C3 = CC(OC)=C(OC)C = C3)C)C |
| 65 | Kadsurenin K | O([C@@]12[C@@H]([C@H]([C@@](C1 = O)(C(= O)C(= C2)CC = C)[H])C3 = CC(OC)=C(O)C = C3)C)C |
| 66 | Kadsurenin L | O([C@@]12[C@@H]([C@H]([C@@]([C@@H]1OC(= O)C)(C(= O)C(= C2)CC = C)[H])C3 = CC(OC)=C(OC)C = C3)C)C |
| 67 | Pipercallosine | C(/C = C/C = C/CCCCC1 = CC = C2C(= C1)OCO2)(= O)NCC(C)C |
| 68 | (S)-1′-methylhexyl caffeate | CCCCC[C@H](C)OC(= O)\C = C\c1ccc(O)c(O)c1 |
| 69 | Futoenone | O1[C@@H]2C[C@@]3([C@@H]([C@H](C2)C4 = CC = 5OCOC5C = C4)C)C1 = CC(= O)C(OC)=C3 |
| 70 | (−)-Cubebin | [H][C@@]1(CO[C@H](O)[C@]1([H])Cc1ccc2OCOc2c1)Cc1ccc2OCOc2c1 |
| 71 | (−)-3,4-Dimethoxy-3,4-desmethylenedioxycubebin | [H][C@@]1(COC(O)[C@]1([H])Cc1ccc2OCOc2c1)Cc1ccc(OC)c(OC)c1 |
| 72 | (S)-1′-methylbutyl caffeate | CCC[C@H](C)OC(= O)\C = C\c1ccc(O)c(O)c1 |
| 73 | (S)-1′-methyloctyl caffeate | CCCCCCC[C@H](C)OC(= O)\C = C\c1ccc(O)c(O)c1 |
| 74 | Futokadsurin B | COc1ccc(cc1OC)[C@@H]1O[C@@H]([C@H](C)[C@H]1C)c1ccc2OCOc2c1 |
| 75 | Futokadsurin C | COc1ccc(cc1OC)[C@H]1O[C@H]([C@H](C)[C@@H]1C)c1ccc2OCOc2c1 |
| 76 | Piperine | O = C(\C = C\C = C\c1ccc2OCOc2c1)N1CCCCC1 |
| 77 | Futokadsurin A | COc1cc(ccc1O)[C@@H]1O[C@@H]([C@@H](C)[C@@H]1C)c1ccc(OC)c(OC)c1 |
| 78 | Burchellin | O1[C@@H]([C@H]([C@]2(C1 = CC(= O)C(OC)=C2)CC = C)C)C3 = CC = 4OCOC4C = C3 |
| 79 | Trans-cinnamic acid | OC(= O)\C = C\c1ccccc1 |
| 80 | Pinocembrin chalcone | Oc1cc(O)c(C(= O)\C = C\c2ccccc2)c(O)c1 |
| 81 | Piperlactam S | O(C = 1C2 = C3C(= CC = 4C2 = CC = CC4)N(C(C3 = CC1O)=O)OC)C |
| 82 | Asebogenin | COc1cc(O)c(C(= O)CCc2ccc(O)cc2)c(O)c1 |
| 83 | (+)-Sesamin | [C@]12([C@@]([C@H](OC1)C3 = CC4 = C(C = C3)OCO4)(CO[C@@H]2C5 = CC6 = C(C = C5)OCO6)[H])[H] |
| 84 | Benzoic acid | OC(= O)c1ccccc1 |
| 85 | N-feruloyltyramine | COc1cc(\C = C\C(= O)NCCc2ccc(O)cc2)ccc1O |
| 86 | Kadsurenin M | O1[C@@H]([C@H](C2 = C1C(OC)=CC(= C2)C(= O)[H])C)C3 = CC(OC)=C(OC)C = C3 |
| 87 | (−)-Epicubenol | [C@]12([C@@H](CC[C@H]([C@]2(CCC(= C1)C)O)C)C(C)C)[H] |
| 88 | Monocerin | [H][C@@]12C[C@H](CCC)O[C@]1([H])c1cc(OC)c(OC)c(O)c1C(= O)O2 |
| 89 | (2S,3aR,9bR)-6,7-dihydroxy-8-methoxy-2-propyl-3,3a-dihydro-2H-furo[3,2-c]isochromen-5(9bH)-one | [H][C@@]12C[C@H](CCC)O[C@]1([H])c1cc(OC)c(O)c(O)c1C(= O)O2 |
| 90 | Fusarentin 6,7-dimethyl ether | CCC[C@H](O)C[C@@H]1Cc2cc(OC)c(OC)c(O)c2C(= O)O1 |
| 91 | Fusarentin 6-methyl ether | CCC[C@H](O)C[C@@H]1Cc2cc(OC)c(O)c(O)c2C(= O)O1 |
| 92 | (3R,4R)-4,8-dihydroxy-3-((R)-2-hydroxypentyl)-6,7-dimethoxyisochroman-1-one | CCC[C@@H](O)C[C@H]1OC(= O)c2c(O)c(OC)c(OC)cc2[C@H]1O |
| 93 | Colletotrialide, (+)- | CCCC(= O)CC[C@H]1OC(= O)c2c(O)c(OC)c(OC)cc12 |
| 94 | Galgravin | C1(= C(C = CC(= C1)[C@@H]2O[C@@H]([C@H]([C@H]2C)C)C3 = CC = C(C(= C3)OC)OC)OC)OC |
| 95 | Piperonal | [H]C(= O)c1ccc2OCOc2c1 |
| 96 | Pinocembrin | Oc1cc(O)c2C(= O)C[C@H](Oc2c1)c1ccccc1 |
| 97 | 1,4-Cineole | C1C[C@]2(CC[C@]1(C)O2)C(C)C |
| 98 | Sodium benzoate | C(C = 1C = CC = CC1)([O-])=O.[Na +] |
| 99 | (−)-Cubenol | [C@]12([C@@H](CC[C@H]([C@@]2(CCC(= C1)C)O)C)C(C)C)[H] |
| 100 | (E,E)-piperic acid | OC(= O)\C = C\C = C\c1ccc2OCOc2c1 |
| 101 | Kavapyrone | COc1cc(oc(= O)c1)[C@@H]1O[C@H]1c1ccccc1 |
| 102 | Alpha-cubebene | C1C[C@H]([C@]2([C@]3([C@@H]1C)CC = C([C@]23[H])C)[H])C(C)C |
| 103 | Acuminatin | O1[C@H]([C@@H](C2 = C1C(OC)=CC(= C2)/C = C/C)C)C3 = CC(OC)=C(OC)C = C3 |
| 104 | Dihydrobetulinic acid | [H][C@]12CC[C@]3([H])[C@@]4(C)CC[C@H](O)C(C)(C)[C@]4([H])CC[C@@]3(C)[C@]1(C)CC[C@]1(CC[C@@H](C(C)C)[C@]21[H])C(O)=O |
| 105 | Platanic acid | [H][C@]12CC[C@]3([H])[C@@]4(C)CC[C@H](O)C(C)(C)[C@]4([H])CC[C@@]3(C)[C@]1(C)CC[C@]1(CC[C@@H](C(C)=O)[C@]21[H])C(O)=O |
| 106 | Canophyllal | [C@]12([C@]([C@]3([C@@](CC1)(CCC(C3)(C)C)C(= O)[H])[H])(CC[C@@]4([C@@]2(CC[C@]5([C@]4(CCC([C@@H]5C)=O)[H])C)[H])C)C)C |
| 107 | Cinnamic acid | [H]C(= Cc1ccccc1)C(O)=O |
| 108 | Methyl linolenate | CC/C = C\C/C = C\C/C = C\CCCCCCCC(= O)OC |
| 109 | Zerumbone | C\C1 = C/CC(C)(C)\C = C\C(= O)\C(C)=C\CC1 |
| 110 | (Z)-3-phenyl-2-propenal | C1 = CC = C(C = C1)/C = C\C = O |
| 111 | (E)-2-methoxycinnamic acid | COC = 1C = CC = CC1/C = C/C(= O)O |
| 112 | Betulinic acid | [H][C@]12CC[C@]3([H])[C@@]4(C)CC[C@H](O)C(C)(C)[C@]4([H])CC[C@@]3(C)[C@]1(C)CC[C@]1(CC[C@@H](C(C)=C)[C@]21[H])C(O)=O |
| 113 | Heptacosan-1-ol | C(CCCCCCCCCCCCCCCCO)CCCCCCCCCC |
| 114 | Pyrethrin I | CC(C)=C[C@@H]1[C@@H](C(= O)O[C@H]2CC(= O)C(C\C = C/C = C)=C2C)C1(C)C |
| 115 | Pyrethrin II | COC(= O)C(\C)=C\[C@@H]1[C@@H](C(= O)O[C@H]2CC(= O)C(C\C = C/C = C)=C2C)C1(C)C |
| 116 | 14-Deoxy-11,12-didehydroandrographolide | [H][C@]12CCC(= C)[C@@H](\C = C\C3 = CCOC3 = O)[C@]1(C)CC[C@@H](O)[C@@]2(C)CO |
| 117 | Andrographolide | [H][C@]12CCC(= C)[C@@H](C\C = C3/[C@H](O)COC3 = O)[C@]1(C)CC[C@@H](O)[C@@]2(C)CO |
| 118 | Dihydroferulic acid | C = 1(C = C(C(= CC1)O)OC)CCC(= O)O |
| 119 | Mesembryanthemoidigenic acid | [C@@]12(C([C@@]3(C[C@@](CO)(C)CC[C@@]3(CC1)C(= O)O)[H])=CC[C@@]4([C@]5(CC[C@@H](C([C@@]5(CC[C@@]24C)[H])(C)C)O)C)[H])C |
| 120 | Methyl N-methylanthranilate | CNC1 = CC = CC = C1C(OC)=O |
| 121 | Delta-elemene | [C@@H]1(C = C(CC[C@@]1(C = C)C)C(C)C)C(C)=C |
| 122 | Decussatin | COc1cc(O)c2c(c1)oc1ccc(OC)c(OC)c1c2 = O |
| 123 | Capensinidin | COc1cc(cc(OC)c1O)-c1[o +]c2cc(O)cc(OC)c2cc1O |
| 124 | Termilignan B | Oc1ccc(CC(= C)C(= C)Cc2ccc3OCOc3c2)cc1 |
| 125 | (Z)-9-hydroxybenzo[c]oxepin-3(1H)-one | Oc1cccc2C = CC(= O)OCc12 |
| 126 | Cyclosordariolone, (rac)- | CC1(O)C(= O)C = Cc2c(CO)c(O)ccc12 |
| 127 | (R)-3-Hydroxy-1-[(R)-4-hydroxy-1,3-dihydroisobenzofuran-1-yl]butan-2-one | C[C@@H](O)C(= O)C[C@H]1OCc2c(O)cccc12 |
| 128 | (R)-3-Hydroxy-1-[(S)-4-hydroxy-1,3-dihydroisobenzofuran-1-yl]butan-2-one | C[C@@H](O)C(= O)C[C@@H]1OCc2c(O)cccc12 |
| 129 | (E)-2-(Hydroxymethyl)-3-(4-hydroxypent-1-enyl)phenol | C[C@@H](O)C\C = C\c1cccc(O)c1CO |
| 130 | 1-(3,9-Dihydroxy-1,3-dihydrobenzo[c]oxepin-3-yl)ethanone, (rac)- | CC(= O)C1(O)OCc2c(O)cccc2C = C1 |
| 131 | Pestalospirane A | C[C@H]1O[C@]2(OCc3c(O)cccc3C = C2)[C@@H](C)O[C@@]11OCc2c(O)cccc2C = C1 |
| 132 | Pestalospirane B | C[C@H]1O[C@]2(OCc3c(O)cccc3C = C2)[C@@H](C)O[C@]11OCc2c(O)cccc2C = C1 |
| 133 | Methyl 3,4,5-trihydroxybenzoate | OC1 = CC(= CC(O)=C1O)C(OC)=O |
| 134 | Ursolic acid | C[C@@H]1CC[C@@]2(CC[C@]3(C)C(= CC[C@@H]4[C@@]5(C)CC[C@H](O)C(C)(C)[C@@H]5CC[C@@]34C)[C@@H]2[C@H]1C)C(O)=O |
| 135 | Arecoline | COC(= O)C1 = CCCN(C)C1 |
| 136 | Propyl benzoate | C = 1C = CC(= CC1)C(= O)OCCC |
| 137 | Plectranthol A | CC(C)c1cc2C = C[C@@]3(C)C(= CCC[C@]3(C)COC(= O)c3ccc(O)c(O)c3)c2c(O)c1O |
| 138 | 11,20-Dihydroxysugiol | C1 = C(C(C)C)C(= C(C2 = C1C(C[C@@]3([C@@]2(CCCC3(C)C)CO)[H])=O)O)O |
| 139 | 11-Hydroxysugiol | C1 = C(C(C)C)C(= C(C2 = C1C(C[C@@]3([C@@]2(CCCC3(C)C)C)[H])=O)O)O |
| 140 | Abietatriene | CC(C)c1ccc2c(CC[C@H]3C(C)(C)CCC[C@]23C)c1 |
| 141 | 1beta-hydroxy arbusculin A | [H][C@@]12CC[C@@]3(C)[C@H](O)CC[C@@](C)(O)[C@]3([H])[C@@]1([H])OC(= O)C2 = C |
| 142 | Alantolactone | [H][C@@]12C[C@@]3(C)CCC[C@H](C)C3 = C[C@]1([H])C(= C)C(= O)O2 |
| 143 | Dehydrocostus lactone | [H][C@@]12CCC(= C)[C@]1([H])[C@H]1OC(= O)C(= C)[C@@H]1CCC2 = C |
| 144 | Costunolide | C\C1 = C/CC\C(C)=C\[C@H]2OC(= O)C(= C)[C@@H]2CC1 |
| 145 | 3-Hydroxyhexane-2,5-dione | CC(CC(C(C)=O)O)=O |
| 146 | Heptacosan-1-ol | C(CCCCCCCCCCCCCCCCO)CCCCCCCCCC |
| 147 | Cyclotetradecane | C1CCCCCCCCCCCCC1 |
| 148 | Cycloeucalenone | [C@]123CCC(= O)[C@@H](C)[C@]1([H])CC[C@]4([H])[C@@]2(C3)CC[C@@]5([C@](CC[C@@]45C)([C@@H](CCC(C(C)C)=C)C)[H])C |
| 149 | Rubimaillin | C12 = CC = CC = C1C3 = C(C(= C2O)C(OC)=O)C = CC(O3)(C)C |
| 150 | Canophyllal | [C@]12([C@]([C@]3([C@@](CC1)(CCC(C3)(C)C)C(= O)[H])[H])(CC[C@@]4([C@@]2(CC[C@]5([C@]4(CCC([C@@H]5C)=O)[H])C)[H])C)C)C |
| 151 | Stepharanine | O(C = 1C2 = C[N +]=3CCC = 4C(C3C = C2C = CC1O)=CC(O)=C(OC)C4)C |
| 152 | (Z)-icos-13-enoic acid | CCCCCC/C = C\CCCCCCCCCCCC(O)=O |
| 153 | Methyl 3,4,5-trihydroxybenzoate | OC1 = CC(= CC(O)=C1O)C(OC)=O |
| 154 | Serratol | C1C(= CCCC(= CCCC(= CC[C@@](C1)(C(C)C)O)C)C)C |
| 155 | Huperzine A | C\C = C1/[C@@H]2Cc3[nH]c(= O)ccc3[C@@]1(N)CC(C)=C2 |
| 156 | Mustakone | CC(C)[C@@H]1CCC2(C)C3C1C2C(C)=CC3 = O |
| 157 | (+)-Nootkatone | [C@@]12(C(CC[C@H](C1)C(C)=C)=CC(C[C@H]2C)=O)C |
| 158 | 1-Icosanoylglycerol | C(CCCCCCCCCCCC(OCC(CO)O)=O)CCCCCCC |
| 159 | Swertisin | O1[C@@H](C2 = C(O)C3 = C(OC(= CC3 = O)C4 = CC = C(O)C = C4)C = C2OC)[C@H](O)[C@@H](O)[C@H](O)[C@H]1CO |
| 160 | Serratol | C1C(= CCCC(= CCCC(= CC[C@@](C1)(C(C)C)O)C)C)C |
| 161 | PLX-4720 | C(CC)S(NC1 = CC = C(C(= C1F)C(C = 2C3 = C(N = CC(= C3)Cl)NC2)=O)F)(= O)=O |
| 162 | Huperzine A | C\C = C1/[C@@H]2Cc3[nH]c(= O)ccc3[C@@]1(N)CC(C)=C2 |
| 163 | N-(2-methoxyethyl)-4-{[6-(pyridin-4-yl)quinazolin-2-yl]amino}benzamide | O = C(C1 = CC = C(NC2 = NC3 = CC = C(C4 = CC = NC = C4)C = C3C = N2)C = C1)NCCOC |
| 164 | Glucotropeolin | [C@H]1(O[C@@H]([C@@H](O)[C@@H]([C@H]1O)O)CO)S/C(= N\OS(O)(= O)=O)/CC = 2C = CC = CC2 |
| 165 | Glucotropeolin(1-) | [C@H]1(O[C@@H]([C@@H](O)[C@@H]([C@H]1O)O)CO)S/C(= N\OS([O-])(= O)=O)/CC = 2C = CC = CC2 |
| 166 | Methyl 12-methyltetradecanoate | O(C(= O)CCCCCCCCCCC(CC)C)C |
| 167 | Carpaine | C[C@@H]1N[C@H]2CC[C@@H]1OC(= O)CCCCCCC[C@@H]1CC[C@H](OC(= O)CCCCCCC2)[C@H](C)N1 |
| 168 | 24-Methylenecycloartanol | [C@]123[C@@]4([C@](C([C@@H](O)CC4)(C)C)(CC[C@]1([C@]5([C@]([C@@]([C@@H](CCC(C(C)C)=C)C)(CC5)[H])(C)CC2)C)[H])[H])C3 |
| 169 | Helvolic acid methyl ester | C = 1[C@@]2([C@@]3(CC[C@@]/4([C@@]([C@]3(C([C@H]([C@]2([C@@H](C(C1)=O)C)[H])OC(= O)C)=O)C)(C[C@@H](\C4 = C(\CCC = C(C)C)/C(= O)OC)OC(= O)C)C)[H])[H])C |
| 170 | Danielone | COc1cc(cc(OC)c1O)C(= O)CO |
| 171 | 2-Isobutylthiazole | N1 = C(SC = C1)CC(C)C |
| 172 | 14-Deoxy-11,12-didehydroandrographolide | [H][C@]12CCC(= C)[C@@H](\C = C\C3 = CCOC3 = O)[C@]1(C)CC[C@@H](O)[C@@]2(C)CO |
| 173 | Andrographolide | [H][C@]12CCC(= C)[C@@H](C\C = C3/[C@H](O)COC3 = O)[C@]1(C)CC[C@@H](O)[C@@]2(C)CO |
| 174 | Dihydroferulic acid | C = 1(C = C(C(= CC1)O)OC)CCC(= O)O |
| 175 | Mesembryanthemoidigenic acid | [C@@]12(C([C@@]3(C[C@@](CO)(C)CC[C@@]3(CC1)C(= O)O)[H])=CC[C@@]4([C@]5(CC[C@@H](C([C@@]5(CC[C@@]24C)[H])(C)C)O)C)[H])C |
| 176 | Methyl N-methylanthranilate | CNC1 = CC = CC = C1C(OC)=O |
| 177 | Delta-elemene | [C@@H]1(C = C(CC[C@@]1(C = C)C)C(C)C)C(C)=C |
| 178 | Decussatin | COc1cc(O)c2c(c1)oc1ccc(OC)c(OC)c1c2 = O |
| 179 | Linalool | CC(C)=CCCC(C)(O)C = C |
| 180 | Nevadensin | COc1ccc(cc1)-c1cc(= O)c2c(O)c(OC)c(O)c(OC)c2o1 |
| 181 | 7-Epi-sesquithujene | C[C@H](CCC = C(C)C)[C@@]12CC = C(C)[C@@H]1C2 |
| 182 | Selina-4(15),7(11)-diene | C1CCC([C@]2([C@]1(CCC(C2) = C(C)C)C)[H]) = C |
| 183 | Codeine | [H][C@]12C = C[C@H](O)[C@@H]3Oc4c(OC)ccc5C[C@H]1N(C)CC[C@@]23c45 |
| 184 | Sulfanilamide | Nc1ccc(cc1)S(N)(= O)=O |
| 185 | Pipercyclobutanamide A(rel) | O = C(\C = C/[C@@H]1[C@@H](\C = C\c2ccc3OCOc3c2)[C@@H]([C@H]1c1ccc2OCOc2c1)C(= O)N1CCCCC1)N1CCCCC1 |
| 186 | (−)-cubebin | [H][C@@]1(CO[C@H](O)[C@]1([H])Cc1ccc2OCOc2c1)Cc1ccc2OCOc2c1 |
| 187 | (−)-3,4-dimethoxy-3,4-desmethylenedioxycubebin | [H][C@@]1(COC(O)[C@]1([H])Cc1ccc2OCOc2c1)Cc1ccc(OC)c(OC)c1 |
| 188 | Piperine | O = C(\C = C\C = C\c1ccc2OCOc2c1)N1CCCCC1 |
| 189 | Pellitorine | CCCCC\C = C\C = C\C(= O)NCC(C)C |
| 190 | Gaudichaudianic acid, (− rac) | CC(C)=CCCC1(C)Oc2c(CC = C(C)C)cc(cc2C = C1)C(O)=O |
| 191 | Isochamanetin | Oc1ccccc1Cc1c(O)cc2O[C@@H](CC(= O)c2c1O)c1ccccc1 |
| 192 | 7-Methoxychamanetin | COc1cc(O)c2C(= O)C[C@H](Oc2c1Cc1ccccc1O)c1ccccc1 |
| 193 | Dichamanetin | Oc1ccccc1Cc1c(O)c(Cc2ccccc2O)c2O[C@@H](CC(= O)c2c1O)c1ccccc1 |
| 194 | 7-Methoxydichamanetin | COc1c(Cc2ccccc2O)c(O)c2C(= O)C[C@H](Oc2c1Cc1ccccc1O)c1ccccc1 |
| 195 | 5″-(2⁗-Hydroxybenzyl)uvarinol | COc1c(Cc2cc(Cc3ccccc3O)ccc2O)c(O)c2C(= O)C[C@H](Oc2c1Cc1cc(Cc2ccccc2O)ccc1O)c1ccccc1 |
| 196 | 2,4-Dodecadienamide | CCCCCCC\C = C\C = C\C(N)=O |
| 197 | 7-Methoxyisochamanetin | COc1cc2O[C@@H](CC(= O)c2c(O)c1Cc1ccccc1O)c1ccccc1 |
| 198 | (2E,4E)-N-[2-(methylsulfinyl)ethyl]-2,4-decadienamide | CCCCC\C = C\C = C\C(= O)NCCS(C)=O |
| 199 | (2E,4E)-N-[(4-hydroxy-3-methoxyphenyl)ethyl]-2,4-decadienamide | CCCCC\C = C\C = C\C(= O)NCCc1ccc(O)c(OC)c1 |
| 200 | 3-(4-Hydroxy-3,5-dimethoxyphenyl)propanoylpyrrole | COc1cc(CCC(= O)n2cccc2)cc(OC)c1O |
| 201 | 3-(3,4,5-Timethoxyphenyl)propanoylpyrrole | COc1cc(CCC(= O)n2cccc2)cc(OC)c1OC |
| 202 | 1-[(2E,4E,6E)-2,4,6-dodecatrienoyl]pyrrolidine | CCCCC\C = C\C = C\C = C\C(= O)N1CCCC1 |
| 203 | 1-[(2E,4Z,8E)-9-(3,4-methylenedioxyphenyl)-2,4,8-nonatrienoyl]pyrrolidine | O = C(\C = C\C = C/CC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 204 | 1-[(2E,4E,10E)-10-(3,4-methylenedioxyphenyl)-2,4,10-undecatrienoyl]pyrrolidine | O = C(\C = C\C = C\CCCC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 205 | 1-[(4E,10E)-11-(3,4-methylenedioxyphenyl)-4,10-undecadienoyl]pyrrolidine | O = C(CC\C = C\CCCC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 206 | 1-[(9E)-10-(3,4-methylenedioxyphenyl)-9-decenoyl]pyrrolidine | O = C(CCCCCCC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 207 | 1-[(2E,4E)-2,4-decadienoyl]pyrrolidine | CCCCC\C = C\C = C\C(= O)N1CCCC1 |
| 208 | 1-[(2E,4E)-2,4-dodecadienoyl]pyrrolidine | CCCCCCC\C = C\C = C\C(= O)N1CCCC1 |
| 209 | 1-[(2E)-7-(3,4-methylenedioxyphenyl)-2-heptenoyl]pyrrolidine | O = C(\C = C\CCCCc1ccc2OCOc2c1)N1CCCC1 |
| 210 | 1-[(2E,4E)-7-(3,4-methylenedioxyphenyl)-2,4-heptadienoyl]pyrrolidine | O = C(\C = C\C = C\CCc1ccc2OCOc2c1)N1CCCC1 |
| 211 | 1-[(2E,8E)-9-(3,4-methylenedioxyphenyl)-2,8-nonadienoyl]pyrrolidine | O = C(\C = C\CCCC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 212 | 1-[(8E)-9-(3,4-methylenedioxyphenyl)-8-nonenoyl]pyrrolidine | O = C(CCCCCC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 213 | 1-[(2E,4E,8E)-9-(3,4-methylenedioxyphenyl)-2,4,8-nonatrienoyl]pyrrolidine | O = C(\C = C\C = C\CC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 214 | 1-[(2E,4E)-11-(3,4-methylenedioxyphenyl)-2,4-undecadienoyl]pyrrolidine | O = C(\C = C\C = C\CCCCCCc1ccc2OCOc2c1)N1CCCC1 |
| 215 | 1-[(2E,10E)-11-(3,4-methylenedioxyphenyl)-2,10-undecadienoyl]pyrrolidine | O = C(\C = C\CCCCCC\C = C\c1ccc2OCOc2c1)N1CCCC1 |
| 216 | (2E,4E)-N-isobutyl-2,4-dodecadienamide | CCCCCCC\C = C\C = C\C(= O)NCC(C)C |
| 217 | (2E,4E)-N-isobutyl-7-(3,4-methylenedioxyphenyl)-hepta-2,4-dienamide | CC(C)CNC(= O)\C = C\C = C\CCc1ccc2OCOc2c1 |
| 218 | (8E)-N-isobutyl-9-(3,4-methylenedioxyphenyl)nona-8-enamide | CC(C)CNC(= O)CCCCCC\C = C\c1ccc2OCOc2c1 |
| 219 | (2E,4E,8E)-N-isobutyl-11-(3,4-methylenedioxyphenyl)undeca-2,4,8-trienamide | CC(C)CNC(= O)\C = C\C = C\CC\C = C\CCc1ccc2OCOc2c1 |
| 220 | N-trans-sinapoyltyramine | COc1cc(cc(OC)c1O)\C = C\C(= O)NCCc1ccc(O)cc1 |
| 221 | Dihydrocubebin, rel- | OC[C@@H](Cc1ccc2OCOc2c1)[C@@H](CO)Cc1ccc2OCOc2c1 |
| 222 | Justiflorinol | OCC(CC(= O)c1ccc2OCOc2c1)C(= O)c1ccc2OCOc2c1 |
| 223 | (−)-Sanguinolignan A | O[C@@H]([C@H]1COC(= O)[C@@H]1C(= O)c1ccc2OCOc2c1)c1ccc2OCOc2c1 |
| 224 | (−)-Sanguinolignan B | O[C@@H]([C@@H]1COC(= O)[C@@H]1C(= O)c1ccc2OCOc2c1)c1ccc2OCOc2c1 |
| 225 | (−)-Sanguinolignan C | COc1ccc(cc1OC)[C@@H](OC(C) = O)[C@H]1COC(= O)[C@@H]1C(= O)c1ccc2OCOc2c1 |
| 226 | (−)-Sanguinolignan D | CC(= O)O[C@@H]([C@@H]1[C@H](COC1 = O)C(= O)c1ccc2OCOc2c1)c1ccc2OCOc2c1 |
| 227 | (7’S)-Parabenzlactone | O[C@@H]([C@H]1COC(= O)[C@@H]1Cc1ccc2OCOc2c1)c1ccc2OCOc2c1 |
| 228 | Flavokawain B | COc1cc(O)c(C(= O)\C = C\c2ccccc2)c(OC)c1 |
| 229 | Methyl 3,4-dihydroxy-5-(3′-methyl-2′-butenyl)benzoate | COC(= O)c1cc(O)c(O)c(CC = C(C)C)c1 |
| 230 | Pipercallosidine | C(/C = C/CCCCC = 1C = C2C(= CC1)OCO2)(NCC(C)C)=O |
| 231 | Kadsurenin C | O([C@@]12[C@@H]([C@H]([C@@]([C@H]1O)(C(= O)C(= C2)CC = C)[H])C3 = CC(OC)=C(OC)C = C3)C)C |
| 232 | Kadsurenin K | O([C@@]12[C@@H]([C@H]([C@@](C1 = O)(C(= O)C(= C2)CC = C)[H])C3 = CC(OC)=C(O)C = C3)C)C |
| 233 | Kadsurenin L | O([C@@]12[C@@H]([C@H]([C@@]([C@@H]1OC(= O)C)(C(= O)C(= C2)CC = C)[H])C3 = CC(OC)=C(OC)C = C3)C)C |
| 234 | Pipercallosine | C(/C = C/C = C/CCCCC1 = CC = C2C(= C1)OCO2)(= O)NCC(C)C |
| 235 | (S)-1′-methylhexyl caffeate | CCCCC[C@H](C)OC(= O)\C = C\c1ccc(O)c(O)c1 |
| 236 | Futoenone | O1[C@@H]2C[C@@]3([C@@H]([C@H](C2)C4 = CC = 5OCOC5C = C4)C)C1 = CC(= O)C(OC)=C3 |
| 237 | (S)-1′-methylbutyl caffeate | CCC[C@H](C)OC(= O)\C = C\c1ccc(O)c(O)c1 |
| 238 | (S)-1′-methyloctyl caffeate | CCCCCCC[C@H](C)OC(= O)\C = C\c1ccc(O)c(O)c1 |
| 239 | Futokadsurin B | COc1ccc(cc1OC)[C@@H]1O[C@@H]([C@H](C)[C@H]1C)c1ccc2OCOc2c1 |
| 240 | Futokadsurin C | COc1ccc(cc1OC)[C@H]1O[C@H]([C@H](C)[C@@H]1C)c1ccc2OCOc2c1 |
| 241 | Futokadsurin A | COc1cc(ccc1O)[C@@H]1O[C@@H]([C@@H](C)[C@@H]1C)c1ccc(OC)c(OC)c1 |
| 242 | Burchellin | O1[C@@H]([C@H]([C@]2(C1 = CC(= O)C(OC)=C2)CC = C)C)C3 = CC = 4OCOC4C = C3 |
| 243 | Piperonal | [H]C(= O)c1ccc2OCOc2c1 |
| 244 | (−)-Cubenol | [C@]12([C@@H](CC[C@H]([C@@]2(CCC(= C1)C)O)C)C(C)C)[H] |
| 245 | (E,E)-piperic acid | OC(= O)\C = C\C = C\c1ccc2OCOc2c1 |
| 246 | Trans-cinnamic acid | OC(= O)\C = C\c1ccccc1 |
| 247 | Pinocembrin chalcone | Oc1cc(O)c(C(= O)\C = C\c2ccccc2)c(O)c1 |
| 248 | Piperlactam S | O(C = 1C2 = C3C(= CC = 4C2 = CC = CC4)N(C(C3 = CC1O)=O)OC)C |
| 249 | Asebogenin | COc1cc(O)c(C(= O)CCc2ccc(O)cc2)c(O)c1 |
| 250 | Epicocconone | C\C = C\C = C\C = C\C(= O)\C = C(O)\C1 = C2C = C3C[C@@H](CO)OC = C3C(= O)[C@@]2(C)OC1 = O |
| 251 | (+)-Sesamin | [C@]12([C@@]([C@H](OC1)C3 = CC4 = C(C = C3)OCO4)(CO[C@@H]2C5 = CC6 = C(C = C5)OCO6)[H])[H] |
| 252 | Pipataline | O1C = 2C = C(\C = C\CCCCCCCCCC)C = CC2OC1 |
| 253 | (−)-Antofine | C = 1C2 = C(C = C(C1OC)OC)C = 3C = C(C = CC3C4 = C2C[C@]5(CCCN5C4)[H])OC |
| 254 | Benzoic acid | OC(= O)c1ccccc1 |
| 255 | Ethyl butyrate | CCCC(= O)OCC |
| 256 | N-feruloyltyramine | COc1cc(\C = C\C(= O)NCCc2ccc(O)cc2)ccc1O |
| 257 | Kadsurenin M | O1[C@@H]([C@H](C2 = C1C(OC)=CC(= C2)C(= O)[H])C)C3 = CC(OC)=C(OC)C = C3 |
| 258 | (−)-Epicubenol | [C@]12([C@@H](CC[C@H]([C@]2(CCC(= C1)C)O)C)C(C)C)[H] |
| 259 | Monocerin | [H][C@@]12C[C@H](CCC)O[C@]1([H])c1cc(OC)c(OC)c(O)c1C(= O)O2 |
| 260 | (2S,3aR,9bR)-6,7-dihydroxy-8-methoxy-2-propyl-3,3a-dihydro-2H-furo[3,2-c]isochromen-5(9bH)-one | [H][C@@]12C[C@H](CCC)O[C@]1([H])c1cc(OC)c(O)c(O)c1C(= O)O2 |
| 261 | Fusarentin 6,7-dimethyl ether | CCC[C@H](O)C[C@@H]1Cc2cc(OC)c(OC)c(O)c2C(= O)O1 |
| 262 | Fusarentin 6-methyl ether | CCC[C@H](O)C[C@@H]1Cc2cc(OC)c(O)c(O)c2C(= O)O1 |
| 263 | (3R,4R)-4,8-dihydroxy-3-((R)-2-hydroxypentyl)-6,7-dimethoxyisochroman-1-one | CCC[C@@H](O)C[C@H]1OC(= O)c2c(O)c(OC)c(OC)cc2[C@H]1O |
| 264 | Colletotrialide, (+)- | CCCC(= O)CC[C@H]1OC(= O)c2c(O)c(OC)c(OC)cc12 |
| 265 | Galgravin | C1(= C(C = CC(= C1)[C@@H]2O[C@@H]([C@H]([C@H]2C)C)C3 = CC = C(C(= C3)OC)OC)OC)OC |
| 266 | 9,10-Epoxy-18-hydroxyoctadecanoic acid | C1(CCCCCCCC(= O)O)C(CCCCCCCCO)O1 |
| 267 | Pinocembrin | Oc1cc(O)c2C(= O)C[C@H](Oc2c1)c1ccccc1 |
| 268 | 1,4-Cineole | C1C[C@]2(CC[C@]1(C)O2)C(C)C |
| 269 | Sodium benzoate | C(C = 1C = CC = CC1)([O-])=O.[Na +] |
| 270 | 9,10,18-Trihydroxyoctadecanoic acid | C(C(C(CCCCCCCCO)O)O)CCCCCCC(= O)O |
| 271 | Kavapyrone | COc1cc(oc(= O)c1)[C@@H]1O[C@H]1c1ccccc1 |
| 272 | Alpha-cubebene | C1C[C@H]([C@]2([C@]3([C@@H]1C)CC = C([C@]23[H])C)[H])C(C)C |
| 273 | Acuminatin | O1[C@H]([C@@H](C2 = C1C(OC)=CC(= C2)/C = C/C)C)C3 = CC(OC)=C(OC)C = C3 |
| 274 | Jerantinine E | [H][C@]12N3CCC[C@@]1(CC)CC(C(= O)OC)=C1Nc4cc(OC)c(O)cc4[C@]21CC3 |
| 275 | Jerantinine F | [H][C@]12CCN3CC[C@@]45C(Nc6cc(OC)c(O)cc46)=C(C[C@@]1(CCO2)[C@]35[H])C(= O)OC |
| 276 | Jerantinine C | [H][C@@]12N3CC[C@]11C(Nc4cc(OC)c(O)cc14)=C(C[C@]2(CC)C = CC3 = O)C(= O)OC |
| 277 | Jerantinine B | [H][C@]12CN3CC[C@@]45C(Nc6cc(OC)c(O)cc46)=C(C[C@](CC)([C@@]1([H])O2)[C@]35[H])C(= O)OC |
| 278 | Jerantinine D | [H][C@@]12O[C@]1([H])[C@@]1(CC)CC(C(= O)OC)=C3Nc4cc(OC)c(O)cc4[C@@]33CCN(C2 = O)[C@@]13[H] |
| 279 | Jerantinine A | [H][C@@]12N3CC[C@]11C(Nc4cc(OC)c(O)cc14)=C(C[C@]2(CC)C = CC3)C(= O)OC |
| 280 | Tabernaemontanine | CC[C@@H]1CN(C)[C@]2(CC = 3C4 = CC = CC = C4NC3C(C[C@@]1([C@@]2(C(= O)OC)[H])[H])=O)[H] |
| 281 | Plectranthol A | CC(C)c1cc2C = C[C@@]3(C)C(= CCC[C@]3(C)COC(= O)c3ccc(O)c(O)c3)c2c(O)c1O |
| 282 | 11,20-Dihydroxysugiol | C1 = C(C(C)C)C(= C(C2 = C1C(C[C@@]3([C@@]2(CCCC3(C)C)CO)[H])=O)O)O |
| 283 | 11-Hydroxysugiol | C1 = C(C(C)C)C(= C(C2 = C1C(C[C@@]3([C@@]2(CCCC3(C)C)C)[H])=O)O)O |
| 284 | Abietatriene | CC(C)c1ccc2c(CC[C@H]3C(C)(C)CCC[C@]23C)c1 |
| 285 | 7-Oxo-10α-cucurbitadienol | [H][C@@]1(CC[C@@]2(C)[C@]3([H])C(= O)C = C4[C@@]([H])(CC[C@H](O)C4(C)C)[C@]3(C)CC[C@]12C)[C@H](C)CCC = C(C)C |
3.4. Target prediction for the phytochemical compounds
Target prediction for the phytochemical compounds is a website that helps to calculate the most likely macromolecular targets of a small bioactive molecule. The SwissTagetPrediction was used to find the target of the compounds that satisfy the Lipinski rule of 5. The overall 24,843 targets were predicted for the 285 compounds (Supplementary Table 1 ). The probability score will be provided for each of the targets. Considering the confidence level, the targets with a probability score of more than 0 were considered for further research (Supplementary Table 2 ).
Supplementary Table 1.
A complete list of protein targets of the phytochemical compounds obtained from the SwissTargetPrediction server.
Supplementary Table 2.
A list of protein targets of the phytochemical compounds with probability score more than 0 obtained from the SwissTargetPrediction server.
3.5. Common target identification
The list of common targets between the COVID-19 infection (339 proteins) and phytochemical targets (24843) were analyzed using the Venn diagram (Fig. 1 ). The Venn diagram was plotted using a web server embedded in Bioinformatics & Evolutionary Genomics. From the analysis, 13 targets, namely, ACE, IMPDH2, EGFR, DPP4, RIPK1, HDAC2, CTSL, POLA1, CTSB, PABPC1, VEGFA, SIGMAR1, and IL6, were identified as potent protein targets.
Fig. 1.
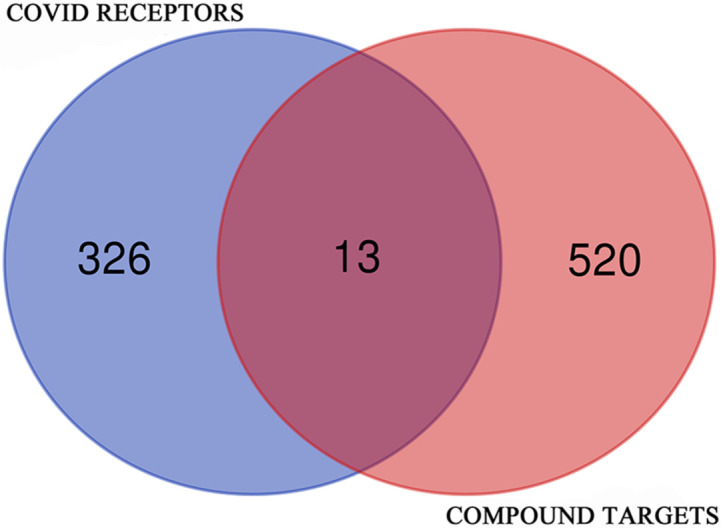
Venn diagram showing the number of COVID-19 receptors, Compound targets and the common genes between the COVID-19 receptors and compound targets.
3.6. Enrichment analysis
BP, CC, biological pathways, and MF were identified using the FunRich tool. Energy pathways, immune response, nucleoside, nucleic acid metabolism, nucleotide, and regulation of nucleobase were identified as the top biological process for the common receptor (Fig. 2A). Nucleus, plasma membrane, cytoplasm, exosome, lysosome, and extracellular were identified as the top cellular component for typical receptors (Fig. 2B). PAR1-mediated thrombin signaling events, IFN-gamma pathway, Nectin adhesion pathway, IL3-mediated pathway events, signaling events mediated by hepatocyte growth factor receptor (c-Met), and PDGF receptor signaling network were identified as the biological pathway for the typical receptors (Fig. 2C). Cytokine activity, transmembrane receptor protein tyrosine kinase activity, peptidase activity, transmembrane receptor activity, DNA-directed DNA polymerase activity, and cysteine-type peptidase activity (Fig. 2D). The complete list of results obtained from the FunRich analysis is tabulated in Supplementary Table 3 .
Fig. 2.
Enrichment analysis using FunRich shows the most significant biological process, cellular component, biological pathway, and molecular function of the identified targets as the input. (A) Biological process for common receptors, (B) Cellular component for common receptors, (C) Biological pathway for common receptors, and (D) Molecular function for common receptors.
Supplementary Table 3.
The binding affinity obtained from the AutoDock Vina between the identified targets and the phytochemical compounds. The binding affinity values are expressed in term of kcal/mol.
3.7. Pathway analysis
Pathway analysis was performed using the Reactome version 76 on 05/06/2021. All the 13 identifiers in the sample were found in Reactome, where 256 pathways were hit by at least one of them. The 25 most relevant significant pathways sorted by P-value are tabulated in Table 6 . TFAP2 (AP-2) family regulates the transcription of growth factors and their receptors. Potential therapeutics for SARS and SARS-CoV (SARS-CoV-1 & SARS-CoV-2) Infections were identified as the top 3 significant pathways with 4/21, 5/84, and 6/203 entities found, respectively (Fig. 3A and B).
Table 6.
The list of 25 most relevant pathways sorted by P value.
| S.·no | Pathway name | Entities found | Ratio | P value | FDRa | Reactions found | Ratio |
|---|---|---|---|---|---|---|---|
| 1 | TFAP2 (AP-2) family regulates transcription of growth factors and their receptors | 4/21 | 0.001 | 2.57E-08 | 6.88E-06 | 4/18 | 0.001 |
| 2 | Potential therapeutics for SARS | 5/84 | 0.006 | 0.000000122 | 0.0000163 | 6/32 | 0.002 |
| 3 | SARS-CoV infections | 6/203 | 0.014 | 0.000000338 | 0.0000301 | 9/254 | 0.019 |
| 4 | Transcriptional regulation by the AP-2 (TFAP2) family of transcription factors | 4/52 | 0.004 | 9.37e-07 | 6.28e-05 | 4/44 | 0.003 |
| 5 | Trafficking and processing of endosomal TLR | 3/16 | 0.001 | 1.75e-06 | 9.29e-05 | 2/7 | 5.21e-04 |
| 6 | Infectious disease | 9/1343 | 0.092 | 0.0000516 | 0.002 | 16/750 | 0.056 |
| 7 | VEGF ligand-receptor interactions | 2/8 | 0.000551 | 0.0000633 | 0.002 | 3/4 | 0.000298 |
| 8 | VEGF binds to VEGFR leading to receptor dimerization | 2/8 | 5.51e-04 | 6.33e-05 | 0.002 | 2/3 | 2.23e-04 |
| 9 | RUNX1 regulates transcription of genes involved in differentiation of keratinocytes | 2/11 | 7.57e-04 | 1.19e-04 | 0.003 | 1/8 | 5.96e-04 |
| 10 | Assembly of collagen fibrils and other multimeric structures | 3/67 | 0.005 | 1.23e-04 | 0.003 | 1/26 | 0.002 |
| 11 | Toll-like receptor cascades | 4/188 | 0.013 | 0.000141 | 0.003 | 14/185 | 0.014 |
| 12 | Disease | 11/2360 | 0.163 | 0.000148 | 0.003 | 99/1591 | 0.119 |
| 13 | Generic transcription pathway | 9/1555 | 0.107 | 0.000163 | 0.003 | 17/824 | 0.061 |
| 14 | Interleukin-4 and interleukin-13 signaling | 4/211 | 0.015 | 2.19e-04 | 0.004 | 2/47 | 0.004 |
| 15 | Regulation of gene expression by hypoxia-inducible factor | 2/15 | 0.001 | 2.21e-04 | 0.004 | 1/7 | 5.21e-04 |
| 16 | RNA polymerase II transcription | 9/1694 | 0.117 | 0.000314 | 0.004 | 17/885 | 0.066 |
| 17 | ERBB2 activates PTK6 signaling | 2/18 | 0.001 | 0.000318 | 0.004 | 2/2 | 0.000149 |
| 18 | ERBB2 regulates cell motility | 2/19 | 0.001 | 0.000354 | 0.005 | 2/2 | 0.000149 |
| 19 | Collagen formation | 3/104 | 0.007 | 0.000444 | 0.006 | 1/77 | 0.006 |
| 20 | PI3K events in ERBB2 signaling | 2/22 | 0.002 | 0.000473 | 0.006 | 5/7 | 0.000521 |
| 21 | Gene expression (transcription) | 9/1855 | 0.128 | 0.000622 | 0.007 | 23/1000 | 0.074 |
| 22 | Signaling by VEGF | 3/137 | 0.009 | 0.000983 | 0.011 | 49/86 | 0.006 |
| 23 | Signaling by ERBB2 KD Mutants | 2/35 | 0.002 | 0.001 | 0.011 | 15/17 | 0.001 |
| 24 | MHC class II antigen presentation | 3/148 | 0.01 | 0.001 | 0.011 | 3/26 | 0.002 |
| 25 | Degradation of the extracellular matrix | 3/148 | 0.01 | 0.001 | 0.011 | 6/105 | 0.008 |
False discovery rate.
Fig. 3.
(A) Pathway shows the transcriptional regulation by the AP-2 (TFAP2) transcription factor family. (B) Pathway showing the potential therapeutics for SARS.
3.8. Gene–gene interaction analysis
The gene–gene interaction analysis helped to prioritize the study of genes in the pathway that helped to understand the underlying mechanism of COVID. The gene–gene interaction analysis was performed using the STRING online server. The number of nodes and edges were found to be 13 and 17, respectively. The average degree of nodes and clustering coefficient of local was found to be 2.62 and 0.566. The anticipated amount of edges was 9 with a PPI enrichment P-value of 0.0115. Except for IMPDH2, PABPC1, SIGMAR1, and POLA1, all the other genes interacted with at least one gene (Fig. 4 ).
Fig. 4.
The gene-gene interaction between the identified targets common to the COVID-19 receptors and compound targets.
3.9. Virtual screening and molecular interaction analysis
The identified targets such as VEGFA, CTSL, CTSB, EGFR, and IL6 have been identified as the target by more than one phytochemical compound (Table 7). Virtual screenings were performed to identify the most suitable phytochemical compound. The compound with the highest binding affinity was taken for the molecular interaction analysis using AutoDock (Supplementary Table 3). We observed that betulinic acid was found to interact best with the ACE protein with the binding energy − 9.32 kcal/mol from the molecular docking results. The 5″-(2⁗-Hydroxybenzyl) uvarinol was found to interact best with Cathepsin L and Cathepsin B & K with the binding energy − 6.71 kcal/mol and − 6.06 kcal/mol, respectively. The (-)-antofine, S)-1′-methyloctyl caffeate, (Z)-3-phenyl-2-propenal, 7-oxo-10α-cucurbitadienol, PLX-4720, and 5″-(2⁗-Hydroxybenzyl) uvarinol were found to interact best to the DPP4, EGFR, HDAC2, IL6, RIPK1, and VEGFA with binding energies − 8.25 kcal/mol, − 4.76 kcal/mol, − 4.39 kcal/mol, − 6.9 kcal/mol, − 5.91 kcal/mol, and − 5.66 kcal/mol respectively. The interactions between the compounds and the proteins are shown in Fig. 5.
Table 7.
The list of selected receptors of the COVID-19 and the identified interacting phytochemical compounds.
| S.·no | Name of the receptor | Name of the compound |
|---|---|---|
| 1 | Angiotensin-converting enzyme | Betulinic acid |
| 2 | Dipeptidyl peptidase IV | (−)-Antofine |
| 3 | Vascular endothelial growth factor A | Isochamanetin |
| 7-Methoxychamanetin | ||
| Dichamanetin | ||
| 7-Methoxydichamanetin | ||
| 5″-(2⁗-Hydroxybenzyl)uvarinol | ||
| 7-Methoxyisochamanetin | ||
| Pinocembrin | ||
| 1,4-Cineole | ||
| 4 | Cathepsin L | Isochamanetin |
| 7-Methoxychamanetin | ||
| Dichamanetin | ||
| 7-Methoxydichamanetin | ||
| 5″-(2⁗-Hydroxybenzyl)uvarinol | ||
| 7-Methoxyisochamanetin | ||
| Pinocembrin chalcone | ||
| Pinocembrin | ||
| 1,4-Cineole | ||
| Carpaine | ||
| 5 | Cathepsin (B and K) | Isochamanetin |
| 7-Methoxychamanetin | ||
| Dichamanetin | ||
| 7-Methoxydichamanetin | ||
| 5″-(2⁗-Hydroxybenzyl)uvarinol | ||
| 7-Methoxyisochamanetin | ||
| Asebogenin | ||
| Pinocembrin | ||
| 1,4-Cineole | ||
| Carpaine | ||
| 6 | Epidermal growth factor receptor erbB1 | 7,4′-dimethylkaempferol |
| Pipataline | ||
| N-trans-sinapoyltyramine | ||
| Flavokawain B | ||
| (S)-1′-methylhexyl caffeate | ||
| (S)-1′-methylbutyl caffeate | ||
| (S)-1′-methyloctyl caffeate | ||
| Trans-cinnamic acid | ||
| Pinocembrin chalcone | ||
| N-feruloyltyramine | ||
| Cinnamic acid | ||
| Dihydroferulic acid | ||
| Capensinidin | ||
| Nevadensin | ||
| 7 | Interleukin-6 | All-cis-octadeca-6,9,12,15-tetraenoic acid |
| 7-Oxo-10α-cucurbitadienol | ||
| 8 | Receptor-interacting serine/threonine-protein kinase 1 | PLX-4720 |
| 9 | Histone deacetylase 2 | (Z)-3-phenyl-2-propenal |
Fig. 5.
Visualization of the best interacting phytochemical with the identified targets. (A) ACE receptor with betulinic acid, (B) Cathepsin L receptor with 5″-(2⁗-Hydroxybenzyl)uvarinol, (C) Cathepsin B & K receptor with 5″-(2⁗-Hydroxybenzyl)uvarinol, (D) DPPR receptor with (−)-antofine, (E) EGFR receptor with S)-1′-methyloctyl caffeate, (F) HDAC2 with (Z)-3-phenyl-2-propenal, (G) IL6 with 7-oxo-10α-cucurbitadienol, (H) RIPK1 with PLX-4720, and (I) VEGFA with 5″-(2⁗-Hydroxybenzyl)uvarinol.
4. Discussion
SARS-CoV-2 belongs to a genetic group of viruses that cause respiratory sickness and was declared a worldwide pandemic on March 11th, 2020 (Cucinotta & Vanelli, 2020). As of September 6th, 2021, the reported cases are more than 220,563,227, and at least 4,565,483 people have died (WHO, 2021b). The number of cases across the globe is gradually increasing and decreasing in terms of the COVID-19 infection wave. The countries in America, Europe, and the South Asian region are highly infected with the second wave of COVID-19 infection. Few countries have started to face the third wave of COVID-19 (Taboada et al., 2021).
On the other hand, drug discovery is still far behind, and only drug repurposing serves as the day's choice (Lamontagne et al., 2020; Wu et al., 2020). Unfortunately, several repurposed drugs that showed promising results in the early studies were failed to treat COVID-19; this also includes hydroxychloroquine (Boulware et al., 2020; Geleris et al., 2020). With this less efficiency of repurposed drugs, the search for the vaccine was on a serious note, and several vaccines were developed across the globe, but none were 100% active. Further, there is always a chance of vaccine failure with the mutations and their evolution in the SARS-CoV-2 (Williams & Burgers, 2021). Developing an immune response against COVID-19 can only be a ray of hope in this scenario.
Traditional medicines have been immune boosters since the ancient days (Ravishankar & Shukla, 2007). Siddha herbal formulations with medicinal value are effective against various causative agents, including influenza, dengue fever, chikungunya, tuberculosis, and others (Jain et al., 2020; Jain, Narayanan, Chaturvedi, Pai, & Sunil, 2018; Jain, Pai, & Sunil, 2018). Currently, the ministry of AYUSH has also approved the use of Kabasura kudineer and Nilavembu kudineer against COVID-19 (Alagu Lakshmi, Shafreen, Priya, & Shunmugiah, 2020; Natarajan et al., 2020). This study is intended to understand the immune-boosting mechanism by the JACOM, Kabasura kudineer, and Nilavembu kudineer against COVID-19. A total of 339 human genes were found to be involved in COVID-19. This list was obtained from the GeneCards database (Table 1). Twenty-five plants were found in the JACOM, Kabasura kudineer, and Nilavembu kudineer formulations (Table 2). A list of 314 phytochemicals was obtained from these 25 plants from the ChEBI database (Table 3). The drug-likeness properties of the 314 phytochemical compounds were evaluated using the online SwissADME server (Table 4). From the analysis, we observed 285 compounds to satisfy the Lipinski rule of 5, and these could be considered drug-like compounds (Table 5). The possible targets for these drug-like compounds were predicted using the online SwissTargetPrediction server. An overall of 24,839 targets was predicted for these 285 compounds (Supplementary Table 1). The targets were ranked based on the probability score. Out of 24,839 targets, 5129 genes (with repeats) were the promising target with a probability score of more than 0 (Supplementary Table 2). A Venn diagram was plotted to find the common targets between the genes involved in the COVID-19 and the compounds target (Table 1, Table 5). From the Venn diagram, 13 targets (ACE, IMPDH2, EGFR, DPP4, RIPK1, HDAC2, CTSL, POLA1, CTSB, PABPC1, VEGFA, SIGMAR1, and IL6) were found to be common between the causative and treatment for COVID-19. Three hundred and twenty-six and 520 genes were COVID-19 receptors and targets for compounds, respectively (Fig. 1). The enrichment analysis was performed to understand the role of identified targets in their BP, CC, biological pathways, and MF. Energy pathways, nucleoside, immune response, nucleotide, regulation of nucleobase, and nucleic acid metabolism were identified as the top biological process for the common receptor (Fig. 2A). Nucleus, plasma membrane, cytoplasm, exosome, lysosome, and extracellular were identified as the top cellular component for common receptors (Fig. 2B). PAR1-mediated thrombin signaling events, Nectin adhesion pathway, IL3-mediated signaling events, IFN-gamma pathway, cellular pathways engaged by hepatocyte growth factor receptor (c-Met), and PDGF receptor signaling network were identified as the biological pathway for the common receptors (Fig. 2C). Cytokine activity, peptidase activity, transmembrane receptor protein tyrosine kinase activity, transmembrane receptor activity, DNA-directed DNA polymerase activity, and cysteine-type peptidase activity (Fig. 2D). Further, the pathways involved in these gene targets were predicted using the Reactome database (Jassal et al., 2020). The pathways such as the TFAP2 (AP-2) family that controls the expression of growth factors and their receptors, Potential therapeutics for SARS, and SARS-CoV Infections were identified as the top 3 pathways out of significant 25 pathways (Table 6). The clear pathway of the TFAP2 (AP-2) family regulates the transcription of growth factors, and their receptors are shown in Fig. 3A. In mammals, there are five transcription factors in the AP-2 (TFAP2) family: TFAP2A, TFAP2B, TFAP2C, TFAP2D, and TFAP2E (AP-2 epsilon). The AP-2 transcription factors have a helix-span-helix motif at the C-terminus, a core basic region, and a transactivation domain at the N-terminus, and are evolutionarily conserved in metazoans (Eckert, Buhl, Weber, Jäger, & Schorle, 2005). EGFR and VEGFA identified genes were found to be involved in this pathway. The identified second most significant pathway was potential therapeutics for SARS. The detailed pathway is shown in Fig. 3B. Based on their efficacy in treating infectious disease with other RNA viruses or in reducing cytokine storms and other illnesses caused by viruses are identical to SARS-CoV-1 and SARS-CoV-2. A significant number of intriguing therapeutic candidates have been found to be also similar. HDAC2, SIGMAR1, IMPDH2, VEGFA, and RIPK1 identified genes were found to be involved in this pathway. SARS-CoV-2 infection pathway is not well annotated. However, the viral infection pathways are curated based on the SARS-CoV-1 and SARS-CoV-2 infection processes and drug responses. Many of the steps in SARS-CoV-1 infections are well studied experimentally correlated with the steps involved in SARS-CoV-2 infection. In comparison with the other two significant pathways, a maximum of 6 targets (CTSL, RIPK1, HDAC2, SIGMAR1, IMPDH2, and VEGFA) are found to be in this pathway (Supplementary Data 1, Supplementary Data 2 ).
Supplementary Data 1.
Pathway analysis report for the identified targets. The analysis was performed against Reactome version 76 on 05/06/2021.
Supplementary Data 2.
A complete enrichment analysis of the identified targets obtained from the FunRich software.
The common genes were subjected to the gene–gene interaction study using the online STRING database to identify the connected genes (Szklarczyk et al., 2019). Out of the identified targets, HDAC2, IL6, EGFR, DPP4, ACE, VEGFA, CTSL, CTSB, and RIPK1 were interlinked with another target (Fig. 4). These targets were already promising targets for COVID-19 treatment in several previous studies. Therefore, activating or suppressing one of these genes could impact the other and improve efficacy.
The Histone deacetylase 2 (HDAC2) plays a crucial role in monocytes' regular immune response function. These monocytes were found to maintain the immune system's regular role when combined with the macrophages. The HDAC2 drugs such as Theophylline, Macrolides, Nortriptyline, and many others have been shown to prevent the pathological response of the inflammatory monocytes and thereby regulate the normal lung function (HDAC2 Regulates Response of Inflammatory Monocytes: A COVID-19 Target, 2020). A study by Liu et al., 2020 has also proposed that the inhibition of HDAC could be a promising target for the COVID-19 infection (Liu et al., 2020).
T-lymphocytes, adipose tissues, and macrophages all produce interleukin-6 (IL-6), a pro-inflammatory cytokine protein. It plays a significant role in atherogenesis and is linked to cardiovascular clinical outcomes (Gabay, 2006). The COVID-19 patients with more severe illnesses had higher inflammatory cytokines linked to pulmonary inflammation, lung destruction, and multiple organ failure. SARS-CoV-2 patients exhibit low concentrations of the modulator of cytokine signaling-3, which again is involved in the stimulation of the IL-6 negative feedback loop. Increased IL 6 has also been discovered to be an indicator of the change from a mild to a severe illness, limiting the severity if caught early (Vatansever & Becer, 2020).
The progression of SARS-CoV induces fibrosis, as well as a review of evidence indicating pulmonary fibrosis is generated by an overactive host response to lung damage driven through epidermal growth factor receptor (EGFR) signaling (Venkataraman & Frieman, 2017). Adeno-associated virus 2 (AAV2) second-strand DNA synthesis and transgene expression are inhibited by FK506-binding protein (FKBP52), which is phosphorylated at tyrosine residues by EGFR-PTK (Yano et al., 2003). Because EGFR is abundantly expressed in various solid tumors and its expression is linked to tumor progression, leading to chemotherapy resistance and poor prognosis. It is a promising strategy for the rational development of new anticancer drugs.
DPP4 is widely known to involve in type 2 diabetic conditions. DPP4 activity affects glucose homeostasis and inflammation in numerous ways. The change in expression levels of DPP4 in the early MERS infections was also studied several years ago (Chan et al., 2015). Currently, the SARS-CoV-2 being the same family of MERS infection, the role of DPP4 in SARS-CoV-2 conditions is also widely studied and found to be a promising target for COVID-19 treatment (Scheen, 2021; Solerte, Di Sabatino, Galli, & Fiorina, 2020).
ACE2 acts as a functional receptor on the cell's surface through which the SARS-CoV-2 can enter the human cell. This ACE2 is expressed highly in the heart, kidney, and lungs. The ACE/ACE2 balance disruption with RAAS (renin-angiotensin-aldosterone system) activation can lead to severe COVID-19, especially in diabetes, cardiovascular diseases, and hypertension (Beyerstedt, Casaro, & Rangel, 2021).
In ICU and non-ICU COVID-19 patients, VEGF concentrations were more significant than in healthy controls. Angiogenesis, neurogenesis, and neuroprotection are all aided by VEGFs, leading to vascular leakiness and permeability. ACE2 inhibits VEGF-A, which reduces vascular permeability in patients with acute lung damage. In the account of SARS-downregulation CoV-2’s of ACE2, the VEGF-A antagonistic effect of ACE2 is expected to be canceled, resulting in overexpression of VEGF and increased vascular permeability and exacerbation of endothelial injury (Yazihan et al., 2021).
The CTSL1 gene codes for a lysosomal cysteine protease involved in intracellular protein degradation. It impacts collagen and elastin, along with alpha-1 protease inhibitor, a key regulator of neutrophil elastase function. CatL is essential in degrading the extracellular matrix, a crucial mechanism for SARS-CoV-2 to enter host cells and is upregulated during chronic inflammation. CatL is also likely involved in the processing of SARS-CoV-2 spike protein. CatL could have been regarded as a valuable therapeutic target because its suppression damages SARS-CoV-2 infection and maybe egress from cells through late stages of infection (Gomes et al., 2020; Pišlar et al., 2020).
Because several patients had greater serum RIPK3 (a family of RIPK1) levels, it is possible that RIPK-3-mediated signaling may lead to necroptosis; it is implicated in the development of COVID-19 pneumonia-related acute lung injury. In ARDS patients, RIPK-3 levels were considerably more significant than in non-ARDS patients. In immunohistochemistry, all epithelial cell samples from COVID-19 individuals were confirmed for active phosphorylated RIPK1. Thus, ICD mediated by RIPK1 may have a part in the progression of SARS-CoV-2 infection and can be a new therapeutic target (Nakamura et al., 2020).
Several compounds targeting these genes are capable of interacting with more than one gene. Virtual screenings were performed to identify the best interaction of the compounds with these targets. From the virtual screening analysis, betulinic acid was found to interact with ACE, 5″-(2⁗-Hydroxybenzyl)uvarinol was found to interact with Cathepsin L, and Cathepsin B & K, -(-)-antofine was found to interact with DPP4, (S)-1′-methyloctyl caffeate was found to interact with EGFR, (Z)-3-phenyl-2-propenal was found to interact with HDAC2, 7-oxo-10α-cucurbitadienol was found to interact with IL6, PLX-4720 was found to interact with RIPK1, and 5″-(2⁗-Hydroxybenzyl)uvarinol was found to interact with VEGFA (Supplementary Table 3). The molecular dockings were performed using the AutoDock, and the interactions were studied using the Discovery Studio (Fig. 5). Thus collectively, these compounds targeting the identified targets could improve the immune system, fighting against the COVID-19 infection.
5. Conclusion
The COVID-19 infection is widely increasing across the globe. The drugs to treat COVID-19 are still in the clinical trials, which might take years for approval. Drugs such as Hydroxychloroquine, Remdesivir, Favipiravir, Lopinavir/ritonavir are widely repurposed to treat the COVID-19 during the earlier outbreak. On the other hand, various vaccine manufacturers such as COVID-19 Vaccine AstraZeneca, COVID-19 Vaccine Janssen, Sputnik, and Covaxin are in the race to develop vaccines, yet none seems to be 100% efficient. In addition, with the evolution of SARS-CoV-2 mutations, the efficiency of the vaccines is becoming a interrogation point every day.
Similarly, these mutations might lead to drug inefficacy and drug-resistant. The promising approach to be safe from COVID-19 is to develop the immune system. The traditional medicines are immune boosters, serving as a promising remedy for various diseases since ancient days. This study used the network pharmacology approach and analyzed the phytochemical compounds in Nilavembu kudineer, Kabasura kudineer, and JACOM and their interacting human protein targets, activating or suppressing the target. These identified compounds can be tested in vivo and in vitro to compare the toxicity and efficiency of the currently available formulations.
Acknowledgments
Conflict of interest
The authors have declared that no conflicts of interest exist.
Acknowledgments
Author contributions
D.T.K., S.U.K., S.P., S.D.M.S., S.R., and C.G.P.D. were involved in the study's design. D.T.K., A.S., A.M., L.M., R.G., M.R., and S.U.K. were involved in the data collection and experimentation. D.T.K., A.S., A.M., L.M., R.G., M.R., and S.U.K. were engaged in the acquisition, analysis, and interpretation results. D.T.K. and S.U.K. drafted the manuscript. C.G.P.D., S.R. and S.P. supervised the entire study and was involved in study design, the acquisition, analysis, understanding of the data, and critically reviewed the manuscript. All authors edited and approved the submitted version of the article.
Funding
No funding agency was involved in the present study.
Acknowledgments
The authors would like to take this opportunity to thank the management of Vellore Institute of Technology (VIT), Vellore, India, and Meenakshi Academy of Higher Education and Research, Chennai, for providing the necessary facilities and encouragement to carry out this work.
References
- Alagu Lakshmi S., Shafreen R.M.B., Priya A., Shunmugiah K.P. Ethnomedicines of Indian origin for combating COVID-19 infection by hampering the viral replication: Using structure-based drug discovery approach. Journal of Biomolecular Structure and Dynamics. 2020 doi: 10.1080/07391102.2020.1778537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyerstedt S., Casaro E.B., Rangel É.B. COVID-19: Angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. European Journal of Clinical Microbiology and Infectious Diseases. 2021;40(5):905–919. doi: 10.1007/s10096-020-04138-6. Springer Science and Business Media Deutschland GmbH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulware D.R., Pullen M.F., Bangdiwala A.S., Pastick K.A., Lofgren S.M., Okafor E.C., et al. A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid-19. New England Journal of Medicine. 2020;383(6):517–525. doi: 10.1056/nejmoa2016638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain D.W., Cidlowski J.A. After 62 years of regulating immunity, dexamethasone meets COVID-19. Nature Reviews Immunology. 2020;20(10):587–588. doi: 10.1038/s41577-020-00421-x. Nature Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Lau S.K.P., To K.K.W., Cheng V.C.C., Woo P.C.Y., Yue K.Y. Middle East Respiratory syndrome coronavirus: Another zoonotic betacoronavirus causing SARS-like disease. Clinical Microbiology Reviews. 2015 doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomedica. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. Mattioli 1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods in Molecular Biology. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- Dhand R., Li J. Coughs and sneezes: Their role in transmission of respiratory viral infections, including SARS-CoV-2. American Journal of Respiratory and Critical Care Medicine. 2020;202(5):651–659. doi: 10.1164/rccm.202004-1263PP. American Thoracic Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert D., Buhl S., Weber S., Jäger R., Schorle H. The AP-2 family of transcription factors. Genome Biology. 2005;6(13):246. doi: 10.1186/gb-2005-6-13-246. BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., et al. The reactome pathway knowledgebase. Nucleic Acids Research. 2016;44(D1):D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay C. Interleukin-6 and chronic inflammation. Arthritis Research and Therapy. 2006;8(Suppl. 2) doi: 10.1186/ar1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geleris J., Sun Y., Platt J., Zucker J., Baldwin M., Hripcsak G., et al. Observational study of hydroxychloroquine in hospitalized patients with Covid-19. New England Journal of Medicine. 2020;382(25):2411–2418. doi: 10.1056/nejmoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller D., Grosdidier A., Wirth M., Daina A., Michielin O., Zoete V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Research. 2014;42(W1):W32–W38. doi: 10.1093/nar/gku293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes C.P., Fernandes D.E., Casimiro F., da Mata G.F., Passos M.T., Varela P., et al. Frontiers in cellular and infection microbiology. Vol. 10. Frontiers Media S.A; 2020. Cathepsin L in COVID-19: From pharmacological evidences to genetics; p. 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings J., De Matos P., Dekker A., Ennis M., Harsha B., Kale N., et al. The ChEBI reference database and ontology for biologically relevant chemistry: Enhancements for 2013. Nucleic Acids Research. 2013;41(D1) doi: 10.1093/nar/gks1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HDAC HDAC2 Regulates response of inflammatory monocytes: a COVID-19 target. 2020. https://www.drugtargetreview.com/article/60593/histone-deacetylase-2-hdac2-regulates-pathological-response-of-inflammatory-monocytes-a-potential-target-of-adjuvant-therapies-for-covid-19-infection/ Retrieved June 12th, 2021, from.
- Jain J., Kumar A., Narayanan V., Ramaswamy R.S., Sathiyarajeswaran P., Shree Devi M.S., et al. Antiviral activity of ethanolic extract of Nilavembu Kudineer against dengue and chikungunya virus through in vitro evaluation. Journal of Ayurveda and Integrative Medicine. 2020;11(3):329–335. doi: 10.1016/j.jaim.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain J., Narayanan V., Chaturvedi S., Pai S., Sunil S. In vivo evaluation of Withania somnifera–based Indian traditional formulation (amukkara choornam), against chikungunya virus–induced morbidity and arthralgia. Journal of Evidence-Based Integrative Medicine. 2018 doi: 10.1177/2156587218757661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain J., Pai S., Sunil S. Standardization of in vitro assays to evaluate the activity of polyherbal siddha formulations against Chikungunya virus infection. VirusDisease. 2018 doi: 10.1007/s13337-018-0421-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., et al. The reactome pathway knowledgebase. Nucleic Acids Research. 2020;48(D1):D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiran G., Karthik L., Shree Devi M.S., Sathiyarajeswaran P., Kanakavalli K., Kumar K.M., et al. In silico computational screening of Kabasura Kudineer—official siddha formulation and JACOM against SARS-CoV-2 spike protein. Journal of Ayurveda and Integrative Medicine. 2020 doi: 10.1016/j.jaim.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D.T., Shaikh N., Kumar S.U., Doss C.G.P., Zayed H. Structure-based virtual screening to identify novel potential compound as an alternative to remdesivir to overcome the RdRp protein mutations in SARS-CoV-2. Frontiers in Molecular Biosciences. 2021;8 doi: 10.3389/fmolb.2021.645216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.U., Kumar D.T., Christopher B.P., Doss C.G.P. The rise and impact of COVID-19 in India. Frontiers in Medicine. 2020;7 doi: 10.3389/fmed.2020.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.U., Kumar D.T., Siva R., Doss C.G.P. Kerala, India's front runner in novel coronavirus disease (COVID-19) Frontiers in Medicine. 2020;7 doi: 10.3389/fmed.2020.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamontagne F., Agoritsas T., MacDonald H., Leo Y.S., DIaz J., Agarwal A., et al. A living WHO guideline on drugs for covid-19. The BMJ. 2020;370:m3379. doi: 10.1136/bmj.m3379. [DOI] [PubMed] [Google Scholar]
- Liu K., Zou R., Cui W., Li M., Wang X., Dong J., et al. Clinical HDAC inhibitors are effective drugs to prevent the entry of SARS-CoV2. ACS Pharmacology and Translational Science. 2020;3(6):1361–1370. doi: 10.1021/acsptsci.0c00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of Computational Chemistry. 1998;19(14):1639–1662. https://doi.org/10.1002/(SICI)1096-987X(19981115)19:14 < 1639::AID-JCC10 > 3.0.CO;2-B. [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H., Kinjo T., Arakaki W., Miyagi K., Tateyama M., Fujita J. Serum levels of receptor-interacting protein kinase-3 in patients with COVID-19. Critical Care. 2020;24(1) doi: 10.1186/s13054-020-03209-6. BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan S., Anbarasi C., Sathiyarajeswaran P., Manickam P., Geetha S., Kathiravan R., et al. The efficacy of Siddha Medicine, Kabasura Kudineer (KSK) compared to Vitamin C & Zinc (CZ) supplementation in the management of asymptomatic COVID-19 cases: A structured summary of a study protocol for a randomised controlled trial. Trials. 2020;21(1) doi: 10.1186/s13063-020-04823-z. BioMed Central Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y.J., Williamson N.A., et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15(15):2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- Patwardhan B., Warude D., Pushpangadan P., Bhatt N. Ayurveda and traditional Chinese medicine: A comparative overview. Evidence-Based Complementary and Alternative Medicine. 2005;2(4):465–473. doi: 10.1093/ecam/neh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pišlar A., Mitrovic A., Sabotič J., Fonovic U.P., Nanut M.P., Jakoš T., et al. The role of cysteine peptidases in coronavirus cell entry and replication: The therapeutic potential of cathepsin inhibitors. PLoS Pathogens. 2020;16(11):e1009013. doi: 10.1371/journal.ppat.1009013. Public Library of Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Mitchell B.M., Plante K.S., Debbink K., Weaver S.C., Menachery V.D. The variant gambit: COVID-19’s next move. Cell Host and Microbe. 2021;29(4):508–515. doi: 10.1016/j.chom.2021.02.020. Cell Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam S., Muthiah K., Parameswaran S., Tamilarasan K., Selvarajan E., Ayyasamy U. Analogy of Kaba Suram with COVID-19 symptoms–A siddha literature review. International Journal of Ayurvedic Medicine. 2020;11(4):616–621. doi: 10.47552/ijam.v11i4.1674. [DOI] [Google Scholar]
- Ravishankar B., Shukla V.J. Indian systems of medicine: A brief profile. African Journal of Traditional, Complementary and Alternative Medicines. 2007;4(3):319–337. doi: 10.4314/ajtcam.v4i3.31226. African Ethnomedicines Network. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebhan M., Chalifa-Caspi V., Prilusky J., Lancet D. GeneCards: Integrating information about genes, proteins and diseases. Trends in Genetics. 1997;13(4):163. doi: 10.1016/S0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- Ross C.L. Integral healthcare: The benefits and challenges of integrating complementary and alternative medicine with a conventional healthcare practice. Integrative Medicine Insights. 2009;2009(4):13–20. doi: 10.4137/IMI.S2239. SAGE Publications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheen A.J. DPP-4 inhibition and COVID-19: From initial concerns to recent expectations. Diabetes and Metabolism. 2021;47(2):101213. doi: 10.1016/j.diabet.2020.11.005. Elsevier Masson s.r.l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solerte S.B., Di Sabatino A., Galli M., Fiorina P. Dipeptidyl peptidase-4 (DPP4) inhibition in COVID-19. Acta Diabetologica. 2020;57(7):779–783. doi: 10.1007/s00592-020-01539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Banu S., Singh P., Sowpati D.T., Mishra R.K. SARS-CoV-2 genomics: An Indian perspective on sequencing viral variants. Journal of Biosciences. 2021;46(1):1–14. doi: 10.1007/s12038-021-00145-7. Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taboada M., González M., Alvarez A., Eiras M., Costa J., Álvarez J., et al. First, second and third wave of COVID-19. What have we changed in the ICU management of these patients? Journal of Infection. 2021;82(6):e14–e15. doi: 10.1016/j.jinf.2021.03.027. W.B. Saunders Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cellular and molecular immunology. 2020;17(6):613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatansever H.S., Becer E. Relationship between IL-6 and COVID-19: To be considered during treatment. Future Virology. 2020;15(12):817–822. doi: 10.2217/fvl-2020-0168. Future Medicine Ltd. [DOI] [Google Scholar]
- Venkataraman T., Frieman M.B. The role of epidermal growth factor receptor (EGFR) signaling in SARS coronavirus-induced pulmonary fibrosis. Antiviral Research. 2017;143:142–150. doi: 10.1016/j.antiviral.2017.03.022. Elsevier B.V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO What is COVID-19 vaccine efficacy? | WHO | Regional Office for Africa. 2021. https://www.afro.who.int/news/what-covid-19-vaccine-efficacy Retrieved June 12th, 2021, from.
- WHO WHO coronavirus (COVID-19) dashboard | WHO coronavirus (COVID-19) dashboard with vaccination data. 2021. https://covid19.who.int/ Retrieved June 8th, 2021, from.
- Williams T.C., Burgers W.A. SARS-CoV-2 evolution and vaccines: Cause for concern? The Lancet Respiratory Medicine. 2021;9(4):333–335. doi: 10.1016/S2213-2600(21)00075-8. Lancet Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Wang L., Kuo H.C.D., Shannar A., Peter R., Chou P.J., et al. An update on current therapeutic drugs treating COVID-19. Current Pharmacology Reports. 2020;6(3):56–70. doi: 10.1007/s40495-020-00216-7. Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano S., Kondo K., Yamaguchi M., Richmond G., Hutchison M., Wakeling A., et al. Distribution and function of EGFR in human tissue and the effect of EGFR tyrosine kinase inhibition. Anticancer Research. 2003;Vol. 23(5A):3639–3650. https://europepmc.org/article/med/14666659 [PubMed] [Google Scholar]
- Yazihan N., Tanacan A., Erol S.A., Anuk A.T., Sinaci S., Biriken D., et al. Comparison of VEGF-A values between pregnant women with COVID-19 and healthy pregnancies and its association with composite adverse outcomes. Journal of Medical Virology. 2021;93(4):2204–2209. doi: 10.1002/jmv.26631. [DOI] [PubMed] [Google Scholar]
- Youns M., Hoheisel J.D., Efferth T. Traditional Chinese Medicines (TCMs) for molecular targeted therapies of tumours. Current Drug Discovery Technologies. 2010;7(1):37–45. doi: 10.2174/157016310791162730. [DOI] [PubMed] [Google Scholar]