Abstract
Twelve human infections with Entamoeba spp. producing uninucleated cysts were studied. DNA was extracted from infected feces and used to amplify part of the ameba small-subunit rRNA gene. Sequence analysis identified four distinct types of Entamoeba, all of which are related to Entamoeba polecki and E. chattoni and two of which have not been reported previously. Whether these genetic types represent different species is unclear. We propose that the agent of all human infections with uninucleated cyst-producing Entamoeba species be reported as “E. polecki-like.”
Human cases of infection with the uninucleated cyst-producing Entamoeba species referred to as Entamoeba polecki are considered to be rare (2, 4), except in Papua New Guinea, where prevalence rates as high as 30% are reported (1, 5), and are often associated with contact with pigs. However, eight cases of human infection with a uninucleated cyst-producing Entamoeba species have been reported; these cases resulted from contact with monkeys, and the agent was identified as E. chattoni (6). The taxonomic status of these uninucleated Entamoeba species over the years has been confusing. They have been identified in various domestic and other animals and have been given separate names, such as E. bovis in cattle, E. ovis in sheep, E. suis and E. polecki in pigs, E. debliecki in pigs and goats, and E. chattoni in monkeys. However, the various species cannot be distinguished from each other morphologically (3), and whether they occur in humans or are even genetically distinct remains to be established. Burrows (3) suggested the use of the name E. polecki for the infectious agent in human cases until it became possible to distinguish one species of uninucleated Entamoeba from another. Other authors prefer to name all of these uninucleated ameba species E. chattoni (6).
During the last 4 years, our laboratory in Leiden, The Netherlands, has received many stool samples (n = 1,229) for species-specific diagnosis of E. histolytica and E. dispar infections. In most cases, E. histolytica/E. dispar-like cysts were found in feces from individuals without clinical signs; a few samples were from patients with clinical signs of amebiasis. From all stool samples, parasite DNA was isolated using spin columns (QIAgen, Hilden, Germany), and PCR–soluble hybridization enzyme-linked assay was performed to identify and differentiate E. histolytica and E. dispar (8, 9). All samples which did not produce a product upon amplification (i.e., were negative) were tested for the presence of inhibitors by spiking individual negative samples with 2 μl (approximately 0.2 ng) of E. dispar DNA and reamplifying with the E. dispar reaction mix. There was no evidence of inhibition in any of the negative samples.
In 15 cases, microscopy revealed uninucleated Entamoeba cysts in which the appearance of the nucleus, inclusions, and chromatoidal bodies suggested that these were unlikely to be immature cysts of E. histolytica or E. dispar. Furthermore, PCR–soluble hybridization enzyme-linked assay reactions for E. histolytica and E. dispar in these samples were negative. We classified such cysts as non-E. histolytica/non-E. dispar cysts, possibly E. polecki or E. chattoni. To confirm the morphological findings, we designed PCR primers based on the known small-subunit rRNA gene sequences for E. polecki and E. chattoni (GenBank accession no. AF149913 and AF149912) such that DNA should be amplified for E. polecki or E. chattoni specifically. The E. polecki-specific primer set consisted of forward primer Epolecki1 (5′-TCG ATA TTT ATA TTG ATT CAA ATG-3′) and reverse primer Epolecki2 (5′-CCT TTC TCC TTT TTT TAT ATT AG-3′), and the E. chattoni-specific primer set consisted of forward primer Echattoni1 (5′-AGG ATT TGT TTT ATA ACA AGT TC-3′) and reverse primer Echattoni2 (5′-TAA ATA ACC TTT CTC CTT TTT CTA TC-3′).
Amplification reactions were performed in a volume of 40 μl containing PCR buffer (10 mM Tris-HCl, [pH 9.0], 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, and 0.01% [wt/vol] gelatin; HT Biotechnology, Cambridge, United Kingdom), each deoxynucleoside triphosphate at 200 μM, 25 pmol of each specific primer, 1 U of Taq polymerase (SuperTaq HC; HT Biotechnology), and 2 μl of the DNA sample. Amplification consisted of 5 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C; and finally 2 min at 72°C. Only 1 sample was positive with the E. polecki primers, and 2 samples were positive with the E. chattoni primers; the other 12 samples remained negative.
To prove that Entamoeba species were indeed present in the negative samples, we designed general Entamoeba primers based on the small-subunit rRNA gene sequences for E. polecki, E. chattoni, E. dispar, E. histolytica, E. hartmanni, and E. coli (GenBank accession no: AF149913, AF149912, Z49256, X64142, AF49906, and AF149915, respectively). Forward primer Entam1 (5′-GTT GAT CCT GCC AGT ATT ATA TG-3′) and reverse primer Entam2 (5′-CAC TAT TGG AGC TGG AAT TAC-3′) were chosen from conserved regions so that DNA of all Entamoeba species would be amplified. Amplification was performed under the conditions described above. In all 15 samples with uninucleated cysts, the expected amplicon of approximately 550 bp was produced. For further analysis, sequencing of the products was performed using the BigDye terminator method (ABI Prism 310 system; Perkin-Elmer, Warrington, United Kingdom). Both strands were sequenced with the primers used for PCR. Sequences were edited with Sequence Navigator software (Perkin-Elmer). Three samples revealed sequences that appeared to be the result of a mixture of different species, even though by microscopy only one type of cyst seemed to be present. The other 12 sequences were aligned using the Multalign program (http://www.toulouse.inra.fr/) with the corresponding regions of the E. polecki, E. chattoni, E. dispar, E. histolytica, E. hartmanni, and E. coli sequences (Fig. 1). The alignment was then used to produce a phylogenetic tree using PAUP* 4.0 (D. L. Swofford, Sinauer Associates, Sunderland, Mass., 1998) (Fig. 2).
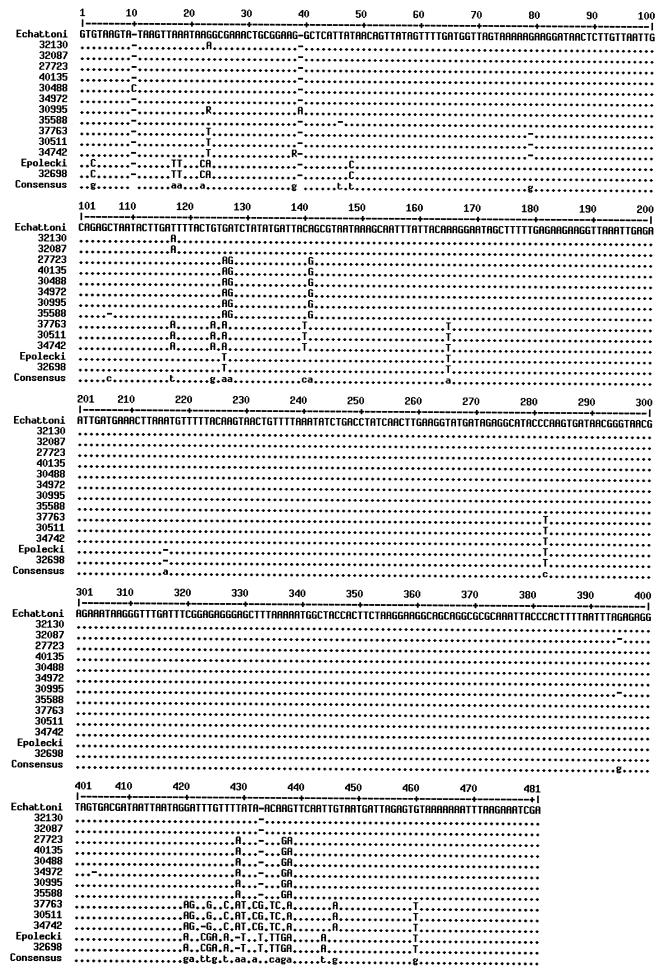
FIG. 1.
Multiple sequence alignment with hierarchical clustering. Dots indicate identity with the E. chattoni sequence (GenBank accession no. AF149912).
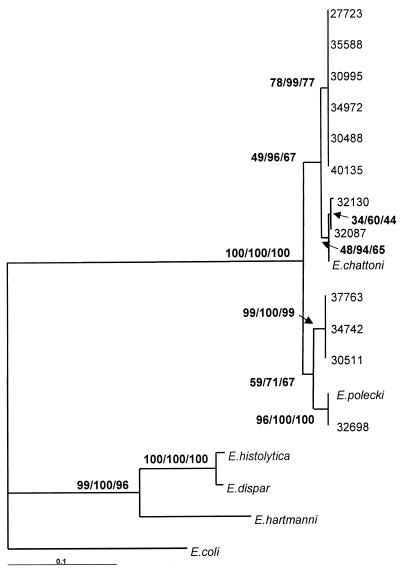
FIG. 2.
Phylogenetic analysis of partial ribosomal DNA sequences. The alignment in Fig. 1 with the added sequences was edited by hand, and phylogenetic analyses were performed using PAUP* 4.0. Maximum likelihood analysis used the HKY model of nucleotide substitution and a transition/transversion ratio of 2, and 100 bootstrap replicates were performed. For both minimum evolution and maximum parsimony analyses, a fast heuristic search was performed with random stepwise addition and 500 bootstrap replicates. Bootstrap support for each analysis is shown at each individual node in the order maximum likelihood, minimum evolution, and maximum parsimony. The scale bar represents the tree distance for the 0.1 changes per site in the sequence.
A large genetic distance exists between the uni-, tetra-, and octanucleated cyst-forming Entamoeba species, as described by Silberman et al. (7). As shown in the phylogenetic tree, all 12 of our sequences cluster with the E. polecki and E. chattoni reference sequences and are widely separated from E. coli on one hand and from E. histolytica, E. dispar, and E. hartmanni on the other. Interestingly, within the uninucleated sequence group, four variants are clearly distinguishable. This is already evident in the alignment and is supported by the phylogenetic tree. The sequence from the sample that produced an amplicon with the E. polecki-specific primers was identical to the corresponding region of the GenBank sequence for E. polecki. The two samples that produced amplicons with the E. chattoni primers were almost identical to the corresponding region of the E. chattoni GenBank sequence. It is likely that the other 12 samples were initially negative for the E. chattoni- and E. polecki-specific reactions due to sequence divergence in one or more of the primer-binding sites.
We have shown that there are (at least) four genetic types of uninucleated cyst-producing Entamoeba species that infect humans. Unfortunately, any mixed infections of uninucleated Entamoeba species with E. histolytica or E. dispar would have been missed in this study because only E. histolytica/E. dispar-negative samples were used. Therefore, the prevalence of the infection cannot be accurately calculated.
At present we do not know whether the E. chattoni-like infections originated from contact with monkeys or whether the E. polecki-like infections came from pigs. What is clear, however, is that humans can undoubtedly be infected with uninucleated cyst-producing Entamoeba species and that more genetic variability exists within this group than previously has been recognized in human infections. Whether the two new uninucleate sequence types correspond to previously described species in other animals remains unknown, as material for comparison has not been available. As there is no consensus on the use of ribosomal sequences to define new species of protozoa, until the species involved can be identified or named, we suggest that the agent of all uninucleated Entamoeba infections in humans be reported as “E. polecki-like.”
Acknowledgments
We acknowledge Erik Claas for introducing Sequence Navigator software and Ronald van Soest for running sequence reactions on the ABI Prism 310 system. We thank Sandra Duivenvoorden for technical assistance.
REFERENCES
- 1.Barnish G, Ashford R W. Occasional parasitic infections of man in Papua New Guinea and Irian Jaya (New Guinea) Ann Trop Med Parasitol. 1989;83:121–135. doi: 10.1080/00034983.1989.11812320. [DOI] [PubMed] [Google Scholar]
- 2.Burrows R B, Klink G E. Endamoeba polecki infections in man. Am J Hyg. 1955;62:156–167. doi: 10.1093/oxfordjournals.aje.a119769. [DOI] [PubMed] [Google Scholar]
- 3.Burrows R B. Morphological differentiation of Entamoeba hartmanni and E. polecki from E. histolytica. Am J Trop Med Hyg. 1959;8:583–589. doi: 10.4269/ajtmh.1959.8.583. [DOI] [PubMed] [Google Scholar]
- 4.Chacin-Bonilla L, Bonilla E, Parra A M, Estevez J, Morales L M, Suarez H. Prevalence of Entamoeba histolytica and other intestinal parasites in a community from Maracaibo, Venuzuela. Ann Trop Med Parasitol. 1992;86:373–380. doi: 10.1080/00034983.1992.11812680. [DOI] [PubMed] [Google Scholar]
- 5.Desowitz R S, Barnish G. Entamoeba polecki and other intestinal protozoa in Papua New Guinea Highland children. Ann Trop Med Parasitol. 1986;80:399–402. doi: 10.1080/00034983.1986.11812040. [DOI] [PubMed] [Google Scholar]
- 6.Sargeaunt P G, Patrick S, O'Keeffe D. Human infections of Entamoeba chattoni masquerade as Entamoeba histolytica. Trans R Soc Trop Med Hyg. 1992;86:633–634. doi: 10.1016/0035-9203(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 7.Silberman J D, Clark C G, Diamond L S, Sogin M L. Phylogeny of the genera Entamoeba and Endolimax as deduced from small-subunit ribosomal RNA sequences. Mol Biol Evol. 1999;16:1740–1751. doi: 10.1093/oxfordjournals.molbev.a026086. [DOI] [PubMed] [Google Scholar]
- 8.Verweij J J, Blotkamp J, Brienen E A T, Aguirre A, Polderman A M. Differentiation of Entamoeba histolytica and Entamoeba dispar cysts using polymerase chain reaction on DNA isolated from faeces with spin columns. Eur J Clin Microbiol Infect Dis. 2000;19:358–361. doi: 10.1007/s100960050494. [DOI] [PubMed] [Google Scholar]
- 9.Verweij J J, van Lieshout L, Blotkamp C, Brienen E A T, van Duivenvoorden S, van Esbroeck M, Polderman A M. Differentiation of Entamoeba histolytica and Entamoeba dispar using PCR-SHELA and comparison of antibody response. Arch Med Res. 2000;31(Suppl. 4):S44–S46. doi: 10.1016/s0188-4409(00)00221-6. [DOI] [PubMed] [Google Scholar]