Abstract
Background
Despite improved outcomes with the introduction of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in the treatment of patients with advanced non-small cell lung cancer (NSCLC) whose tumors harbor EGFR-activating mutations, unfortunately most patients eventually develop drug resistance. We and others recently reported that AXL activation confers acquired and intrinsic EGFR TKI resistance and represents a bypass resistance mechanism analogous to MET amplification in a subset of patients. This study aims to better assess the mechanisms whereby specific AXL inhibitors overcome such EGFR TKI resistance in NSCLC.
Methods
AXL inhibitors including MGCD265 (glesatinib), MGCD516 (sitravatinib) and R428 (BGB-324) alone or in combination with erlotinib were used to test the inhibitory effect on EGFR TKI resistant NSCLC cells. Subsequently, the effects of single or combinational treatment on cell cycle and apoptosis were assessed. Then, RNA sequencing study was conducted to evaluate the dynamic gene expression profile changes and consequently based on key cellular pathway alterations studies of migration and EMT were pursued.
Results
Administration of AXL inhibitors in combination with erlotinib significantly inhibited the growth of erlotinib-resistant NSCLC cells through potently inducing G2-M cell cycle arrest and enhancing apoptosis, relative to single agent treatment. RNA-sequencing analysis identified that several groups of genes enriched in cell survival inhibition or apoptosis promotion were upregulated, whereas genes enriched in DNA replication and repair, cell cycle and cell division were downregulated in cells treated with the combination of erlotinib and AXL inhibitor. Lastly, in line with pathway alterations indicating impaired migration, experiments showed reduced migration and EMT upon combination therapy.
Conclusions
Our results indicate that effective blockade of the AXL pathway may represent a novel strategy to overcome EGFR TKI resistance for the treatment of biomarker-selected subsets of NSCLC patients.
Keywords: Epidermal growth factor receptor (EGFR), tyrosine kinase inhibitor (TKI) resistance, AXL receptor tyrosine kinase, RNA-sequencing, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
Introduction
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer and accounts for ~85% of the cases (1,2). Activating epidermal growth factor receptor (EGFR) mutations, most commonly exon 19 deletions and the L858R mutation of exon 21 can be identified in 10–35% NSCLC patients and have been shown to play a driver oncogenic role in malignant transformation and progression (1-5). EGFR mutations are usually heterozygous, with the mutant allele also commonly showing gene amplification (4,5). These mutations increase constitutive activation of the receptor without ligand binding, leading to hyperactivation of downstream pro-survival signaling pathways (4,5). The over-activation of growth-promoting signaling is associated with tumor progression and metastasis.
EGFR targeted therapy using tyrosine kinase inhibitors (TKIs) has shown improved outcomes in patients with advanced NSCLC harboring EGFR mutations and is the established standard of care for front-line management (2-5). Erlotinib is an FDA-approved first-line EGFR TKI that has demonstrated superiority over standard platinum-based chemotherapy (6-9). While outcomes are clearly improved with the use of EGFR TKIs, a major challenge is that tumors inevitably acquire resistance to these drugs and disease progression ensues (2-4,10,11). Secondary mutations in EGFR (most commonly T790M) have been revealed conferring resistance to first and second generation EGFR TKIs in >50% of the NSCLC patients. MET amplification presents the second most common validated resistance mechanism occurring in another 5–10% (3,10). With the recent introduction of the highly potent EGFR T790M inhibitor, osimertinib in front-line management, bypass resistance mechanisms, such as MET amplification are noted with increasing frequency (12). We and others recently demonstrated that overexpression and activation of AXL can also confer resistance to EGFR TKI treatment such as erlotinib and the 3rd generation agent, osimertinib, including acquired bypass resistance and intrinsic resistance leading to the emergence of EGFR TKI tolerant cells (13,14). AXL is a receptor tyrosine kinase (RTK) that has been demonstrated to be overexpressed and activated in many human cancers (such as lung, breast, and pancreatic cancer) and correlated with poor prognosis, epithelial-to-mesenchymal transition (EMT), metastasis and drug resistance (15). Interestingly, emerging evidence has demonstrated that blockade of AXL in various model systems with specific small molecule kinase inhibitors can lead to restored drug sensitivity and improved therapeutic efficacy, defining AXL as a promising novel treatment target in a multitude of settings (14,16-18).
Based on the aforementioned information of AXL in therapeutic resistance, we here present a series of studies aimed at understanding the pathways governed by AXL activation and to determine whether AXL inhibition could help restore sensitivity to EGFR TKI therapy. We tested the effect of 3 AXL inhibitors including MGCD265, MGCD516 and R428 on a well-characterized erlotinib-resistant NSCLC cell model. MGCD265 and MGCD516 are multi-targeted TKIs which bind to and inhibit the phosphorylation of several RTKs, including the MET receptor (hepatocyte growth factor receptor), the Tie-2 receptor, vascular endothelial growth factor receptor (VEGFR), and AXL (19-21). In several preclinical studies, MGCD265 and MGCD516 demonstrated tumor regression in multiple human xenograft tumor models in mice (20,21). R428 is a highly selective inhibitor that specifically targets AXL. R428 administration has been reported to reduce metastatic burden and to extend survival in mouse models of breast cancer metastasis (22). The results we obtained indeed indicate that AXL inhibition at least partially restored the sensitivity of erlotinib via downregulation of MAPK and PI3K/Akt pathways resulting in induction of cell cycle arrest and apoptosis. We also pursued RNA-sequencing analyses and identified that several key groups of genes involved in cell survival inhibition or apoptosis promotion were upregulated, whereas a number of downregulated genes were involved in cell cycle, DNA replication and repair in cells treated with different agents. Our results further identify AXL as a promising therapeutic target to overcome drug resistance and cancer progression.
Methods
Antibodies and reagents
Antibodies against total and phosphorylated AXL, EGFR, MET, AKT, ERK1/2, and anti-GAPDH antibody were purchased from Cell Signaling Technologies (Danvers, MA). Erlotinib, MGCD265, MGCD516 and R428 were purchased from Selleck Chemicals (Houston, TX). Bromodeoxyuridine (BrdU) and 7-amino-actinomycin D (7-AAD) were purchased from BD Biosciences (San Jose, CA).
Cell culture
HCC827 human lung cancer cell lines were purchased from the American Type Culture Collection (Manassas, VA). HCC827-ER3 cells were established as previously described (13). HCC827 and HCC827-ER3 cells were maintained in RPMI 1640 medium supplemented with 10% FBS and 1× Antibiotic/Antimycotic (Life Technologies, Carlsbad, CA). All cell lines were recently tested and authenticated using DNA fingerprinting Short Tandem Repeat (STR) analysis at the Genomics Core in the Department of Genetics, Albert Einstein College of Medicine.
Cell viability assay
Cell viability assays were performed as described previously (13).
Western blot
The cell lysate preparation, SDS-PAGE electrophoresis and nitrocellulose membrane transfer, and primary antibody incubation were performed as described previously (23). For detection, the membranes were incubated with a horseradish peroxidase-conjugated secondary antibody for 1 h and the image was visualized with an ECL detecting kit (Amersham Biosciences, Piscataway, NJ).
Cell cycle analysis
The cells were harvested, fixed, and stained with BrdU and 7-aminoactinomycin-D (7-AAD) following the protocol provided by the manufacturer. The cell cycle data was collected via FACS Calibur (BD Biosciences) and analysis was accomplished with FlowJo (Tree Star, Inc.).
Apoptosis assay
Cellular apoptosis was analyzed using an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Life Technologies, Carlsbad, CA). Briefly, the cells were treated with relevant agents at described concentrations for 72 h. The cells were mixed with 5 µL Annexin V-FITC and 10 µL of 20 µg/mL PI reagents, and then incubated at room temperature in the dark for 20 min. After adding 400 µL PBS, the cells were immediately subjected to flow cytometry analysis using FACS Canto II flow cytometer (BD Biosciences).
Wound healing assay
Wound healing assay was performed to measure two-dimensional cancer cell movement. Briefly, cells were grown to full confluence in six-well plates. A scratch was made on the cell monolayer using a sterile 200 µL pipette tip. The monolayer was washed twice and incubated in drug-containing medium for 24 and 48 h, respectively. Cells were observed and images were taken using a light microscope (Olympus IX-71).
RNA sequencing
Total RNA was extracted from H827-ER3 cells treated with indicated agents according to the manufacturer’s protocol (RNeasy mini kit, Qiagen). The purified mRNA was used for RNA-seq library construction and whole transcriptome analysis. Libraries were sequenced on the Illumina HiSeq 2500 platform. The raw sequence reads were aligned to the human transcriptome (version GRCh37.75) using the Salmon software (version 2.1.0) and subsequently processed with R (version 3.5.1)/Bioconductor (version 3.8) for import into R and aggregation of transcript-level abundance estimates to the gene level. Gene-level differential expression analysis between sample groups was performed using DESeq2 (version 1.7.3) according to recommended practices. The adjusted p-value cutoff [false detection rate (FDR)] was set to 0.05.
Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
The function of the differentially-expressed genes (DEGs) was subjected to GO analysis according to the principles of GO, which organizes genes into hierarchical categories and reveals the gene regulatory networks on the basis of biological process and molecular function (24). To further understand the function of these DEGs, pathway analysis was performed to examine the significant pathways of the DEGs according to the KEGG databases. An online Bioinformatics enrichment tool (DAVID; david.ncifcrf.gov) was used to perform the GO and pathway analysis in the present study. Biological processes of GO terms were illustrated in the GO analysis. The p-value indicated the significance of GO term and pathway term enrichment in the differentially-expressed mRNA list (P<0.05 was considered to be statistically significant).
Quantitative RT-PCR
Quantitative RT-PCR was performed to validate gene expression changes identified by RNA sequencing. Briefly, total RNA was extracted from drug-treated HCC827-ER3 cells using the RNeasy mini kit (Qiagen, Valencia, CA). cDNA was synthesized with SuperScript III reverse transcriptase using oligo(dT) primers (Life Technologies, Carlsbad, CA), and RT-PCR was performed on a LightCycler with SYBR Green probes (Thermo Scientific) according to the manufacturer’s protocol. Ratios of the expression level of each gene to that of the reference gene were then calculated. Sequences for the primers used for quantitative RT-PCR for the selected genes are listed in Table S1.
Table S1. Primer sequences used for qPCR.
| Gene name | Sense primer sequence | Antisense primer sequence |
|---|---|---|
| GAPDH | CTT TGG TAT CGT GGA AGG ACT C | GTA GAG GCA GGG ATG ATG TTC |
| SIAH1 | TCC ACC TTC TCT GTA CTC CTG | TGG ACA CTT CGA GGT ACC G |
| RPS6KA5 | TGA GTA AGG AGT TTG TGG CTG | ACT GCC TTG TCA TGT CCT G |
| JDP2 | GCT GAA ATA CGC TGA CAT CCG | TTT TCC TTC GCT CCT CTT CC |
| MKNK2 | AGA AAC CAG CCG AAC TTC AG | CCA AAG TCA GAG TCT CCG TG |
| TRIB3 | TGA TCT CAA GCT GTG TCG C | AGT ATC TCA GGT CCC ACG TAG |
| PLEC1 | CAG TAC ATC AAC GCC ATC AAG | GAC TGG ACC TTG GGC TTC |
| DDIT4 | GTT TGA CCG CTC CAC GAG | GTG TTC ATC CTC AGG GTC AC |
| SESN2 | GCC TCA CCT ACA ATA CCA TCG | CAC CTC CCC ATA ATC ATA GTC ATC |
| HSPA8 | GGA CAA GAG TAC GGG AAA AGA G | GTC CCT CTG CTT CTC ATC TTC |
| MASTL | TCA ATC CCA CAC CTT CAT ATC C | TTT CAA CTG CAT TCC AAC TCA TC |
| MTFP1 | GCG GAT GCC ATT GAC AAA G | CTA GAG CCT GCC ATA CAA AGG |
| NBEAL2 | GCC ACT TCA TCG ACA AAC AG | TGT CAT AGC AGG CAT TCC AG |
| NDOR1 | GCT CTT TGA ATA CTG CAA CCG | GAT GAG GTC CAA CAG GTA GTC |
Statistical analysis
Data were presented as the mean ± standard deviation. Data was analyzed using a two-tailed t-test. P<0.05 was considered to be statistically significant.
Results
AXL inhibition partially but significantly restored erlotinib sensitivity in HCC827-ER3 cells, an erlotinib-resistant cell line
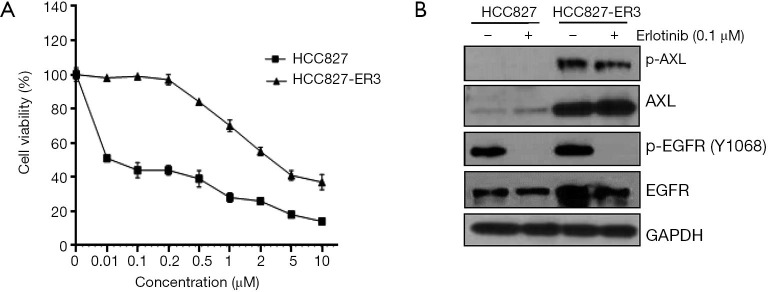
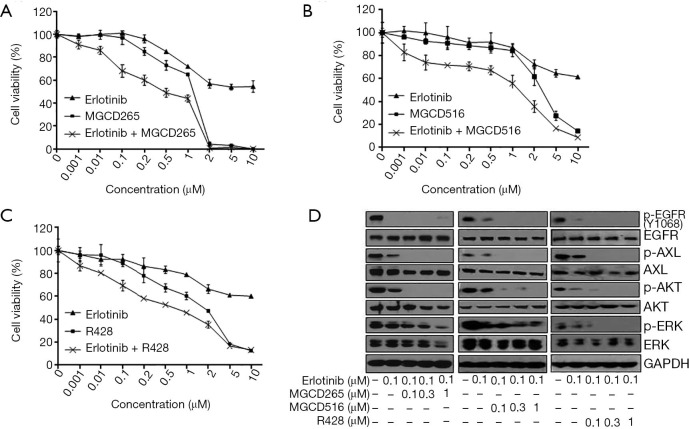
We previously reported that increased expression and activation of AXL conferred resistance to erlotinib treatment and AXL inhibition may overcome acquired resistance to erlotinib in biomarker-selected patient (13). Thus in the current study, we tested a variety of distinct AXL inhibitors to determine whether they could exert such a role and inhibit erlotinib-resistant tumor cell growth. The erlotinib-resistant HCC827-ER3 cells used in this study were established previously (13). As shown in Figure 1A,B, HCC827-ER3 cells exhibit robust resistance to erlotinib treatment, with significant upregulation of AXL and EGFR (Figure 1B). Erlotinib treatment significantly repressed EGFR activation in HCC827-ER3 cells, whereas the phosphorylation of AXL was not affected (Figure 1B). Subsequently, we tested the effect of AXL inhibitors: MGCD265, MGCD516 and R428 on HCC827-ER3 cells. We found that MGCD265, MGCD516 and R428 exhibited modest inhibitory effects when used as a single agent, although more potent compared to erlotinib alone in HCC827-ER3 cells (Figure 2A,B,C). More importantly, the combination of AXL inhibitor using any of the three agents and erlotinib more significantly repressed cell growth than single agent treatment (Figure 2A,B,C). These results indicate that blockade of AXL signaling partially but significantly restore erlotinib sensitivity in NSCLC cells. The subsequent signaling pathway analysis demonstrated that the combination of erlotinib and MGCD265 significantly decreased EGFR, AXL and downstream AKT and ERK1/2 phosphorylation whereas erlotinib alone only showed a modest inhibitory effect (Figure 2D). Taken together, these results indicate that AXL inhibitor and erlotinib combination treatment can at least partially reverse the acquired resistance of erlotinib in AXL-overexpressing NSCLC cells.
Figure 1.
Upregulation of AXL confers acquired resistance to erlotinib in HCC827-ER3 cells. (A) HCC827 and HCC827-ER3 cells were treated with indicated concentrations of erlotinib for 72 hours and viability was measured by MTS cell viability assay. Results are from three independent experiments and are expressed as percent viability of vehicle-treated cells. Data are shown as the mean ± SD. (B) Cells were treated with or without indicated dose of erlotinib for 6 hours, after which cell lysates were prepared and subjected to immunoblot analysis with antibodies to phosphorylated (p) or total forms of AXL, EGFR and GAPDH (loading control). EGFR, epidermal growth factor receptor.
Figure 2.
Inhibition of AXL activation in combination with erlotinib treatment synergistically inhibited the cell growth of HCC827-ER3 cells. (A,B,C) HCC827-ER3 cells were treated with indicated concentrations of erlotinib or the AXL inhibitors MGCD265 (A), MGCD516 (B) or R428 (C) or the combination of erlotinib with the indicated AXL inhibitor for 72 hours. Cell viability was measured using the MTS cell viability assay kit. Data are expressed as percent viability of vehicle-treated cells. Results are expressed as the mean ± SD. (D) HCC827-ER3 cells were treated with the indicated dose of erlotinib or a combination of erlotinib and one of the AXL inhibitors: MGCD265, MGCD516 or R428 for 6 hours. The cell lysates were prepared and subjected to immunoblot analysis with antibodies to phosphorylated (p) or total forms of EGFR, AXL, MET, AKT, ERK and GAPDH.
Combination of AXL inhibition and erlotinib treatment significantly delayed S phase, led to a G2-M phase cell cycle arrest and enhanced cellular apoptosis
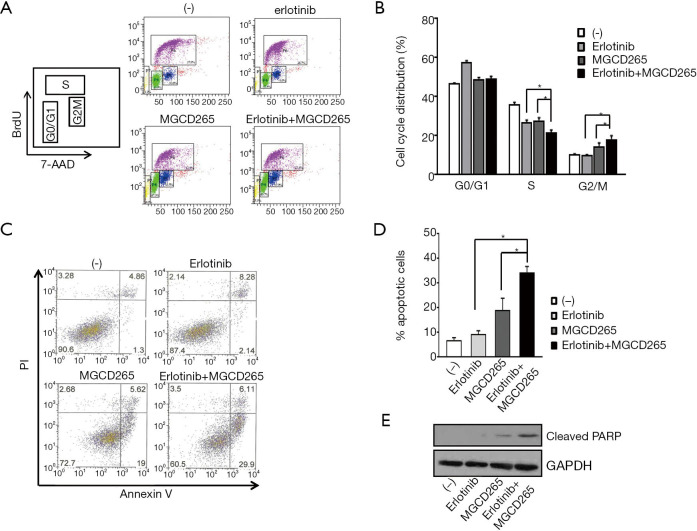
We next attempted to dissect the intrinsic mechanisms by which AXL inhibition restores erlotinib sensitivity. First, a cell cycle analysis was performed to test if the induction of cell cycle arrest could contribute to the anti-proliferative potency of AXL inhibition in HCC827-ER3 cells. We used the model inhibitor, MGCD265 to perform our subsequent experiments. As shown in Figure 3A,B, relative to erlotinib or MGCD265 single agent treatment which showed some inhibitory effect on S phase cell cycle, the combination of erlotinib and MGCD265 treatment led to more significant repression of S phase. Meanwhile, we also found that MGCD265 alone treatment slightly increased G2-M arrest compared to untreated control or erlotinib treatment in HCC827-ER3 cells (Figure 3B). Importantly, MGCD265 and erlotinib combination treatment not only significantly repressed S phase, but further induced stronger and more extended G2-M arrest relative to either MGCD265 or erlotinib alone treatment. These results clearly indicate that inhibition of cell proliferation by the combination of erlotinib and MGCD265 may be mediated at least partially through inhibiting S phase entry and inducing G2-M phase cell cycle arrest.
Figure 3.
Combination of AXL inhibition and erlotinib treatment significantly delayed S phase, led to G2-M phase cell cycle arrest, and enhanced cellular apoptosis. HCC827-ER3 cells were treated with indicated concentrations of erlotinib, MGCD265 or combination of erlotinib and MGCD265 for 72 hours. Cells were stained with BrdU and 7-AAD and cell cycle distribution was analyzed using flow cytometry. Representative flow cytometric plots of BrdU versus 7-AAD are shown in panel (A) and percentages of cell cycle distributions in each treated group of cells were shown in panel (B). The results are presented as the mean ± SD. *, P<0.05. (C) Cell apoptosis was determined by flow cytometry with Annexin V/PI dual-staining; and (D) the percentage of apoptotic cells in each treated group is shown. All data were expressed as the mean ± SD of three experiments. *, P<0.05. (E) HCC827-ER3 cells were treated as described above. Cell lysates were prepared and subjected to immunoblot analysis with anti-cleaved PARP and GAPDH antibodies.
Subsequently, the effect of MGCD265 and/or erlotinib on cellular apoptosis was evaluated by staining with FITC Annexin V and PI followed by flow cytometry analysis. As shown in Figure 3C,D, erlotinib treatment induced a modest increase in cell death whereas MGCD265 alone treatment caused a remarkable increase of cellular apoptosis relative to untreated control cells in HCC827-ER3 cells. Of note, the combination of the two compounds showed significant enhancement of apoptosis compared to single agent treatment. Cleaved PARP has been used widely as a validated marker for apoptosis (25). Thus, we next tested the effect of MGCD265 and/or erlotinib treatment on cleaved PARP expression. We found that MGCD265 alone treatment significantly increased cleaved-PARP levels to a higher degree than noted in erlotinib-treated cells. Relative to MGCD265, combination treatment with the two compounds yielded much more cleaved-PARP than either agent alone (Figure 3E). Taken together, these results provide clear evidence that erlotinib and MGCD265 combination therapy induced cellular apoptosis more significantly than either MGCD265 or erlotinib alone treatment.
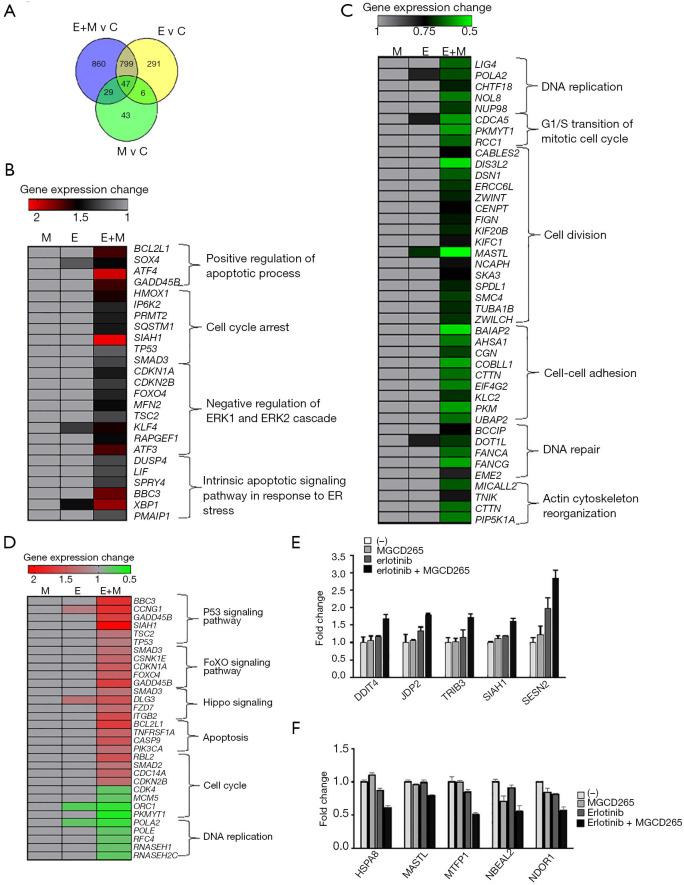
GO and KEGG pathway analysis of differentially expressed genes (DEGs) in HCC827-ER3 cells treated with combination of erlotinib and AXL inhibitor
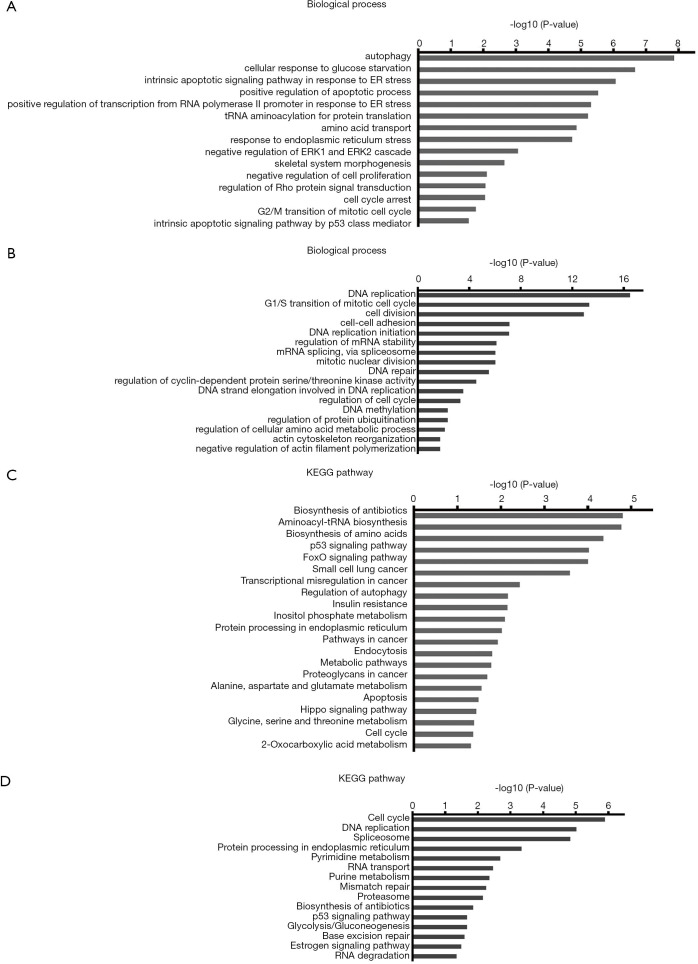
To gain further insights into the biological mechanisms through which EGFR/AXL inhibitor treatment represses cell functions, RNA-sequencing was conducted to assess the global gene expression profile under different treatment conditions. RNA-sequencing revealed that in total 875 DEGs were upregulated and 872 DEGs were downregulated in the combinational treatment with erlotinib and MGCD265 as compared with untreated control samples. In order to arrive at functional interpretations for the identified DEGs, the online DAVID database was applied to assess the GO categories and KEGG pathways of these DEGs. The top enriched GO terms with respect to biological processes for up-regulated and down-regulated DEGs are shown in Figure 4A,B, respectively. In this regard, it is relevant to note that most up-regulated DEGs were enriched in the biological processes of cell growth inhibition, such as apoptosis promotion, negative regulation of cell proliferation, and cell cycle arrest; whereas the down-regulated DEGs were mostly enriched in DNA replication and repair, cell cycle and cell division.
Figure 4.
GO function and KEGG pathway enrichment analysis of DEGs identified in HCC827-ER3 cells treated with combination of erlotinib and MGCD265. (A,C) The top biological process terms of GO analysis and the top KEGG signaling pathways enriched amongst the upregulated genes. (B,D) The top biological process terms and the top KEGG signaling pathways enriched amongst the downregulated genes. The y-axis indicates functional groups by GO category. The x-axis indicates –log (P value) of GO category. Significance is expressed as the P value calculated using Fisher’s exact test (P<0.05). GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially-expressed genes.
The KEGG pathway analyses for up-regulated and down-regulated DEGs are shown in Figure 4C,D, respectively. Notably, among the pathways that upregulated DEGs were enriched in, multiple pro-apoptotic pathways including p53 signaling, FoxO signaling, apoptosis and Hippo signaling pathway were highly ranked (Figure 4C); on the contrary, the top ranked pathways for those downregulated DEGs are signaling pathways regulating cell cycle and DNA replication and repair (Figure 4D). These results provide an accurate functional description as to how the global transcriptome is modulated by combinatorial treatment with erlotinib and AXL inhibitor leading to significant inhibition of erlotinib-resistant cell growth through promoting apoptosis and impairing cell cycle.
Comparison and validation of DEGs identified in HCC827-ER3 cells treated with EGFR/AXL inhibitors
Our RNA-seq data revealed that 1,143 genes were significantly differentially expressed with erlotinib, 125 genes with MGCD265 and 1,735 genes with the combinational treatment of erlotinib and MGCD265, in comparison to untreated control samples (Figure 5A). Our previous results demonstrated that combination treatment has a more robust effect on G2-M phase cell cycle arrest and apoptosis induction relative to erlotinib or MGCD265 alone treatment. In order to achieve a rational interpretation of the global transcriptomic changes with different treatments, we next extracted the DEGs that were found in erlotinib and MGCD265 combinational treatment and that were shown to be enriched in multiple biological process and KEGG pathways, and then compared the expression level of these DEGs in the combinational treatment samples with that in single erlotinib or MGCD265 treatment. Through comparing the level of those DEGs in different treatment conditions (erlotinib plus MGCD265 vs. erlotinib vs. MGCD265), we found that the expression levels of most upregulated DEGs with combination treatment were indeed higher than that in the erlotinib or MGCD265 alone treatment group (Figure 5B and Table S2); whereas for the downregulated DEGs enriched in regulating cell cycle arrest, DNA repair, and cell division, we observed that the levels of most DEGs were lower with combination treatment than in the erlotinib or MGCD265 alone treatment groups (Figure 5C and Table S3). Similarly, the expression level of most upregulated DEGs enriched in KEGG pathways were found to be higher with combination as opposed to single agent treatment, whereas the levels of downregulated DEGs were lower in combinational treatment than in single agent treatment (Figure 5D and Tables S4,S5). Next, we further focused on the DEGs (fold change >2; Table S6) in the group treated with combination of erlotinib and MDCG265 versus erlotinib alone as this is the group potentially defining the contribution of AXL inhibition to treatment response. To further verify the differential gene expression of these DEGs, we performed qPCR on 5 of the most upregulated genes including: DDIT4, JDP2, TRIB3, SIAH1, and SESN2; and total 5 of the most downregulated genes: HSPA8, MASTL, MTFP1, NBEAL2, and NDOR1. We confirmed that the combination of erlotinib and MDCG265 significantly increased the expression levels of the 5 upregulated genes whereas repressed the level of the selected 5 downregulated genes relative to that in the erlotinib or MDCG265 alone treated cohorts (Figure 5E,F). These results indicated that the combination of erlotinib and MGCD265 induced more significant changes in the transcriptome profile compared to single erlotinib or MGCD265 treatment, and these significant differences in transcriptomic changes promoted enhanced apoptosis and cell growth arrest in the combination-treated HCC827-ER3 cells.
Figure 5.
RNA-seq-based transcriptome comparison between different treatment conditions and validation of DEGs identified in these treatments. (A) Venn diagram showing the total number of DEGs (P<0.05) identified in HCC827-ER3 cells treated with erlotinib, MGCD265, or combination of erlotinib and MGCD265 (E, erlotinib; M, MGCD265; and E + M: erlotinib plus MGCD265). (B,C) The expression levels of representative upregulated DEGs (B) and downregulated DEGs (C) that enriched in multiple biological processes in HCC827-ER3 cells with indicated treatments. (D) The expression levels of representative DEGs that enriched in multiple KEGG pathways in HCC827-ER3 cells with indicated treatment. (E,F) The gene expression for 5 of the most highly upregulated (fold change >2) and 5 downregulated (fold change >2) genes were validated by qPCR. DEGs, differentially-expressed genes.
Table S2. Expression level of upregulated DEGs (vs. control) enriched in multiple biological processes.
| Gene symbol | Fold change (treatment vs. control) | ||
|---|---|---|---|
| MGCD265 + erlotinib | Erlotinib | MGCD265 | |
| Positive regulation of apoptotic process | |||
| BCL6 | 1.570810516 | 1.500691193 | 1 |
| BNIP3L | 1.396316619 | 1 | 1.280790364 |
| BCL2L1 | 1.693605198 | 1 | 1 |
| ARHGEF2 | 1.858789066 | 1.631498432 | 1 |
| SOS2 | 1.433273931 | 1.245688175 | 1 |
| SOX4 | 1.529794862 | 1.246951616 | 1 |
| TIAM | 1.219890263 | 1 | 1 |
| ATF4 | 1.99619867 | 1 | 1 |
| APBB2 | 1.290583732 | 1 | 1 |
| ANKRD1 | 1.599681138 | 1 | 1.5471155 |
| APH1A | 1.333287179 | 1 | 1 |
| CDK19 | 1.33659376 | 1 | 1 |
| DUSP6 | 1.567727508 | 1.465150443 | 1 |
| ECT2 | 1.300534572 | 1 | 1 |
| GADD45B | 1.678462013 | 1 | 1 |
| HMOX1 | 1.606346423 | 1 | 1 |
| HMGB1 | 1.398352809 | 1.429730499 | 1 |
| HIP1R | 1.271504711 | 1 | 1 |
| IP6K2 | 1.423977161 | 1 | 1 |
| JMY | 1.312008281 | 1.349909538 | 1 |
| NET1 | 1.256360135 | 1.211107554 | 1 |
| NUPR1 | 3.565986854 | 3.455794632 | 1 |
| PNMA2 | 1.317620231 | 1 | 1 |
| PMAIP1 | 1.273440546 | 1 | 1 |
| PSEN1 | 1.321673666 | 1.231496166 | 1 |
| PTGS2 | 1.509385178 | 1.501834979 | 1 |
| PRMT2 | 1.453487499 | 1 | 1 |
| RPS6KA2 | 1.597029169 | 1.604806177 | 1 |
| SAV1 | 1.200108843 | 1 | 1 |
| SQSTM1 | 1.470799563 | 1 | 1 |
| SIAH1 | 2.066825829 | 1 | 1 |
| TXNIP | 1.999955437 | 1.855142834 | 1 |
| TP53 | 1.230938025 | 1 | 1 |
| Cell cycle arrest | |||
| DDIT3 | 2.649910503 | 2.099035989 | 1 |
| HBP1 | 1.801522281 | 1.713758259 | 1 |
| SMAD3 | 1.310052368 | 1 | 1 |
| APBB2 | 1.290583732 | 1 | 1 |
| CDKN1A | 1.463461144 | 1 | 1 |
| CDKN2B | 1.346878936 | 1 | 1 |
| ERN1 | 1.514346277 | 1.424297261 | 1 |
| FOXO4 | 1.429176125 | 1 | 1 |
| JMY | 1.312008281 | 1.349909538 | 1 |
| MFN2 | 1.514552582 | 1 | 1 |
| PPP1R15A | 1.291899504 | 1.206875857 | 1 |
| TSC2 | 1.304248887 | 1 | 1 |
| TP53 | 1.230938025 | 1 | 1 |
| MYC | 1.410985277 | 1.266748014 | 1 |
| Negative regulation of ERK1 and ERK2 cascade | |||
| ERRFI1 | 1.261886742 | 1.289822014 | 1 |
| KLF4 | 1.599439716 | 1.347331821 | 1 |
| RAPGEF1 | 1.521917605 | 1 | 1 |
| XBP1 | 1.869085583 | 1.5056869 | 1 |
| ATF3 | 1.708230933 | 1 | 1 |
| DUSP4 | 1.296108342 | 1 | 1 |
| DUSP6 | 1.567727508 | 1.465150443 | 1 |
| EZR | 1.476568899 | 1.544670372 | 1.510260914 |
| LIF | 1.295467052 | 1 | 1 |
| SPRY4 | 1.334091553 | 1 | 1 |
| Intrinsic apoptotic signaling pathway in response to ER stress | |||
| DDIT3 | 2.649910503 | 2.099035989 | 1 |
| HBP1 | 1.801522281 | 1.713758259 | 1 |
| SMAD3 | 1.310052368 | 1 | 1 |
| APBB2 | 1.290583732 | 1 | 1 |
| CDKN1A | 1.463461144 | 1 | 1 |
| CDKN2B | 1.346878936 | 1 | 1 |
| ERN1 | 1.514346277 | 1.424297261 | 1 |
| FOXO4 | 1.429176125 | 1 | 1 |
| JMY | 1.312008281 | 1.349909538 | 1 |
| MFN2 | 1.514552582 | 1 | 1 |
| PPP1R15A | 1.291899504 | 1.206875857 | 1 |
| TSC2 | 1.304248887 | 1 | 1 |
| TP53 | 1.230938025 | 1 | 1 |
| MYC | 1.410985277 | 1.266748014 | 1 |
Table S3. Expression level of downregulated DEGs (vs. control) enriched in multiple biological processes.
| Gene symbol | Fold change (treatment vs. control) | ||
|---|---|---|---|
| MGCD265 + erlotinib | Erlotinib | MGCD265 | |
| Cell division | |||
| ATAD3B | 0.699796587 | 0.717925923 | 1 |
| CABLES2 | 0.750020498 | 1 | 1 |
| DIS3L2 | 0.53274437 | 1 | 1 |
| LIG4 | 0.650349446 | 1 | 1 |
| DSN1 | 0.659154236 | 1 | 1 |
| ERCC6L | 0.696356995 | 1 | 1 |
| FBXO5 | 0.609049566 | 0.695204352 | 1 |
| HAUS6 | 0.824159456 | 0.780089206 | 1 |
| NSUN2 | 0.757933959 | 0.774571282 | 1 |
| PDS5B | 0.793715408 | 0.767906154 | 1 |
| PHF13 | 0.770399369 | 0.781579307 | 1 |
| ZWINT | 0.714788883 | 1 | 1 |
| CDC20 | 0.787424854 | 1 | 1 |
| CDC25A | 0.598816259 | 0.65588207 | 1 |
| CDC6 | 0.638133071 | 0.724462682 | 1 |
| CDC7 | 0.819192058 | 0.824801908 | 1 |
| CDCA2 | 0.724016995 | 0.683917853 | 1 |
| CDCA5 | 0.598816259 | 0.789532223 | 1 |
| CENPT | 0.747000771 | 1 | 1 |
| CCNA2 | 0.796728486 | 0.828706336 | 1 |
| CCND3 | 0.597693581 | 0.689911532 | 1 |
| CCNE2 | 0.601208611 | 0.627989732 | 1 |
| CCNF | 0.811518208 | 1 | 1 |
| CDK2 | 0.747170127 | 0.785909293 | 1 |
| CDK4 | 0.809001405 | 1 | 1 |
| CDK6 | 0.779894059 | 0.796641826 | 1 |
| FAM64A | 0.778219456 | 1 | 1 |
| FIGN | 0.726964207 | 1 | 1 |
| HELLS | 0.763870624 | 0.770328066 | 1 |
| KLHL42 | 0.799134241 | 0.821392492 | 1 |
| KIF18B | 0.833116897 | 1 | 1 |
| KIF20B | 0.711042036 | 1 | 1 |
| KIFC1 | 0.770301058 | 1 | 1 |
| MASTL | 0.496263385 | 0.704866858 | 1 |
| MCM5 | 0.809011572 | 1 | 1 |
| NCAPG | 0.774262647 | 0.754577509 | 1 |
| NCAPH | 0.770217973 | 1 | 1 |
| NCAPD3 | 0.748339557 | 0.798079695 | 1 |
| NUDC | 0.77975868 | 1 | 1 |
| PRPF40A | 0.784792564 | 1 | 1 |
| PRKCE | 0.808702222 | 1 | 1 |
| RCC1 | 0.653393194 | 1 | 1 |
| SEPT | 0.79834066 | 0.796708043 | 1 |
| SKA3 | 0.750522021 | 1 | 1 |
| SPDL1 | 0.716440826 | 1 | 1 |
| SMC4 | 0.687627705 | 1 | 1 |
| TACC3 | 0.820450543 | 1 | 1 |
| TUBA1B | 0.707870107 | 1 | 1 |
| USP37 | 0.801775898 | 0.829695094 | 1 |
| VRK1 | 0.813398638 | 0.7946735 | 1 |
| VPS4A | 0.830147228 | 1 | 1 |
| ZWILCH | 0.701666802 | 1 | 1 |
| DNA replication | |||
| BARD1 | 0.637203915 | 0.759313554 | 1 |
| BRIP1 | 0.630692829 | 0.652681375 | 1 |
| LIG4 | 0.650349446 | 1 | 1 |
| POLA2 | 0.673108697 | 0.802900219 | 1 |
| POLD3 | 0.793045387 | 0.822547354 | 1 |
| POLE | 0.808760222 | 1 | 1 |
| DSCC1 | 0.672814491 | 0.705769428 | 1 |
| GINS2 | 0.635446077 | 0.730979491 | 1 |
| GINS3 | 0.767525935 | 0.784890792 | 1 |
| RAD1 | 0.784182393 | 0.781117533 | 1 |
| RBM14 | 0.726254055 | 0.751910676 | 1 |
| TICRR | 0.739873815 | 0.775313361 | 1 |
| CDC25A | 0.598816259 | 0.65588207 | 1 |
| CDC45 | 0.78138931 | 0.786718155 | 1 |
| CDC6 | 0.638133071 | 0.724462682 | 1 |
| CDC7 | 0.819192058 | 0.824801908 | 1 |
| CHAF1A | 0.725619903 | 0.749045337 | 1 |
| CHAF1B | 0.695921621 | 0.73503896 | 1 |
| CHTF18 | 0.720097815 | 1 | 1 |
| CLSPN | 0.515666821 | 0.545984201 | 1 |
| CDK2 | 0.747170127 | 0.785909293 | 1 |
| DTL | 0.671447943 | 0.728623805 | 1 |
| DUT | 0.808708758 | 1 | 1 |
| EXO1 | 0.600585149 | 0.668348878 | 1 |
| FEN1 | 0.646919176 | 0.673411842 | 1 |
| MCM10 | 0.579587004 | 0.673882478 | 1 |
| MCM4 | 0.796587629 | 0.786586647 | 1 |
| MCM5 | 0.809011572 | 1 | 1 |
| NOL8 | 0.628457322 | 1 | 1 |
| NUP98 | 0.692971474 | 1 | 1 |
| ORC1 | 0.651124396 | 0.75489963 | 1 |
| ORC6 | 0.74897745 | 0.713235537 | 1 |
| PCNA | 0.730196232 | 0.752830213 | 1 |
| RTEL1 | 0.818759095 | 0.816647045 | 0.795584455 |
| RFC4 | 0.825145898 | 1 | 1 |
| RFC5 | 0.657860132 | 0.739748932 | 1 |
| RRM1 | 0.785388749 | 0.832808076 | 1 |
| RRM2 | 0.637552411 | 0.685144139 | 1 |
| DNA repair | |||
| BCCIP | 0.748416646 | 1 | 1 |
| POLE | 0.808760222 | 1 | 1 |
| POLQ | 0.77025084 | 0.794883047 | 1 |
| DOT1L | 0.6937146 | 0.793343611 | 1 |
| FANCA | 0.644724544 | 1 | 1 |
| FANCG | 0.588956357 | 1 | 1 |
| FANCI | 0.799174667 | 0.829467802 | 1 |
| PDS5B | 0.793715408 | 0.767906154 | 1 |
| RAD1 | 0.784182393 | 0.781117533 | 1 |
| RAD18 | 0.75267768 | 0.716666737 | 1 |
| RAD51AP1 | 0.695368416 | 1 | 0.697240398 |
| RAD54L | 0.721212552 | 0.728997562 | 1 |
| RBM14 | 0.726254055 | 0.751910676 | 1 |
| TICRR | 0.739873815 | 0.775313361 | 1 |
| CHAF1A | 0.725619903 | 0.749045337 | 1 |
| CLSPN | 0.515666821 | 0.751910437 | 1 |
| CDK2 | 0.747170127 | 0.785909293 | 1 |
| EME2 | 0.799044431 | 1 | 1 |
| EXO1 | 0.600585149 | 0.668348878 | 1 |
| FEN1 | 0.646919176 | 0.673411842 | 1 |
| MCRS1 | 0.830340008 | 1 | 1 |
| MSH6 | 0.666630119 | 0.663940906 | 1 |
| NABP1 | 0.617556529 | 0.567306751 | 1 |
| RTEL1 | 0.818759095 | 0.816647045 | 0.795584455 |
| RFC5 | 0.657860132 | 0.739748932 | 1 |
| RFWD3 | 0.803087075 | 0.816565228 | 1 |
| TEX15 | 0.775624104 | 0.779854234 | 1 |
| UBE2T | 0.748803457 | 0.720974576 | 1 |
| Actin cytoskeleton reorganization | |||
| BCCIP | 0.748416646 | 1 | 1 |
| POLE | 0.808760222 | 1 | 1 |
| FARP2 | 0.811762735 | 1 | 1 |
| MICALL2 | 0.661973595 | 1 | 1 |
| TNIK | 0.780659561 | 1 | 1 |
| CTTN | 0.641550617 | 1 | 1 |
| EZR | 0.803732687 | 0.757486198 | 1 |
| FLNA | 0.826785759 | 1 | 1 |
| PIP5K1A | 0.626792059 | 1 | 1 |
Table S4. Expression level of upregulated DEGs (vs. control) enriched in multiple KEGG pathways.
| Gene symbol | Fold change (treatment vs. control) | ||
|---|---|---|---|
| MGCD265 + erlotinib | Erlotinib | MGCD265 | |
| P53 signaling pathway | |||
| BBC3 | 1.775598694 | 1 | 1 |
| CASP8 | 1.50509795 | 1.714538463 | 1 |
| CASP9 | 1.513227018 | 1 | 1 |
| CCNG1 | 1.775598694 | 1.233975191 | 1 |
| CCNG2 | 2.263213411 | 1.974321134 | 1 |
| CDKN1A | 1.463461144 | 1 | 1 |
| GADD45B | 1.678462013 | 1 | 1 |
| PMAIP1 | 1.273440546 | 1 | 1 |
| RRM2B | 1.353963073 | 1 | 1 |
| SESN2 | 6.626958929 | 5.933017006 | 1 |
| SIAH1 | 2.066825829 | 1 | 1 |
| TSC2 | 1.304248887 | 1 | 1 |
| TP53 | 1.230938025 | 1 | 1 |
| Apoptosis | |||
| BCL2L1 | 1.693605198 | 1 | 1 |
| CFLAR | 1.322209947 | 1 | 1 |
| TNFRSF1A | 1.392397204 | 1 | 1 |
| CASP8 | 1.50509795 | 1.714538463 | 1 |
| CASP9 | 1.513227018 | 1 | 1 |
| IKBKB | 1.529761794 | 1.37112148 | 1.457373682 |
| PIK3CA | 1.31888819 | 1 | 1 |
| TP53 | 1.230938025 | 1 | 1 |
| Cell cycle | |||
| BUB1 | 1.211254334 | 1 | 1 |
| RBL2 | 1.524049945 | 1 | 1 |
| SMAD2 | 1.294697918 | 1 | 1 |
| SMAD3 | 1.310052368 | 1 | 1 |
| CDC14A | 1.39217711 | 1 | 1 |
| CDC14B | 1.205490294 | 1.217160462 | 1 |
| CDKN1A | 1.463461144 | 1 | 1 |
| CDKN2B | 1.346878936 | 1 | 1 |
| GADD45B | 1.678462013 | 1 | 1 |
| STAG2 | 1.233069452 | 1 | 1 |
| TP53 | 1.230938025 | 1 | 1 |
| MYC | 1.410985277 | 1.266748014 | 1 |
| FoXO signaling pathway | |||
| BCL6 | 1.570810516 | 1.500691193 | 1 |
| FBXO25 | 1.442974414 | 1.42794837 | 1 |
| FBXO32 | 1.604190926 | 1.466903076 | 1 |
| GABARAPL1 | 1.587151943 | 1.524525299 | 1 |
| RBL2 | 1.524049945 | 1 | 1 |
| SMAD2 | 1.294697918 | 1 | 1 |
| SMAD3 | 1.310052368 | 1 | 1 |
| SOS2 | 1 | 1.245688175 | 1 |
| CSNK1E | 1.380889371 | 1 | 1 |
| CCNG2 | 2.263213411 | 1.974321134 | 1 |
| CDKN1A | 1.463461144 | 1 | 1 |
| FOXO4 | 1.429176125 | 1 | 1 |
| GADD45B | 1.678462013 | 1 | 1 |
| IKBKB | 1.529761794 | 1.37112148 | 1.457373682 |
| IRS2 | 1.385004242 | 1.331280376 | 1 |
| PIK3CA | 1.31888819 | 1 | 1 |
| PCK2 | 3.853122264 | 4.037367457 | 1 |
| Hippo signaling | |||
| BBC3 | 1.775598694 | 1 | 1 |
| SMAD2 | 1.294697918 | 1 | 1 |
| SMAD3 | 1.310052368 | 1 | 1 |
| AJUBA | 1.873377925 | 1.67385878 | 1 |
| AXIN1 | 1.250607347 | 1 | 1 |
| CSNK1E | 1.380889371 | 1 | 1 |
| CTGF | 1.538758842 | 1 | 1.428814977 |
| DLG3 | 1.512565821 | 1.296332083 | 1 |
| FZD7 | 1.342673277 | 1 | 1 |
| ITGB2 | 1.572901939 | 1 | 1 |
| PARD3 | 1.250711844 | 1 | 1 |
| SAV1 | 1.200108843 | 1 | 1 |
| MYC | 1.410985277 | 1.266748014 | 1 |
Table S5. Expression level of downregulated DEGs (vs. control) enriched in multiple biological processes.
| Gene symbol | Fold change (treatment vs. control) | ||
|---|---|---|---|
| MGCD265 + erlotinib | Erlotinib | MGCD265 | |
| Cell cycle | |||
| E2F2 | 0.698264583 | 0.765341746 | 0.829617942 |
| SKP2 | 0.666279155 | 0.735078961 | 1 |
| CDC20 | 0.787424854 | 1 | 1 |
| CDC25A | 0.598816259 | 0.65588207 | 1 |
| CDC45 | 0.78138931 | 0.786718155 | 1 |
| CDC6 | 0.638133071 | 0.724462682 | 1 |
| CDC7 | 0.819192058 | 0.824801908 | 1 |
| CCNA2 | 0.796728486 | 0.828706336 | 1 |
| CCND3 | 0.597693581 | 0.689911532 | 1 |
| CCNE2 | 0.601208611 | 0.627989732 | 1 |
| CDK2 | 0.747170127 | 0.785909293 | 1 |
| CDK4 | 0.809001405 | 1 | 1 |
| CDK6 | 0.779894059 | 0.796641826 | 1 |
| CDKN2C | 0.699493903 | 0.740163861 | 1 |
| MCM4 | 0.796587629 | 0.786586647 | 1 |
| MCM5 | 0.809011572 | 1 | 1 |
| ORC1 | 0.651124396 | 0.75489963 | 1 |
| ORC6 | 0.74897745 | 0.713235537 | 1 |
| PCNA | 0.730196232 | 0.752830213 | 1 |
| PKMYT1 | 0.594922683 | 1 | 1 |
| SFN | 0.816845946 | 0.70916061 | 1 |
| TGFB2 | 0.830374389 | 0.812733345 | 1 |
| DNA replication | |||
| POLA2 | 0.673108697 | 0.802900219 | 1 |
| POLD3 | 0.793045387 | 0.822547354 | 1 |
| POLE | 0.808760222 | 1 | 1 |
| FEN1 | 0.646919176 | 0.673411842 | 1 |
| MCM4 | 0.796587629 | 0.786586647 | 1 |
| MCM5 | 0.809011572 | 1 | 1 |
| PCNA | 0.730196232 | 0.752830213 | 1 |
| RFC4 | 0.825145898 | 1 | 1 |
| RFC5 | 0.657860132 | 0.739748932 | 1 |
| RNASEH1 | 0.826004445 | 1 | 1 |
| RNASEH2C | 0.793225524 | 1 | 1 |
Table S6. Expression level of upregulated DEGs (combination of erlotinib and MGCD265 vs. control; fold change >2).
| Gene symbol | Fold change (treatment vs. control) | ||
|---|---|---|---|
| MGCD265 + erlotinib | Erlotinib | MGCD265 | |
| Upregulated DEGs | |||
| PLEC | 13.29248342 | 10.8312812 | 11.75436746 |
| SESN2 | 6.626958928 | 5.933017343 | 1 |
| CHAC1 | 5.707103105 | 5.753205488 | 1 |
| DDIT4 | 4.702788471 | 4.219327288 | 1 |
| SLC7A11 | 4.054158255 | 3.498450309 | 1 |
| CTH | 4.004821716 | 2.771962999 | 1.417387273 |
| SLC6A9 | 3.895412031 | 4.138867318 | 1 |
| PCK2 | 3.853122264 | 4.037367742 | 1 |
| ASNS | 3.708656207 | 1.661348801 | 1 |
| NUPR1 | 3.565986853 | 3.455793471 | 1 |
| TRIB3 | 3.436248991 | 2.664563103 | 1 |
| JDP2 | 3.34918592 | 2.657128502 | 1 |
| SLC1A5 | 3.25953761 | 1.699564365 | 1 |
| ULK1 | 2.715179648 | 2.062475526 | 1 |
| CBS | 2.680069964 | 2.302972206 | 1 |
| DDIT3 | 2.649910504 | 2.099036131 | 1 |
| SARS | 2.641990297 | 1.655447203 | 1 |
| NCOA7 | 2.583821563 | 2.339772077 | 1 |
| MARS | 2.545254693 | 1.618080069 | 1 |
| PSAT1 | 2.500856424 | 1.712593791 | 1 |
| CEBPB | 2.462880354 | 2.163868758 | 1 |
| HERPUD1 | 2.412907008 | 1.848697272 | 1 |
| MBNL1 | 2.406773612 | 1.57328823 | 1 |
| YIPF4 | 2.387163111 | 1 | 2.056901916 |
| GPT2 | 2.335734456 | 1.858376737 | 1 |
| KLHL24 | 2.320515458 | 1.885448834 | 1 |
| MOCOS | 2.279153704 | 1.576747887 | 1 |
| CCNG2 | 2.263213411 | 1.656921209 | 1 |
| PIM1 | 2.247915381 | 1.81422954 | 1 |
| SCEL | 2.236888876 | 1 | 1 |
| PLAT | 2.213839953 | 1.824981703 | 1 |
| CREBRF | 2.208256826 | 1.723335985 | 1 |
| SLC3A2 | 2.206340833 | 1.250882259 | 1 |
| SHMT2 | 2.197397554 | 1.43195683 | 1 |
| RPS6KA5 | 2.179750059 | 1.216449981 | 1 |
| HKDC1 | 2.176221503 | 1.783259921 | 1 |
| UPP1 | 2.167799403 | 1.385538692 | 1 |
| MKNK2 | 2.164672463 | 1.539592982 | 1 |
| SLC1A4 | 2.10600201 | 2.031761415 | 1 |
| MLPH | 2.07707234 | 1 | 1 |
| SIAH1 | 2.066825829 | 1 | 1 |
| CARS | 2.059623213 | 1.737241112 | 1 |
| Downregulated DEGs | |||
| HSPA8 | 0.496264101 | 0.704867015 | 1 |
| MASTL | 0.495580044 | 0.656329871 | 1 |
| MTFP1 | 0.494410059 | 0.739380551 | 1 |
| NBEAL2 | 0.473310883 | 0.721503851 | 1 |
| NDOR1 | 0.473156713 | 0.569417604 | 1 |
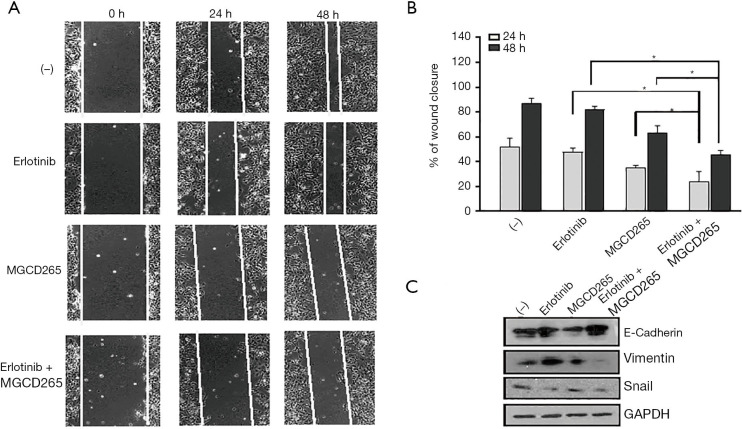
Combination of AXL inhibition and erlotinib treatment significantly repressed HCC827-ER3 cell migration
We previously demonstrated that upregulation of AXL plays an important role in cell migration (13). A group of the above noted downregulated DEGs are indeed enriched in the biological process of cell-cell adhesion and actin cytoskeleton reorganization as well as membrane ruffling formation, actin organization and focal adhesion formation—key aspects of cellular migration. Accordingly, we further evaluated the effect of erlotinib and/or MGCD265 on HCC827-ER3 cell migration and the cellular EMT phenotype. As compared to erlotinib alone treatment which modestly reduced cell migration (vs. control), MGCD265 alone treatment showed a clear inhibitory effect on cell migration (Figure 6A,B). Importantly, treatment of erlotinib in combination with MGCD265 more significantly impaired scratch closure than either erlotinib or MGCD265 alone treatment (Figure 6A,B). The process of epithelial-mesenchymal transition (EMT) is defined as a key initiating step for tumor cell migration, and we have reported that AXL overexpression and activation induced EMT in HCC827-ER3 cells (13). So we next assessed whether and how erlotinib and/or MGCD265 treatment affect EMT in these cells. The experiments demonstrated that the combination of erlotinib and MGCD265 significantly increased the expression of E-cadherin while repressing the expression of Vimentin and Snail in HCC827-ER3 cells (Figure 6C). Taken together, these results suggest that the combination of erlotinib and MGCD265 treatment synergistically represses EMT and inhibits the motility competence of HCC827-ER3 cells.
Figure 6.
Combination of AXL inhibition and erlotinib treatment significantly repressed the migration of HCC827-ER3 cells. (A) Representative images from in vitro scratch wound healing assays in HCC827-ER3 cells treated with erlotinib and/or MDCG625. (B) The bar graph illustrates the percentage of wound closure at indicated time points during the scratch wound assay. *, P<0.05. (C) HCC827-ER3 cells were treated as described above. Cell lysates were prepared and subjected to immunoblot analysis with EMT marker antibodies including E-cadherin, vimentin, Snail and GAPDH.
Discussion
EGFR targeted therapy has become the standard of care for patients with advanced EGFR-mutated NSCLC and leads to significantly improved outcomes as compared to traditional chemotherapy (26). However, acquired resistance remains a significant issue and with the more recent introduction of the third-generation EGFR inhibitor, osimertinib in first-line management, alternative non-EGFR mediated resistance mechanisms, such as bypass resistance, EMT transition and small cell transformation are increasingly becoming key issues in further improving outcomes (12,26,27). We and others recently reported AXL overexpression and activation as a novel mechanism that can lead to bypass resistance to EGFR TKI treatment as well as intrinsic resistance inducing treatment tolerance to the novel third generation EGFR inhibitor, osimertinib (13,14). The findings have been confirmed by others and it also appears that AXL overactivation might confer resistance to EGFR-targeted therapy in a wide range of cancer types (16-18,28). Therefore, further development of effective AXL-targeted therapeutic strategies is anticipated to bring significant therapeutic benefits to appropriately selected patient subsets.
In the current study, we define the efficacy of a range of AXL inhibitors to overcome resistance mediated by AXL activation. Our results demonstrate that administration of AXL inhibitors in combination with EGFR TKI therapy partially but significantly restores erlotinib sensitivity and inhibits the viability of erlotinib-resistant HCC827-ER3 cells. Along with the results reported in a recently published study which demonstrated that activated AXL confers intrinsic resistance and induces the emergence of osimertinib-tolerant cells, while the combination of AXL inhibition and osimertinib remarkably repressed tumor growth in EGFR-mutated lung cancer (14), our results strongly indicate that such AXL inhibitors might be potent candidates for targeting AXL-mediated resistance to erlotinib in NSCLC patients.
The Gas/AXL signaling axis plays important roles in tumorigenesis, EMT, invasion and metastasis, and acquired resistance to chemotherapy-resistant cancers (15,29). PI3K/AKT and MAPK signaling cascades are the major pathways mediating Gas6/AXL signaling that regulate the aforementioned phenotypes (15,29). Overexpression of AXL leading to constitutive activity of these 2 cascades in the HCC827-ER3 cells promoted cell proliferation and migration, and eventually led to development of acquired resistance to erlotinib (13). Notably, our data clearly demonstrates that AXL inhibition in combination with erlotinib treatment dramatically reduces PI3K/AKT and MAPK activity, which leads to cell growth arrest and death, and therefore contributes to overcoming erlotinib resistance. The activation of MAPK and PI3K/AKT pathways tightly regulate numerous cell functions, such as proliferation, apoptosis, and cell migration (30). Our results demonstrate that the combination of the AXL inhibitor, MGCD265 and erlotinib induces cell cycle arrest and apoptosis, which is at least partially mediated via downregulation of MAPK and PI3K/Akt activation. In our study, MGCD265 plus erlotinib significantly reduced S phase entry and induced G2-M cell cycle arrest. S phase involves DNA replication and DNA repair, and G2-M phase regulates cell mitosis and division (31,32). The impairment of S phase entry and concomitant G2-M phase logically interferes with cell cycle progression and eventually inhibits cell survival. Simultaneously, we observe that the combination of erlotinib and AXL inhibitor shows a synergistical induction of apoptosis further demonstrating that AXL inhibition can restore EGFR TKI sensitivity.
The regulation of cell cycle and apoptosis are highly complicated processes and tightly regulated by multiple pathways. In our model systems, the downregulation of MAPK and PI3K/Akt pathways may only partially account for the overall regulation of these cell functions. In order to gain further insights, we performed an analysis of global transcriptional changes using RNA-sequencing indeed highlighting additional pathways that might participate in the inhibition of cell growth. In this study, in line with the above we specifically found that a group of upregulated DEGs, such as ATF4, SIAH1 and BBC3 (Figure 5B), were mainly enriched in biological process in positive regulation of apoptotic process and negative regulation of cell proliferation. On the contrary, the downregulated DEGs were enriched in the regulation of DNA replication and repair, cell cycle regulation and cell division. As the representative DEGs found in our study, ATF4 or BBC3 have been identified to promote the induction of apoptosis under persistent stress (33,34), and SIAH1 plays a crucial role in cell cycle arrest and the induction of apoptosis (35,36). The downregulated DEG, Mastl (Figure 5C) is a serine/threonine kinase that plays a key role in M phase by acting as a regulator of mitosis entry and maintenance (37) and it may therefore play a negative role in regulation of cell cycle and cell division. These DEGs enriched in the biological process of cell cycle, cell division and apoptosis provide a more complete picture of the broad array of functional changes as a consequence of AXL inhibition. In addition, the KEGG pathway analysis also indicated that a number of upregulated DEGs were enriched in apoptosis pathway, p53 signaling pathway, FoXO and Hippo pathway, whereas the top ranked pathways that the downregulated DEGs were enriched in included the cell cycle and the DNA replication pathway. Besides the well-known pro-apoptotic role of the p53 pathway (38), FoXO signaling may also promote cell growth inhibition and/or apoptosis by either inducing expression of pro-apoptotic members of the Bcl2-family and death receptor ligands such as Fas, or enhancing levels of various cyclin-dependent kinase inhibitors (CDKIs) (39). In addition, the Hippo signaling pathway also has been shown to be responsible for cell proliferation inhibition and promoting apoptosis (40). These results show that the majority of enriched DEGs are involved in the signaling pathways of cell cycle and cell death, which therefore provides a reasonable interpretation that the combination of AXL inhibitor and erlotinib treatment regulates the cell cycle and apoptosis, and therefore restores drug sensitivity to erlotinib and inhibits erlotinib-resistant cell proliferation.
Additionally, a group of downregulated DEGs, such as BAIAP2, PIP5K1A and CTTN, have been found that were enriched in the biological process of cell-cell adhesion and actin cytoskeleton reorganization. BAIAP2, PIP5K1A and CTTN are necessary for membrane ruffling formation, actin organization and focal adhesion formation during directional cell migration (41-43), therefore downregulation of these genes is anticipated to negatively regulate cell migration and induce EMT. Therefore, these transcriptomic alterations provide a distinct genetic and biochemical background by which the combination of erlotinib with AXL inhibitors inhibits erlotinib-resistant cell migration.
In conclusion, our study provides clear evidence that treatment with AXL inhibitors might be an effective therapeutic strategy to overcome the acquired resistance to erlotinib in appropriately biomarker-selected patient subsets. Future studies will need to further investigate the in vivo effects of the combination of erlotinib and AXL inhibitor on erlotinib-resistant animal model, in models that might demonstrate the efficacy of the combination to prevent the emergence of resistance and ultimately in clinical trials to show clinical validity of these observations.
Acknowledgments
Funding: This work was supported by funds to Dr. Balazs Halmos from the American Cancer Society, Uniting Against Lung Cancer and the Pelka Memorial Fund.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional ethical approval and individual informed consent were waived due to the nature of the study.
Footnotes
Conflicts of Interest: BH serves the unpaid editorial board member of Translational Cancer Research from Jan 2019 to Dec 2020. Dr. Halmos has received clinical research support and consulting fees from Merck, Mirati, BMS, Guardant Health, Foundation Medicine, Pfizer, Novartis, Spectrum, Genentech, Astra-Zeneca, Boehringer-Ingelheim, GSK, AbbVie, Takeda. The other authors have no conflicts of interest to declare.
References
- 1.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. 10.1038/nature25183 [DOI] [PubMed] [Google Scholar]
- 2.Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. 10.1038/nrc2088 [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer 2010;10:760-74. 10.1038/nrc2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 2009;28 Suppl 1:S24-31. 10.1038/onc.2009.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol 2011;6:49-69. 10.1146/annurev-pathol-011110-130206 [DOI] [PubMed] [Google Scholar]
- 6.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 7.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 2005;353:123-32. 10.1056/NEJMoa050753 [DOI] [PubMed] [Google Scholar]
- 8.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. 10.1016/S1470-2045(11)70184-X [DOI] [PubMed] [Google Scholar]
- 9.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958-67. 10.1056/NEJMoa0904554 [DOI] [PubMed] [Google Scholar]
- 10.Stewart EL, Tan SZ, Liu G, et al. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations-a review. Transl Lung Cancer Res 2015;4:67-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackman D, Pao W, Riely GJ, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol 2010;28:357-60. 10.1200/JCO.2009.24.7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piotrowska Z, Thress KS, Mooradian M, et al. MET amplification (amp) as a resistance mechanism to osimertinib. J Clin Oncol 2017;35:abstr 9020.
- 13.Zhang Z, Lee JC, Lin L, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet 2012;44:852-60. 10.1038/ng.2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taniguchi H, Yamada T, Wang R, et al. AXL confers intrinsic resistance to osimertinib and advances the emergence of tolerant cells. Nat Commun 2019;10:259. 10.1038/s41467-018-08074-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paccez JD, Vogelsang M, Parker MI, et al. The receptor tyrosine kinase Axl in cancer: biological functions and therapeutic implications. Int J Cancer 2014;134:1024-33. 10.1002/ijc.28246 [DOI] [PubMed] [Google Scholar]
- 16.Byers LA, Diao L, Wang J, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res 2013;19:279-90. 10.1158/1078-0432.CCR-12-1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer AS, Miller MA, Gertler FB, et al. The receptor AXL diversifies EGFR signaling and limits the response to EGFR-targeted inhibitors in triple-negative breast cancer cells. Sci Signal 2013;6:ra66. 10.1126/scisignal.2004155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giles KM, Kalinowski FC, Candy PA, et al. Axl mediates acquired resistance of head and neck cancer cells to the epidermal growth factor receptor inhibitor erlotinib. Mol Cancer Ther 2013;12:2541-58. 10.1158/1535-7163.MCT-13-0170 [DOI] [PubMed] [Google Scholar]
- 19.Do KT, MacConaill L, Dubuc A, et al. Evaluation of the MET/Axl receptor tyrosine kinase (RTK) inhibitor MGCD265 in a patient with metastatic non-small cell lung cancer (NSCLC) harboring Axl amplification. International Association for the Study of Lung Cancer (IASLC). Oral Presentation at the 16th World Conference on Lung Cancer 2015. [Google Scholar]
- 20.Beaulieu N, Beaulieu C, Dupont I, et al. Preclinical development of MGCD265, a potent orally active c-Met/VEGFR multi-target kinase inhibitor. AACR Annual Meeting 2008. [Google Scholar]
- 21.Patwardhan PP, Ivy KS, Musi E, et al. Significant blockade of multiple receptor tyrosine kinases by MGCD516 (Sitravatinib), a novel small molecule inhibitor, shows potent anti-tumor activity in preclinical models of sarcoma. Oncotarget 2016;7:4093-109. 10.18632/oncotarget.6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holland SJ, Pan A, Franci C, et al. R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res 2010;70:1544-54. 10.1158/0008-5472.CAN-09-2997 [DOI] [PubMed] [Google Scholar]
- 23.Wang F, Yamauchi M, Muramatsu M, et al. RACK1 regulates VEGF/Flt1-mediated cell migration via activation of a PI3K/Akt pathway. J Biol Chem 2011;286:9097-106. 10.1074/jbc.M110.165605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aredia F, Scovassi AI. Poly(ADP-ribose): a signaling molecule in different paradigms of cell death. Biochem Pharmacol 2014;92:157-63. 10.1016/j.bcp.2014.06.021 [DOI] [PubMed] [Google Scholar]
- 26.Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res 2015;4:36-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minari R, Bordi P, Tiseo M. Third-generation epidermal growth factor receptor-tyrosine kinase inhibitors in T790M-positive non-small cell lung cancer: review on emerged mechanisms of resistance. Transl Lung Cancer Res 2016;5:695-708. 10.21037/tlcr.2016.12.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brand TM, Iida M, Stein AP, et al. AXL mediates resistance to cetuximab therapy. Cancer Res 2014;74:5152-64. 10.1158/0008-5472.CAN-14-0294 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 29.Wu X, Liu X, Koul S, et al. AXL kinase as a novel target for cancer therapy. Oncotarget 2014;5:9546-63. 10.18632/oncotarget.2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci 2011;36:320-8. 10.1016/j.tibs.2011.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee N, Siede W. Replicating damaged DNA in eukaryotes. Cold Spring Harb Perspect Biol 2013;5:a019836. 10.1101/cshperspect.a019836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaufmann WK, Kaufman DG. Cell cycle control, DNA repair and initiation of carcinogenesis. FASEB J 1993;7:1188-91. 10.1096/fasebj.7.12.8375618 [DOI] [PubMed] [Google Scholar]
- 33.Han J, Back SH, Hur J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 2013;15:481-90. 10.1038/ncb2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han J, Flemington C, Houghton AB, et al. Expression of bbc3, a pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc Natl Acad Sci U S A 2001;98:11318-23. 10.1073/pnas.201208798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roperch JP, Lethrone F, Prieur S, et al. SIAH-1 promotes apoptosis and tumor suppression through a network involving the regulation of protein folding, unfolding, and trafficking: identification of common effectors with p53 and p21(Waf1). Proc Natl Acad Sci U S A 1999;96:8070-3. 10.1073/pnas.96.14.8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Stevens J, Rote CA, et al. Siah-1 mediates a novel beta-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol Cell 2001;7:927-36. 10.1016/S1097-2765(01)00241-6 [DOI] [PubMed] [Google Scholar]
- 37.Voets E, Wolthuis RM. MASTL is the human orthologue of Greatwall kinase that facilitates mitotic entry, anaphase and cytokinesis. Cell Cycle 2010;9:3591-601. 10.4161/cc.9.17.12832 [DOI] [PubMed] [Google Scholar]
- 38.Schuler M, Green DR. Mechanisms of p53-dependent apoptosis. Biochem Soc Trans 2001;29:684-8. 10.1042/bst0290684 [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Tang N, Hadden TJ, et al. Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta 2011;1813:1978-86. 10.1016/j.bbamcr.2011.03.010 [DOI] [PubMed] [Google Scholar]
- 40.Mo JS, Park HW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep 2014;15:642-56. 10.15252/embr.201438638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miki H, Yamaguchi H, Suetsugu S, et al. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 2000;408:732-5. 10.1038/35047107 [DOI] [PubMed] [Google Scholar]
- 42.Doughman RL, Firestone AJ, Wojtasiak ML, et al. Membrane ruffling requires coordination between type Ialpha phosphatidylinositol phosphate kinase and Rac signaling. J Biol Chem 2003;278:23036-45. 10.1074/jbc.M211397200 [DOI] [PubMed] [Google Scholar]
- 43.Helwani FM, Kovacs EM, Paterson AD, et al. Cortactin is necessary for E-cadherin-mediated contact formation and actin reorganization. J Cell Biol 2004;164:899-910. 10.1083/jcb.200309034 [DOI] [PMC free article] [PubMed] [Google Scholar]