Abstract
Background
Overexpression of the tripartite motif containing 6 (TRIM6) is associated with dismal prognosis in cancer patients, but its exact roles in lung adenocarcinoma (LUAD) have not been reported.
Methods
The roles of TRIM6 are identified by using The Cancer Genome Atlas (TCGA), TIMER2, Gene Expression Omnibus (GEO), etc., and the regulatory networks and related-prognostic biomarkers of TRIM6 are identified via the ENCORI and LNCAR databases in the LUAD progression.
Results
TRIM6 expression level in LUAD tissues was significantly increased. TRIM6 over-expression level in LUAD patients was associated with smoking, clinical stage, histological type, lymph node metastasis, TP53 mutation and dismal prognosis, and related to prognosis-related age, race, sex, clinical stage and tumor purity of LUAD patients. TRIM6 overexpression was associated with the levels of CD8+ T cells, macrophages, neutrophils and myeloid dendritic cells, and correlated with the levels of LUAD immune cell markers CD8A, IRF5, CD163, VSIG4, MS4A4A, ITGAM, HLA-DPA1, NRP1, ITGAX, etc. TRIM6 might influence the progression of LUAD by regulating homologous recombination, oocyte meiosis, and ubiquitin-mediated proteolysis. LUAD patients with overexpression of miR-101-3p, miR-335-5p, miR-374a-3p, miR-628-5p, and NEAT1 had a poor prognosis.
Conclusions
NEAT1-miR-101-3p/335-5p/374a-3p/628-5p-TRIM6 network, which we constructed from our results, might be an important factor in the dismal prognosis of LUAD patients.
Keywords: NEAT1, overall survival (OS), disease-free survival (DFS), lung adenocarcinoma (LUAD), tripartite motif containing 6 (TRIM6)
Introduction
New cases of lung cancer (LC) are a high proportion of the total number of cancers globally, and the mortality rate is high (1,2). In recent years, there have been some novel molecules expected to delay the progression of LC and improve the prognosis of patients (3-6). For example, atezolizumab is a monoclonal antibody that binds to programmed death-ligand 1 (PD-L1) and blocks the interaction between PD-L1 and the programmed cell death-1 (PD-1) and B7.1 receptor. Compared with chemotherapy alone, combination therapy of atezolizumab and chemotherapy has been more effective in patients with advanced non-small cell lung cancer (NSCLC), with improved overall survival (OS) (3). In pStage IA1–IB high-malignant subtype and Stage IIA–IIIA patients, epidermal growth factor receptor (EGFR)-positive patients have worse 5-year recurrence-free survival (RFS) than EGFR-negative patients. Positive EGFR mutation status is significantly related to the patient's RFS (4). However, there is still a need to identify more valuable biomarkers to extend OS for cancer patients.
During the development of NSCLC, some molecules are often expressed abnormally, including genes, micro RNAs (miRNAs), long non-coding RNAs (lncRNAs), etc. (7-11). For example, lncRNA TRPM2-AS is overexpressed in NSCLC tissues and cells. The proliferation, migration and invasive ability of NSCLC cells that interfered with the expression of TRPM2-AS were inhibited, and apoptosis was significantly increased. TRPM2-AS increases the expression level of EGFR by regulating the expression of sponge miR-138-3p, thereby promoting the proliferation, migration and invasion of NSCLC cells, and aggravating tumor growth and migration (7). lncRNA LCAT1 is significantly upregulated in LC tissues and associated with a dismal prognosis. Interfering with LCAT1 expression inhibits the growth and invasion of LC cells and tumor development in nude mice both in vitro and in vivo. LCAT1 plays a role as the competitive endogenous RNAs (ceRNAs) of miR-4715-5p, which leads to the upregulation of RAC1 activity of the endogenous target RAC family. Inhibitory effects on the expression of RAC1 improved the efficacy of paclitaxel monotherapy against LC cells in vitro (11). The expression levels of lncRNA DNAH17-AS1 and CCNA2 were significantly upregulated in NSCLC tissues and cells, while the expression level of miR-877-5p was significantly decreased. Overexpression of DNAH17-AS1 is associated with the TNM stage, distant metastasis, short OS, and disease-free survival (DFS) of NSCLC patients. Interfering with the expression of DNAH17-AS1 inhibits the proliferation, migration and invasion of NSCLC cells, and promotes cell apoptosis. DNAH17-AS1 could upregulate CCNA2 to played a carcinogenic role by acting as a sponge for miR-877-5p (12). These results suggest that the ceRNA network plays an important role in delaying the progression of NSCLC.
The tripartite motif containing 6 (TRIM6) also plays a crucial role in carcinogenesis (13-15). It is upregulated in colorectal cancer (CRC) and its expression level is an independent predictor of dismal prognosis in CRC patients. Knocking down TRIM6 expression inhibits the proliferation and cell cycle arrest of CRC cells, and increases the sensitivity of cells to 5-fluorouracil and oxaliplatin. TRIM6 promotes CRC progression through the TiS21/FOXM1 mechanism (13), and its expression level is significantly enhanced in breast cancer (BC) cells and tissues. Overexpression of TRIM6 promotes BC progression by increasing YAP1, while knocking out TRIM6 has the opposite effect. TRIM6 promotes ubiquitination-mediated STUB1 degradation to promote YAP1 signaling transduction. Overexpression of STUB1 reduces the promoting effect of TRIM6 on BC cell growth (10). However, the roles of TRIM6 in the progression of NSCLC and its adenocarcinoma (LUAD) subtypes has not been reported. Therefore, the roles of and potential mechanisms of TRIM6 in the progression of LUAD were comprehensively analyzed in this study based on The Cancer Genome Atlas (TCGA), TIMER2, and Gene Expression Omnibus (GEO) databases, and the upstream regulatory network of TRIM6 was constructed to provide new candidate prognostic biomarkers for the treatment of LUAD patients. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-2181).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
TCGA database
Gene expression of 594 LUAD FKPM and 522 clinical data were downloaded from the TCGA database, and TRIM6 expression data were extracted for analysis in LUAD tissues. In the analysis of TRIM6 expression and survival of NSCLC patients, patients with missing prognostic information were excluded.
GEO database
The series matrix data files for the GSE1037, GSE18842, GSE19188 and GSE30219 datasets were downloaded from the GEO database website. GSE1037 dataset platform was GPL962: CHUGAI 41K. The platform of GSE18842, GSE19188 and GSE30219 datasets was GPL570: [HG-U133_PLUS_2] Affymetrix Human Genome U133 Plus 2.0 Array.
UALCAN database
The UALCAN database was used to analyze the cancer gene expression level and patient prognosis or clinicopathological characteristics of LUAD patients. TRIM6 expression level and the clinical values of TRIM6 in LUAD were investigated using the UALCAN database.
Lung cancer explorer (LCE) database
The LCE database, which includes multicenter data, was used to show the relationship between gene expression and survival in LC patients. In this database, the TRIM6 expression level and its prognostic value in LUAD were demonstrated by meta-analysis.
GEPIA2 database
This is a secondary database developed in recent years based on cancer gene expression data from the TCGA and GTEX databases. The GEPIA2 website has the advantage of being simple to operate and can be used to investigate the TRIM6 expression in cancer tissues and its value in the OS and DFS of LUAD patients.
TIMER2 database
The TIMER2 database displays gene expression levels in pan-cancer tissue by using cancer data from the TCGA database, and identifies the relationship between gene expression level and cancer immune purity, immune cells and cell markers. The TRIM6 expression level in pan-cancer tissues and its potential value in immunity and prognosis were analyzed.
Creation of lncRNA-miRNA-gene network
The upstream binding miRNAs of TRIM6 and miRNA-binding lncRNAs were screened in the ENCORI database. The expression of TRIM6-binding miRNAs in LUAD tissues and the role in prognosis were verified in miRNA expression level and prognosis module, and the clinical value of miRNA-binding lncRNAs was identified in the LNCAR database and literature reports. In our study, the value of NEAT1 in the prognosis of LUAD patients was explored using meta-analysis of the literature in recent years. From this, a lncRNA-miRNA-gene network was constructed.
Meta-analysis
A systematic search from 6 relevant databases including PubMed, Embase, Cochrane Library, Web of Science, Google Scholar and CNKI was executed to get all related papers. The software of Review Manager 5.3 was applied to analyze the prognostic data. HRs with 95% CI were used to inquiry the correlation between aberrant NEAT1 expression and the OS via meta-analysis (16).
TRIM6-related signaling pathways
In 539 LUAD tissues, the median expression value of TRIM6 was grouped, and gene set enrichment analysis (GSEA) was conducted to investigate the potential mechanisms of high and low TRIM6 expression in LUAD progression (2). The model was run 1,000 times in GSEA software.
Statistical analysis
The mean levels of TRIM6 and NEAT1 in LUAD tissues were determined by t test. The relationship between TRIM6 expression level and the survival time of LUAD patients was assessed via the survival analysis. P<0.05 was considered statistically significant.
Results
Expression of TRIM6 in pan-cancer tissues
In the TIMER2 database, TRIM6 was overexpressed in cholangiocarcinoma, esophageal carcinoma, glioblastoma multiforme, liver hepatocellular carcinoma and LUAD tissues, and was under-expressed significantly in breast invasive carcinoma, colon adenocarcinoma, head and neck squamous cell carcinoma, kidney chromophobe, kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, prostate adenocarcinoma, rectal adenocarcinoma and uterine corpus endometrial carcinoma tissues (Figure S1).
Overexpression of TRIM6 in LUAD tissues
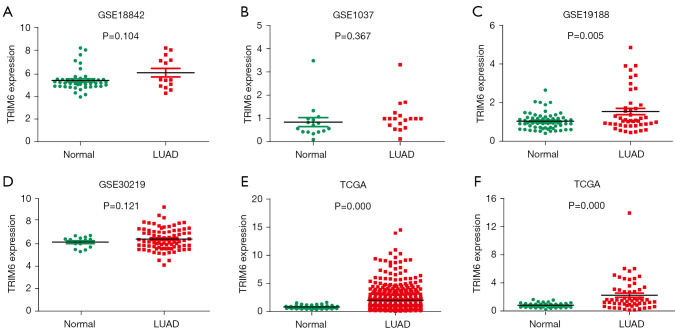
In the TCGA and GEO databases, the TRIM6 expression level was significantly increased in LUAD tissues. The GSE18842, GSE1037, GSE19188, and GSE30219 datasets showed TRIM6 overexpression in LUAD tissues. The result for the GSE19188 dataset was statistically significant (Figure 1A-1D). In the TCGA database, TRIM6 was overexpressed in unpaired LUAD tissues (Figure 1E), and in 59 paired LUAD tissues (Figure 1F). Meta-analysis of the data from TCGA and GEO databases revealed that TRIM6 was overexpressed in LUAD tissues (Figure S2).
Figure 1.
TRIM6 expression level significantly increased in LUAD tissues. (A-D) GSE18842, GSE1037, GSE19188, and GSE30219 datasets; (E,F) unpaired and paired LUAD tissues in TCGA database. TRIM6, tripartite motif containing 6; LUAD, lung adenocarcinoma. TCGA, The Cancer Genome Atlas.
Relation of TRIM6 expression level to smoking, clinical stage, histological type, lymph node metastasis and TP53 mutation in LUAD patients
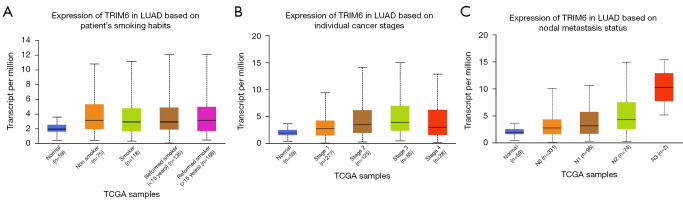
In the UALCAN database, the TRIM6 expression level was associated with smoking, clinical stage, histological type, lymph node metastasis and TP53 mutation in LUAD patients according to correlation analysis (Figure 2). TRIM6 expression level was related to smoking (Nonsmoker vs. Reformed smoker 2, P=1.173520E-02), clinical stage (Stage 1 vs. Stage 2, P=8.863900E-03; Stage 1 vs. Stage 3, P=3.498400E-03), histological type (Not Otherwise Specified, NOS) vs. Papillary, P=4.411700E-02), lymph node metastasis (N0 vs. N2, P=4.879600E-03), and TP53 mutation (TP53-Mutant vs. TP53-NonMutant, P=4.360600E-04) in LUAD patients. In the TIMER2 database, TRIM6 expression showed statistical significance in both the wild type and mutant type (Figure S3).
Figure 2.
Association of TRIM6 expression level with (A) smoking, (B) clinical stage, and (C) lymph node metastasis of LUAD patients. TRIM6, tripartite motif containing 6; LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas.
Overexpression of TRIM6 and dismal prognosis in LUAD patients
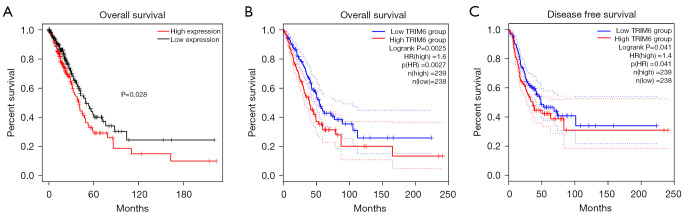
Survival analysis showed that TRIM6 overexpression was associated with poor prognosis in patients with LUAD (Figure 3 and Figure S3). In the TCGA database, TRIM6 overexpression was associated with shorter OS in LUAD patients compared with the group with lower TRIM6 expression (Figure 3A). In the GEPIA2 database, TRIM6 overexpression was associated with shorter OS and DFS in LUAD patients compared with the group with low TRIM6 expression (Figure 3B,3C). Meta-analysis of the data from TCGA and GEO databases revealed that TRIM6 overexpression was a risk factor for poor prognosis in LUAD patients (Figure S4).
Figure 3.
Correlation of TRIM6 overexpression level with LUAD short overall survival (A,B) and disease-free survival (C). TRIM6, tripartite motif containing 6; LUAD, lung adenocarcinoma.
Correlation of overexpression of TRIM6 with prognosis-related age, race, sex, clinical stage, and tumor purity in LUAD patients
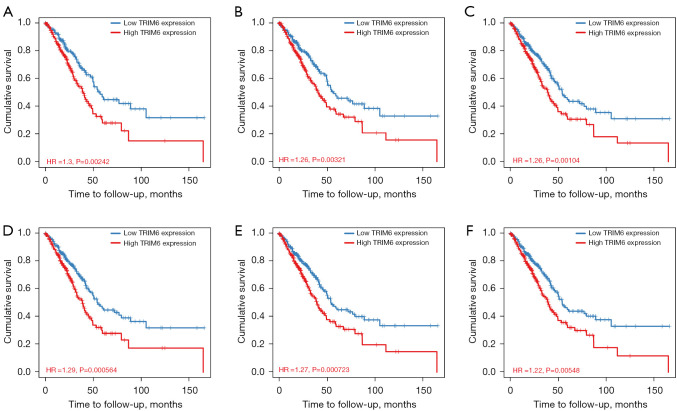
TRIM6 overexpression was associated with prognosis-related age, race, sex, clinical stage, and tumor purity in LUAD patients in the survival analysis (Figure 4). In the TIMER2 database, TRIM6 overexpression was associated with dismal prognosis in LUAD patients (Figure 4A). TRIM6 overexpression was associated with prognosis-related tumor purity, age, race, sex, and clinical stage in LUAD patients via subgroup analysis (Figure 4B-4F).
Figure 4.
Association of TRIM6 overexpression level with dismal prognosis, age, race, sex, clinical stage, and tumor purity in LUAD patients. (A) Overall survival; (B-F) tumor purity, age, race, sex, and clinical stage. TRIM6, tripartite motif containing 6; LUAD, lung adenocarcinoma.
Relation of TRIM6 overexpression to levels of CD8+ T cells, macrophages, neutrophils and myeloid dendritic cells
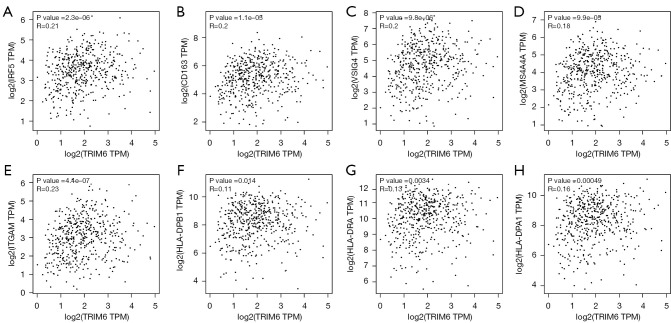
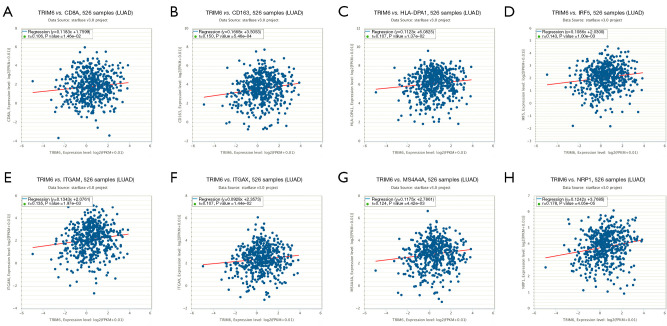
In the TIMER2 database, the TRIM6 overexpression level was related to the levels of CD8+ T cells, macrophages, neutrophils and myeloid dendritic cells (Table 1). In the LUAD tissues of the GEPIA2 database, the TRIM6 expression level correlated with the levels of LUAD immune cell markers IRF5 (r=0.21), CD163 (r=0.2), VSIG4 (r=0.2), MS4A4A (r=0.18), ITGAM (r=0.23), HLA-DPB1 (r=0.11), HLA-DRA (r=0.13), HLA-DPA1 (r=0.16), NRP1 (r=0.29), and ITGAX (r=0.16), all of which were statistically significant (Figure 5, Table 2). In the ENCORI database, the TRIM6 expression level correlated with the levels of LUAD immune cell markers CD8A (r=0.106), IRF5 (r=0.143), CD163 (r=0.15), VSIG4 (r=0.123), MS4A4A (r=0.124), ITGAM (r=0.135), HLA-DPA1 (r=0.107), NRP1 (r=0.178) and ITGAX (r=0.107), and all were statistically significant (Figure 6, Table 3).
Table 1. Relationship of TRIM6 overexpression to the immune cells of LUAD.
| Infiltrate | r | P | Adjusted P |
|---|---|---|---|
| CD8+ T cells | 0.226788294 | 3.60E-07 | 8.99E-06 |
| Neutrophils | 0.163253153 | 0.000272579 | 0.001447699 |
| Macrophages | 0.133174932 | 0.003049891 | 0.009530909 |
| Myeloid dendritic cells | 0.180194277 | 5.73E-05 | 0.000584439 |
TRIM6, tripartite motif containing 6; LUAD, lung adenocarcinoma.
Figure 5.
Correlation of TRIM6 with the levels of LUAD immune cell markers in the GEPIA2 database (A-H). TRIM6, tripartite motif containing 6; LUAD, lung adenocarcinoma.
Table 2. Relationship of TRIM6 overexpression to the markers of immune cells in LUAD tissues from the GEPIA2 database.
| Cell type | Markers | r | P |
|---|---|---|---|
| CD8+ T cell | CD8A | 0.089 | 0.051 |
| CD8B | –0.02 | 0.67 | |
| Macrophage | INOS (NOS2) | 0.049 | 0.28 |
| IRF5 | 0.21 | 2.3e-06 | |
| COX2 (PTGS2) | 0.08 | 0.079 | |
| CD163 | 0.2 | 1.1e-05 | |
| VSIG4 | 0.2 | 9.8e-06 | |
| MS4A4A | 0.18 | 9.9e-05 | |
| Neutrophil | CD66b (CEACAM8) | 0.081 | 0.075 |
| CD11b (ITGAM) | 0.23 | 4.4e-07 | |
| Dendritic cell | HLA-DPB1 | 0.11 | 0.014 |
| HLA-DQB1 | 0.063 | 0.17 | |
| HLA-DRA | 0.13 | 0.0034 | |
| HLA-DPA1 | 0.16 | 0.00049 | |
| BDCA-1 (CD1C) | 0.033 | 0.47 | |
| BDCA-4 (NRP1) | 0.29 | 4.7e-11 | |
| CD11c (ITGAX) | 0.16 | 0.00045 |
TRIM6, tripartite motif containing 6; LUAD, lung adenocarcinoma.
Figure 6.
Correlation of TRIM6 with the levels of LUAD immune cell markers in the ENCORI database (A-H). TRIM6, tripartite motif containing 6; LUAD, lung adenocarcinoma.
Table 3. Relationship of TRIM6 overexpression to the markers of immune cells in LUAD tissues from the ENCORI database.
| Cell type | Markers | r | p |
|---|---|---|---|
| CD8+ T cell | CD8A | 0.106 | 0.0146 |
| CD8B | 0.006 | 0.9 | |
| Macrophage | INOS (NOS2) | –0.024 | 0.582 |
| IRF5 | 0.143 | 0.001 | |
| COX2 (PTGS2) | 0.041 | 0.348 | |
| CD163 | 0.15 | 5.46e-4 | |
| VSIG4 | 0.123 | 4.81e-3 | |
| MS4A4A | 0.124 | 4.42e-3 | |
| Neutrophil | CD66b (CEACAM8) | 0.039 | 0.372 |
| CD11b (ITGAM) | 0.135 | 1.97e-03 | |
| Dendritic cell | HLA-DPB1 | 0.05 | 0.254 |
| HLA-DQB1 | 0.052 | 0.23 | |
| HLA-DRA | 0.084 | 0.0545 | |
| HLA-DPA1 | 0.107 | 0.0137 | |
| BDCA-1 (CD1C) | –0.024 | 0.577 | |
| BDCA-4 (NRP1) | 0.178 | 4.05e-5 | |
| CD11c (ITGAX) | 0.107 | 0.0144 |
TRIM6, tripartite motif containing 6; LUAD, lung adenocarcinoma.
Possible involvement of TRIM6 in the biological functions and signaling mechanisms of LUAD
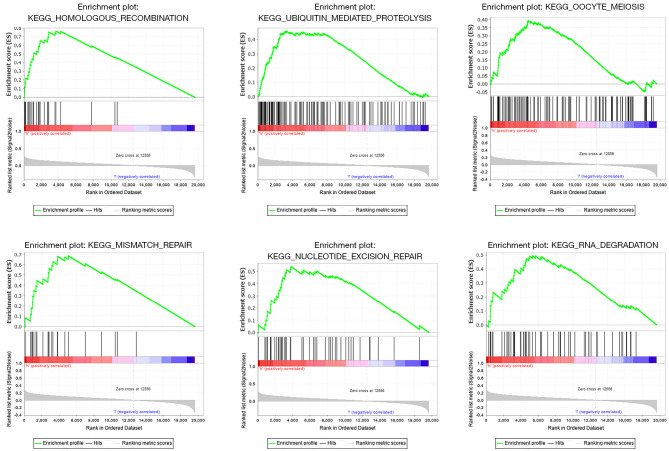
The GSEA results suggested that increased TRIM6 might be involved in sister chromatid cohesion, cellular carbohydrate catabolic process, meiotic cell cycle process, positive regulation of DNA replication, DNA recombination, cytokinesis and other biological processes (Figure S5, Table S1), and participate in the regulation of homogeneous recombination, ubiquitin mediated proteolysis, mismatch repair, nucleotide excision repair, pyrimidine metabolism, RNA degradation, RIG I like receptor signaling pathway, and oocyte meiosis (Figure 7).
Figure 7.
Possible involvement of TRIM6 in the signaling mechanisms of LUAD via gene set enrichment analysis. TRIM6, tripartite motif containing 6; LUAD, lung adenocarcinoma.
NEAT1-miRNAs-TRIM6 network
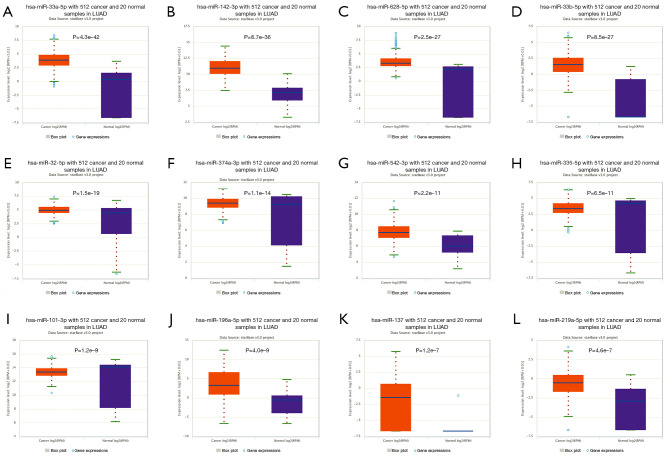
In the ENCORI database, the miRNAs that TRIM6 bound were miR-23a-3p, miR-25-3p, miR-32-5p, miR-33a-5p, miR-92A-3p, miR-101-3p, miR-196a-5p, miR-219a-5p, miR-23b-3p, and miR-137, miR-142-3p, miR-146a-5p, miR-361-5p, miR-363-3p, miR-376c-3p, miR-379-5p, miR-381-3p, miR-383-5p, miR-337-3p, miR-335-5p, miR-196b-5p, miR-146b-5p, miR-495-3p, miR-524-5p, miR-520d-5p, miR-520g-3p, miR-520h, miR-508-3p, miR-92b-3p, miR-33b-5p, miR-641, miR-542-3p, miR-758-3p, miR-374a-3p, miR-628-5p, miR-300, miR-513c-5p, miR-1321, miR-2115-3p, miR-514b-5p and miR-5688. miR-513c-5p, miR-514b-5p, miR-25-3p, miR-376c-3p, miR-520d-5p, miR-196b-5p, miR-337-3p, miR-146a-5p, miR-374a-3p, miR-137, MIR-101-3p, miR-146b-5p, miR-32-5p, miR-758-3p, miR-542-3p, miR-628-5p, miR-196a-5p, miR-33a-5p, miR-219a-5p, miR-335-5p, miR-33b-5p, and miR-142-3p, which were all overexpressed in LUAD tissues (P<0.05). miR-520G-3p, miR-524-5p, miR-383-5p, and miR-520h showed low expression in LUAD tissues (P<0.05, Figure 8).
Figure 8.
Abnormal expression of the microRNAs that TRIM6 bound in LUAD tissues from the ENCORI database. LUAD, lung adenocarcinoma; TRIM6, tripartite motif containing 6; LUAD, lung adenocarcinoma.
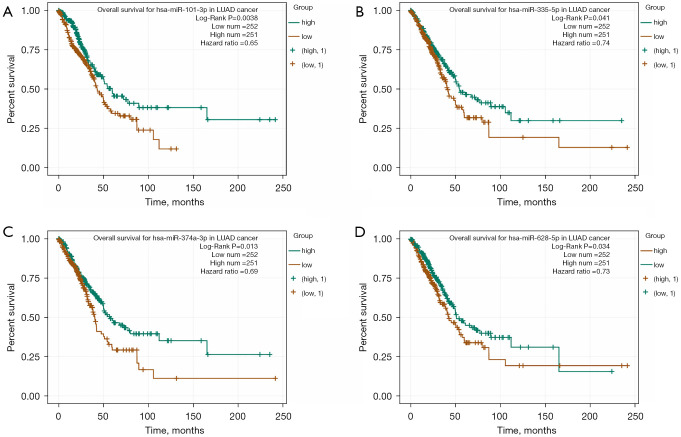
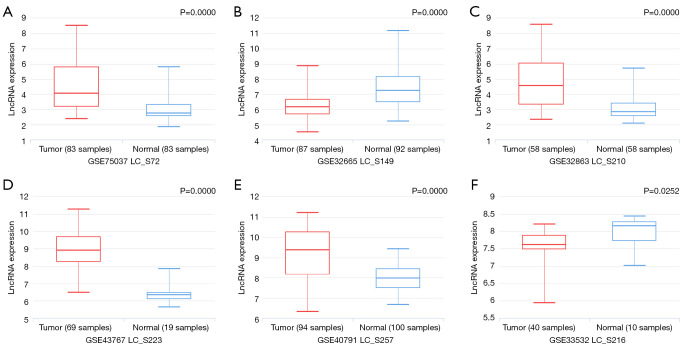
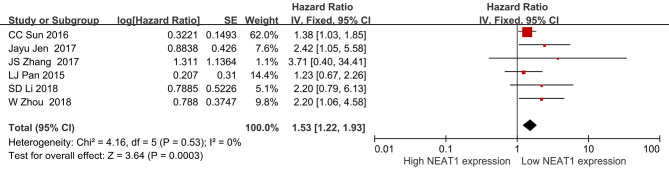
In addition, the role of miRNAs in the prognosis of LUAD patients was analyzed by survival analysis, and the results showed that LUAD patients with overexpression of miR-101-3p, miR-335-5p, miR-374a-3p and miR-628-5p had a poor prognosis, with statistical significance (Figure 9). It was found that lncRNA NEAT1 might regulate miR-101-3p, miR-335-5p, miR-374a-3p and miR-628-5p through MRE via the miRNA–23), and NEAT1 (LC_S257) was overexpressed in LUAD tissues in the LNCAR database. The expression levels of NEAT1 (LC_S149) and NEAT1 (LC_S216) in LUAD tissues were significantly decreased (Figure 10). Figure 11 shows that NEAT1 overexpression in LUAD tissues was related to dismal prognosis of LUAD patients via meta-analysis (14-19). The constructed TRIM6 network mechanism provides a new regulatory mechanism for LUAD progression (Figure S6).
Figure 9.
Dismal prognosis of LUAD patients with overexpression of (A) miR-101-3p, (B) miR-335-5p, (C) miR-374a-3p and (D) miR-628-5p. LUAD, lung adenocarcinoma.
Figure 10.
NEAT1 expression levels in LUAD tissues. (A) LC_S72; (B) LC_S149; (C) LC_S210; (D) LC_S223; (E) LC_S257; (F) LC_S216. LUAD, lung adenocarcinoma.
Figure 11.
Relationship of NEAT1 to poor prognosis of LUAD patients via meta-analysis. LUAD, lung adenocarcinoma.
Discussion
In recent years, more and more studies have reported the involvement of ceRNA networks in regulating the progression of LUAD (17-19). Compared with adjacent normal tissues, LINC00942 is up-regulated in LUAD tissues. Overexpression of LINC00942 is associated with dismal survival. Low expression of LINC00942 inhibits the proliferation, migration and invasion of A549 and H1299 cells, and promotes cell apoptosis. LINC00942 up-regulates the expression of FZD1 by combining miR-5006-5p to participate in the growth and migration of LUAD (17). circ-AASDH is highly expressed in LUAD tissues and cells, and is related to tumor size, clinical stage and dismal prognosis. Interfering with the expression of circ-AASDH can inhibit the proliferation and migration of LUAD cells and promote cell apoptosis. circ-AASDH can act as a sponge of miR-140-3p to promote E2F7 expression (19). In this study, we found that TRIM6 was excessively elevated in LUAD tissues from a multi-database analysis. TRIM6 overexpression was associated with smoking, clinical stage, histological type, lymph node metastasis, TP53 mutation, and dismal prognosis in LUAD patients, and was associated with prognosis-related age, race, sex, clinical stage, and tumor purity in LUAD patients. We constructed the NEAT1-miR-101-3p/miR-335-5p/miR-374a-3p/miR-628-5p-TRIM6 signaling axis, which is related to LUAD progression. Recent multicenter data suggest that NEAT1 acts as an oncogenic factor in NSCLC progression (20-25). NEAT1 is highly expressed in NSCLC tissues and cells, and closely associated with advanced clinical stage, lymph node metastasis, distant metastasis, and dismal prognosis in NSCLC patients (25,26). Knocking down NEAT1 expression inhibits NSCLC and LUAD cell growth and migration (26,27). NEAT1 is highly expressed in LUAD tissues. NEAT1 overexpression is related to the late TNM stage, lymph node metastasis and OS in LUAD patients, and is a risk factor for dismal prognosis in LUAD patients. Knockout of NEAT1 expression in LUAD A549 and H1299 cells can delay tumor cell proliferation and migration (21). NEAT1 is associated with drug resistance in NSCLC patients (28-30). Compared with NSCLC cells and normal bronchial epithelial cells, NEAT1 was overexpressed in paclitaxel-resistant NSCLC cells (28). Elimination of NEAT1 reversed paclitaxel resistance by increasing PARP and caspase-3 expression and inducing apoptosis, and was associated with activation of the Akt/mTOR resistance signaling pathway (29). Sorafenib treatment of A549 cells and PC9 cells reduced cell proliferation and induced cell apoptosis, and the proliferation effect of NEAT1 silencing was significantly increased (30).
In addition, miR-101-3p is low expressed in NSCLC cells. Overexpression of miR-101-3p can directly bind to and inhibit the expression of MALAT-1, thereby inhibiting the proliferation, migration and invasion of NSCLC cells (31). miR-335-5p is down-regulated in NSCLC tissues. Overexpression of miR-335-5p inhibits TGF-β1-mediated NSCLC migration and invasion (32). KCNMB2-AS1 silencing hinders the proliferation, migration and invasion of NSCLC cells, promotes cell apoptosis, and inhibits tumor growth in vivo. KCNMB2-AS1 acts as a sponge for endogenous miR-374a-3p to increase ROCK1 expression (33). These results indicate that our constructed network was closely related to the metastasis and drug resistance of NSCLC. However, the roles of NEAT1-miR-101-3p/miR-335-5p/miR-374a-3p/miR-628-5p-TRIM6 signaling axis in LUAD needs to be further confirmed by constructing the cell models.
It is well known that the immune microenvironment plays an important role in LUAD progression. The tumor immune microenvironment is composed of tumor cells, immune cells, mesenchymal cells and extracellular components, which are related to genetic status. SRGN is overexpressed in TTF-1 negative LUAD cells. The expression level of SRGN in LUAD is associated with the dismal prognosis. SRGN regulates the expression of PD-L1, interleukin 6, interleukin 8 and CXC motif chemokine 1 in LUAD cells, increases the migration and invasion ability of LUAD and fibroblasts, and enhances angiogenesis. SRGN can trigger the aggressive and immunosuppressive phenotype of TTF-1 negative LUAD cells (34). Our study found that TRIM6 overexpression was related to levels of CD8+ T cells, macrophages, neutrophils and myeloid dendritic cells. The TRIM6 expression level correlated with the levels of LUAD immune cell markers CD8A, IRF5, CD163, VSIg4, MS4A4a, ITGAM, HLA-DPA1, NRP1 and ITGAX. CD8A is a marker of CD8+ T cells; IRF5, CD163, VSIG4 and MS4A4A are markers of macrophages; ITGAM is a neutrophil marker; and HLA-DPA1, NRP1 and ITGAX are markers of dendritic cells. Studies have shown that these markers have tumor suppressor or tumor promoting effects in cancer (35-38). High CD8A expression is associated with T stage, lymph node metastasis, and prognosis in LUAD patients (35). Overexpression of CD163 in glioma specimens was associated with adverse prognosis, and loss of CD163 expression inhibited cell cycle progression and proliferation (36). CD163 silencing reduced the activity of the Akt/GSK3β/β-catenin/cyclin D1 pathway by regulating CK2 expression, but CD163 was upregulated in CD133-positive glioma stem cells (GSC) (36). Low CD163 expression resulted in altered expressions of GSC markers CD133, ALDH1A1, NANOG, and OCT4 (36). The relationship between TRIM6 and immune cells and their markers needs further confirmation.
Our study had a large sample size and utilized a variety of databases. However, it LUADked basic research demonstration, which is a future research plan.
Conclusions
TRIM6 overexpression was associated with smoking, clinical stage, histological type, lymph node metastasis, TP53 mutation, and dismal prognosis, and related to prognosis-related age, race, sex, clinical stage, and tumor purity in LUAD patients. The overexpression level of TRIM6 correlated with the levels of LUAD immune cells and their markers. NEAT1–miR-101-3p/miR-335-5p/miR-374a-3p/miR-628-5p-TRIM6 signaling network might be an important factor in the prognosis of patients with LUAD.
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-2181
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-2181). The authors have no conflicts of interest to declare.
(English Language Editor: K. Brown)
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Guo Q, Ke XX, Fang SX, et al. PAQR3 Inhibits Non-small Cell Lung Cancer Growth by Regulating the NF-κB/p53/Bax Axis. Front Cell Dev Biol 2020;8:581919. 10.3389/fcell.2020.581919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamaguchi T, Hida T. Addition of atezolizumab to nab-paclitaxel plus carboplatin is a new standard option for the first-line treatment for non-squamous non-small cell lung cancer. Transl Cancer Res 2019;8:E11-4. 10.21037/tcr.2019.12.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito M, Miyata Y, Tsutani Y, et al. Positive EGFR mutation status is a risk of recurrence in pN0-1 lung adenocarcinoma when combined with pathological stage and histological subtype: A retrospective multi-center analysis. Lung Cancer 2020; 141:107-113. 10.1016/j.lungcan.2020.01.018 [DOI] [PubMed] [Google Scholar]
- 5.Kagimoto A, Tsutani Y, Izaki Y, et al. Prediction of Lymph Node Metastasis Using Semiquantitative Evaluation of PET for Lung Adenocarcinoma. Ann Thorac Surg 2020;110:1036-1042. 10.1016/j.athoracsur.2020.03.032 [DOI] [PubMed] [Google Scholar]
- 6.Guo Q, Ke XX, Liu Z, et al. Evaluation of the Prognostic Value of STEAP1 in Lung Adenocarcinoma and Insights into Its Potential Molecular Pathways via Bioinformatic Analysis. Front Genet, 2020;11:242. 10.3389/fgene.2020.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui D, Feng Y, Shi K, et al. Long non-coding RNA TRPM2-AS sponges microRNA-138-5p to activate epidermal growth factor receptor and PI3K/AKT signaling in non-small cell lung cancer. Ann Transl Med 2020;8:1313. 10.21037/atm-20-6331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li ZW, Zhang TY, Yue GJ, et al. Small nucleolar RNA host gene 22 (SNHG22) promotes the progression of esophageal squamous cell carcinoma by miR-429/SESN3 axis. Ann Transl Med 2020;8:1007. 10.21037/atm-20-5332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ke D, Guo Q, Fan TY, et al. Analysis of the Role and Regulation Mechanism of hsa-miR-147b in Lung Squamous Cell Carcinoma Based on The Cancer Genome Atlas Database. Cancer Biother Radiopharm 2021;36:280-91. 10.1089/cbr.2020.4187 [DOI] [PubMed] [Google Scholar]
- 10.Xiao B, Zhang W, Chen L, et al. Analysis of the miRNA-mRNA-lncRNA network in human estrogen receptor-positive and estrogen receptor-negative breast cancer based on TCGA data. Gene 2018;658:28-35. 10.1016/j.gene.2018.03.011 [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Qiu Q, Qian X, et al. Long noncoding RNA LCAT1 functions as a ceRNA to regulate RAC1 function by sponging miR-4715-5p in lung cancer. Mol Cancer 2019;18:171. 10.1186/s12943-019-1107-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du LJ, Mao LJ, Jing RJ. Long noncoding RNA DNAH17-AS1 promotes tumorigenesis and metastasis of non-small cell lung cancer via regulating miR-877-5p/CCNA2 pathway. Biochem Biophys Res Commun 2020;533:565-72. 10.1016/j.bbrc.2020.09.047 [DOI] [PubMed] [Google Scholar]
- 13.Zheng S, Zhou C, Wang Y, et al. TRIM6 promotes colorectal cancer cells proliferation and response to thiostrepton by TIS21/FoxM1. J Exp Clin Cancer Res 2020;39:23. 10.1186/s13046-019-1504-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei C, Wu J, Liu W, et al. Tripartite motif-containing protein 6 facilitates growth and migration of breast cancer through degradation of STUB1. Eur J Histochem 2021;65:3214. 10.4081/ejh.2021.3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Sun W, Qiao G, et al. The Expression of Tripartite Motif Protein 36 and β-Catenin Correlates with the Prognosis of Esophageal Cancer. Gastroenterol Res Pract 2020;2020:7641761. 10.1155/2020/7641761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang SX, Chen C, Guo Q, et al. High lncSNHG15 expression may predict poor cancer prognosis: a meta-analysis based on the PRISMA and the bio-informatics analysis. Biosci Rep 2020;40:BSR20194468. 10.1042/BSR20194468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R, Wang X, Zhang J, et al. LINC00942 Promotes Tumor Proliferation and Metastasis in Lung Adenocarcinoma via FZD1 Upregulation. Technol Cancer Res Treat 2021;20:1533033820977526. 10.1177/1533033820977526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Zhong L, Nie K, et al. Identification of LINC00665-miR-let-7b-CCNA2 competing endogenous RNA network associated with prognosis of lung adenocarcinoma. Sci Rep 2021;11:4434. 10.1038/s41598-020-80662-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Wo Y, Lu T, et al. Circ-AASDH functions as the progression of early stage lung adenocarcinoma by targeting miR-140-3p to activate E2F7 expression. Transl Lung Cancer Res 2021;10:57-70. 10.21037/tlcr-20-1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan LJ, Zhong TF, Tang RX, et al. Upregulation and clinicopathological significance of long non-coding NEAT1 RNA in NSCLC tissues. Asian Pac J Cancer Prev 2015;16:2851-5. 10.7314/APJCP.2015.16.7.2851 [DOI] [PubMed] [Google Scholar]
- 21.Zhou W, Chen X, Hu Q, et al. Galectin-3 activates TLR4/NF-κB signaling to promote lung adenocarcinoma cell proliferation through activating lncRNA-NEAT1 expression. BMC Cancer 2018;18:580. 10.1186/s12885-018-4461-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Li Y, Dong M, et al. Long non-coding RNA NEAT1 regulates E2F3 expression by competitively binding to miR-377 in non-small cell lung cancer. Oncol Lett 2017;14:4983-8. 10.3892/ol.2017.6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jen J, Tang YA, Lu YH, et al. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol Cancer 2017;16:104. 10.1186/s12943-017-0674-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun C, Li S, Zhang F, et al. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget 2016;7:51784-814. 10.18632/oncotarget.10108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Yang J, Xia Y, et al. Long Noncoding RNA NEAT1 Promotes Proliferation and Invasion via Targeting miR-181a-5p in Non-Small Cell Lung Cancer. Oncol Res 2018;26:289-96. 10.3727/096504017X15009404458675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao L, Bi M, Zhang H, et al. Downregulation of NEAT1 Suppresses Cell Proliferation, Migration, and Invasion in NSCLC Via Sponging miR-153-3p. Cancer Biother Radiopharm 2020;35:362-70. 10.1089/cbr.2019.3119 [DOI] [PubMed] [Google Scholar]
- 27.Xiong DD, Li ZY, Liang L, et al. The LncRNA NEAT1 Accelerates Lung Adenocarcinoma Deterioration and Binds to Mir-193a-3p as a Competitive Endogenous RNA. Cell Physiol Biochem 2018;48:905-18. 10.1159/000491958 [DOI] [PubMed] [Google Scholar]
- 28.Li B, Gu W, Zhu X. NEAT1 mediates paclitaxel-resistance of non-small cell of lung cancer through activation of Akt/mTOR signalling pathway. J Drug Target 2019;27:1061-7. 10.1080/1061186X.2019.1585437 [DOI] [PubMed] [Google Scholar]
- 29.Zang F, Rao Y, Zhu X, et al. Shikonin suppresses NEAT1 and Akt signaling in treating paclitaxel-resistant non-small cell of lung cancer. Mol Med 2020;26:28. 10.1186/s10020-020-00152-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mu L, Zhao H, Yang Y, et al. Long noncoding RNA NEAT1 aggravates sorafenib-resistance in non-small cell lung cancer via regulating miRNA-335/c-Met. J BUON 2021;26:345-52. [PubMed] [Google Scholar]
- 31.Zhang X, He X, Liu Y, et al. MiR-101-3p inhibits the growth and metastasis of non-small cell lung cancer through blocking PI3K/AKT signal pathway by targeting MALAT-1. Biomed Pharmacother 2017;93:1065-1073. 10.1016/j.biopha.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 32.Du W, Tang H, Lei Z, et al. miR-335-5p inhibits TGF-β1-induced epithelial-mesenchymal transition in non-small cell lung cancer via ROCK1. Respir Res 2019; 20:225. 10.1186/s12931-019-1184-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H, Wang Z, Wang Z. Long Noncoding RNA KCNMB2-AS1 Increases ROCK1 Expression by Sponging microRNA-374a-3p to Facilitate the Progression of Non-Small-Cell Lung Cancer. Cancer Manag Res 2020;12:12679-12695. 10.2147/CMAR.S270646 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Tanaka I, Dayde D, Tai MC, et al. SRGN-Triggered Aggressive and Immunosuppressive Phenotype in a Subset of TTF-1-Negative Lung Adenocarcinomas. J Natl Cancer Inst 2021;undefined:undefined.. [DOI] [PMC free article] [PubMed]
- 35.Ma K, Qiao Y, Wang H, et al. Comparative expression analysis of PD-1, PD-L1, and CD8A in lung adenocarcinoma. Ann Transl Med 2020;8:1478. 10.21037/atm-20-6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen T, Chen J, Zhu Y, et al. CD163, a novel therapeutic target, regulates the proliferation and stemness of glioma cells via casein kinase 2. Oncogene 2019;38:1183-99. 10.1038/s41388-018-0515-6 [DOI] [PubMed] [Google Scholar]
- 37.Ma LL, Guo LL, Luo Y, et al. Cdc42 subcellular relocation in response to VEGF/NRP1 engagement is associated with the poor prognosis of colorectal cancer. Cell Death Dis 2020;11:171. 10.1038/s41419-020-2370-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vescarelli E, Gerini G, Megiorni F, et al. MiR-200c sensitizes Olaparib-resistant ovarian cancer cells by targeting Neuropilin 1. J Exp Clin Cancer Res 2020;39:3. 10.1186/s13046-019-1490-7 [DOI] [PMC free article] [PubMed] [Google Scholar]