Abstract
Background
To evaluate the clinical value of microRNA (miR) and circulating tumor RNA (ctDNA) in the diagnosis of epithelial ovarian cancer (EOC) by meta-analysis and indirect comparison based on common reference criteria.
Methods
The PubMed, EMBASE, MEDLINE, Cochrane Library, Chinese biology medicine (CBM), China national knowledge infrastructure (CNKI), Wanfang, and Chinese Weipu (VIP) databases were searched by computer. The retrieval time limit was from the date of establishment of the database to September 2020. Two researchers independently screened the literature and extracted the basic data according to the inclusion and exclusion criteria formulated in advance, and evaluated the literature quality according to the quality assessment of diagnostic accuracy research (quadas-2). The Meta disc 1.4 and Stata 12.0 software programs were used for meta-analysis to calculate the combined sensitivity, combined specificity, combined positive likelihood ratio, combined negative likelihood ratio and combined diagnostic odds ratio (DOR). The summary receiver operating characteristic (SROC) curve was drawn using Revman 5.3 software, and the stability of the results was evaluated by sensitivity analysis. The publication bias was evaluated by Deek’s funnel asymmetric test. The relative diagnostic odds ratio (RDOR) results of indirect comparison between microRNA and ctDNA were obtained using R software.
Results
Nineteen articles were included, including a total of 1,351 EOC patients and 1,194 controls. The heterogeneity test showed that there was obvious heterogeneity caused by non-threshold effect. The random effects model was used for meta-analysis of microRNA in the diagnosis of EOC. The results showed that there was no significant difference between microRNA and ctDNA in the accuracy of EOC diagnosis. The asymmetric test of Deek’s funnel chart showed that there was no significant publication bias.
Discussion
There are some limitations in this study, there is no blind diagnostic test, and the intensity of indirect comparison evidence is lower than that of direct comparison evidence. The accuracy of diagnostic tests and the imperfection of mesh meta-analysis statistical methods. MicroRNA and ctDNA have similar clinical diagnostic value for EOC.
Keywords: Epithelial ovarian cancer (EOC), microRNA, ctDNA, indirect comparison, meta-analysis
Introduction
Over the past decade, the incidence and mortality rates of ovarian cancer have increased significantly (1). According to statistics, approximately 22,240 patients in the United States were diagnosed with invasive epithelial ovarian cancer (EOC) in 2013, of which about 14,000 patients with EOC died (2). EOC is the most common histopathological type of ovarian cancer, accounting for about 80–90%, which is the highest among all gynecological malignancies (3). Since the early symptoms of EOC are hidden, as well as atypical and lack of effective early diagnostic indicators, most patients are not diagnosed until later stage. In addition, the recurrence of the disease and drug resistance significantly reduce the survival rate of patients with advanced EOC. In the early stage of EOC, the mass is limited to the ovary, and the patients are more likely to be cured completely. Therefore, biomarkers that can screen and diagnose EOC in the early stage are urgently needed to improve the overall survival rate.
In recent years, the detection of molecular biological tumor markers has played an important role in the diagnosis of many malignant tumors. It has been found that microRNAs play an important role in regulating biological processes such as cell growth, apoptosis, metastasis, drug resistance, and invasion of ovarian cancer (4). At present, numerous studies have reported that peripheral blood circulating microRNAs have potential diagnostic value for a variety of tumor-related diseases; however, their diagnostic value for EOC remains unclear. This is mainly because the research design, tumor histopathological classification, sample type, microRNA type, detection method, and other factors are not exactly the same, resulting in an inconsistent diagnostic value for EOC. In addition to microRNAs, multiple studies have confirmed that ctDNA can be used as a marker for tumor dynamic monitoring (5). ctDNA is a single stranded or double stranded DNA fragment existing in the peripheral blood of tumor patients, and its molecular biological characteristics are still unclear. In addition to oncogene mutations, ctDNA also carries a variety of other tumor related molecular features, such as methylation changes, loss of heterozygotes and tumor derived viral gene sequences. In addition, the biological characteristics of ctDNA include: (I) the length of ctDNA is shorter than that of normal circulating free DNA, less than 150 bp; (II) the proportion of ctDNA in cfdna can be as low as 0.01%, up to more than 90%; (III) the main sources of ctDNA are as follows: apoptotic and necrotic tumor cells, surviving tumor cells and circulating tumor cells; (IV) short half-life: about 2 hours. These studies have shown that compared with the traditional tumor marker CA125, the detection of individualized ctDNA markers in EOC patients can be used to detect tumor residual lesions and dynamically predict the treatment effect (6). CA125 and HFA are important indicators for post-treatment monitoring of ovarian cancer. Studies have shown that the concentration of ctDNA in patients with ovarian cancer decreases gradually after operation. The area under the working characteristic curve (AUC) is 0.917 and the sensitivity is 88.9%. The AUC and sensitivity are higher than CA125 (0.724, 75%) and HE4 (0.743, 80.6%). The high level of ctDNA in patients with ovarian cancer before chemotherapy and recurrent ovarian cancer may be caused by residual tumors, CtDNA may be an ideal biomarker to monitor the progression and predict the prognosis of ovarian cancer.In the clinical diagnosis of EOC, clinicians are faced with the problem of how to choose the best diagnostic method. At present, although many clinical studies have used postoperative histopathology as the gold standard to evaluate the diagnostic value of microRNA and ctDNA in EOC, there has been no comparison between them. At present, many studies have reported the potential diagnostic value of miRNA and ctDNA in esophageal cancer, head and neck cancer, gastric cancer, endometrial cancer and other tumors. However, its value in the diagnosis of ovarian cancer is not clear. Due to the different research design, specimen type, miRNA and ctDNA, the results are not consistent, and different research conclusions are not completely consistent. Based on this, under the condition of insufficient direct comparative study evidence of microRNA and ctDNA detection, this study makes use of histopathological examination to conduct an indirect comparison of the diagnostic value of microRNA and ctDNA for EOC. The results are reported as follows.
We present the following article in accordance with the PRISMA reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-2609).
Methods
Literature retrieval strategy and data source
Search strategy
Based on the target disease, the diagnostic test to be evaluated, and the diagnostic accuracy index, the search terms were formulated and combined with free words. Finally, the Chinese search words were ovarian cancer, epithelial ovarian cancer, ovarian tumor, ovarian malignant tumor, epithelial ovarian malignant tumor, free DNA, circulating tumor DNA, cell free DNA, peripheral blood circulation DNA, peripheral blood circulation microRNA, microRNA, Mi-RNA, microRNA, diagnosis, sensitivity, specificity, sensitivity, and specificity. The English search terms were “epithelial oval cancer”, “carcinoma oval epic”, “epithelial carcinoma”, “oval cancer”, “circulating tumor DNA”, “tumor DNA”, “cell free tumor DNA”, “cell free tumor DNA”, “cell free nuclear acids”, “nuclear acids”, “circulating cell free nuclear acids”, “cfdna”, “cirdna”, “miRNA”, “microRNA”, “diagnoses”, “sensitivity”, and “specificity”.
Data sources
We performed a literature search of the PubMed, EMBASE, MEDLINE, the Cochrane Library, Chinese Biology Medicine (CBM), China National Knowledge Infrastructure (CNKI), Wanfang, and Chinese Weipu (VIP) databases. The retrieval time limit was from the date of establishment of the database to September 2020, and the languages were Chinese and English. At the same time, other relevant documents were searched manually, and the references included in the documents were tracked to supplement and obtain more documents.
Literature inclusion, exclusion criteria and diagnosis
The literature inclusion criteria were as follows: (I) study type: diagnostic test to study the diagnostic accuracy of EOC with histopathological examination as the gold standard; (II) study object: after histopathological examination, the tissue type was clearly EOC patients, and the control type as clear, without limitation on age, etiology, or race; (III) diagnostic test method: the expression of microRNA or ctDNA in peripheral blood circulation studying the diagnostic value for EOC; and (IV) outcome indicators: combined sensitivity, combined specificity, combined positive likelihood ratio, combined negative likelihood ratio, combined diagnostic odds ratio, as well as construction of a summary receiver operating characteristic (SROC) curve and calculating the area under curve (AUC). Relative diagnostic odds ratio (RDOR) result of indirect comparison between ctDNA and microRNA.
The literature exclusion criteria were as follows: (I) the case group was patients with malignant tumors of other tissues and organs, and the control group was the study of recurrent or metastatic ovarian cancer; (II) the included cases were patients with ovarian cancer, but the EOC four grid table data could not be obtained; (III) the study of qualitative analysis based on ctDNA methylation; (IV) studies with incorrect data, unable to obtain four-grid table data or research with incomplete data; and (V) repeated research, duplicate publication, and repeated collection of research conducted by the same author or research unit on the same population.
Diagnosis with miR and ctDNA for EOC: mainly include real-time PCR (RT-PCR), digital PCR (dPCR) and next generation sequencing (NGS).
Literature screening, data extraction, and quality evaluation
One author independently screened and extracted the data by reading the literature one-by-one. The literature included by both sides was simultaneously evaluated by the quality of diagnostic accuracy study (quadas-2). The second author of this paper decided whether to include literature that had differences and was difficult to determine whether it satisfied the inclusion criteria. The extracted data mainly included the first author, country where the research object was located, number of cases in the experimental and control groups, age of study subjects, microRNA and ctDNA sample type, microRNA, ctDNA detection method and microRNA content type, cut-off value, four grid table data, true positive (TP), false positive (FP), false negative (FN), and true negative (TN). The risk of bias was evaluated using the quadas-2 tool. All studies were evaluated independently by the two authors, and the results were consistent.
Statistical method
Meta-disc1.4 software (The Ramóny Cajal Hospital in Madrid, Spain) was used for meta-analysis to analyze the threshold effect between the studies of microRNA and ctDNA, and the Spearman correlation coefficient between the logarithm of sensitivity and the logarithm of “1-specificity” was tested, indicating the existence of a threshold effect with P<0.05. The heterogeneity among the studies was analyzed by Meta-disc1.4 software, and the diagnostic odds ratio (DOR) was calculated. The Cochran-Q value and I2 value were used to test the heterogeneity. P>0.05 and I2≤50% indicated that there was no statistical heterogeneity, and the fixed effects model was used to merge the effect quantity. However, P<0.05 and I2>50% indicated that there was statistical heterogeneity, and the random effects model was used to merge the effect quantity, including merge sensitivity, merge specificity, merge positive likelihood ratio, merge negative likelihood ratio, and merge DOR. “netmeta” of R software (Parametric Technology Corporation, USA) was used to calculate the area under the SROC curve (AUC). The installation package displayed the DOR results of indirect comparison of microRNA and ctDNA. This study is an indirect comparison of microRNA and ctDNA based on the gold standard of histopathological diagnosis. It belongs to the star mesh meta-analysis in the mesh meta type, which meets the similarity conditions, and there is no data for direct comparison, so there is no need for consistency test. When the confidence interval of the RDOR result contains 1, it indicates that the difference between the two diagnostic tests is not statistically significant. If A vs. B; when RDOR >1 and the confidence interval does not include 1, the diagnostic value of A is greater than that of B. Alternatively, when RDOR <1 and the confidence interval does not include 1, the diagnostic value of A is smaller than that of B, suggesting that the difference between the two diagnostic tests is statistically significant. Meta regression was used to analyze the source of heterogeneity, and Stata 12.0 software was used to draw the asymmetry of the Deek’s funnel test and evaluate whether there was publication bias. The difference was statistically significant when P<0.05, indicating that there was publication bias.
Results
Literature search results and basic information of included literature
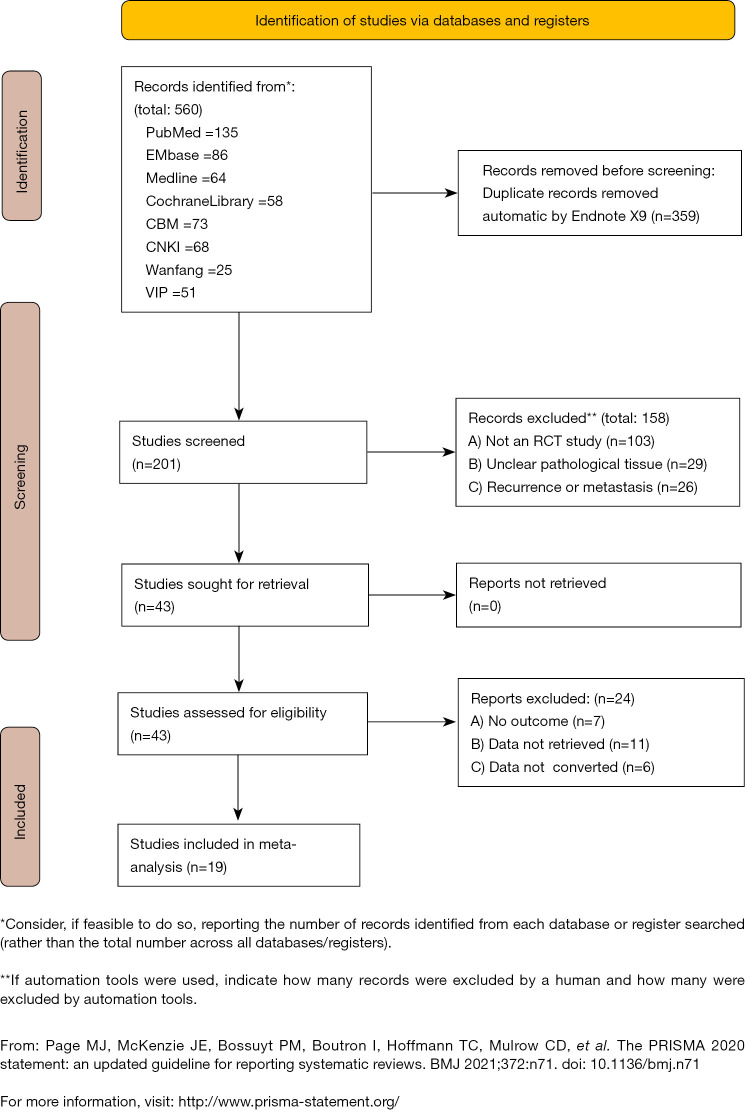
A total of 554 literatures were obtained through preliminary screening, and six additional articles were obtained through follow-up references. Firstly, 359 duplicate literatures that were included in different databases were excluded. Secondly, 158 articles were excluded after reading titles and abstracts. Also, after reading the full texts of the remaining articles, the case reports, clinical symptom analyses, reviews, and abstracts of meeting contents where the full text could not be obtained were screened according to the inclusion criteria (7). Finally, 19 articles were included (see Figure 1). There were 11 original studies on the diagnosis of EOC by microRNA detection (three in Chinese and eight in English, with 842 patients in the EOC group and 643 subjects in the control group). There were also eight original articles on the diagnosis of EOC by ctDNA detection (3 in Chinese and 5 in English, with 509 patients in the EOC group and 551 subjects in the control group). Taken together, a total of 1,351 patients with EOC and 1,194 controls were included in this study (see Tables 1,2 for basic data).
Figure 1.
Flowchart of literature inclusion and exclusion.
Table 1. Basic data of the literature on the diagnosis of EOC by microRNA detection.
| First author | Country | Case group (EOC) | Control group | Sample type | Test method | microRNA/ ctDNA |
Cut off value | TP | FP | FN | TN | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of cases | Age (years) | Number of cases | Age (years) | |||||||||||
| Chung (8) | Republic of Korea | 18 | 57.5 [42–74] | 12 | Unknown | Serum | qRT-PCR | miR-132 | 0.0001 | 12 | 1 | 6 | 11 | |
| miR-26a | 0.0041 | 18 | 3 | 0 | 9 | |||||||||
| let-7b | 0.066 | 15 | 0 | 3 | 12 | |||||||||
| miR-145 | 0.0011 | 9 | 0 | 9 | 12 | |||||||||
| Guo (9) | China | 50 | 54.5±2.4 | 50 | 51.4±3.2 | Serum | qRT-PCR | miR-92 | Unknown | 40 | 12 | 10 | 38 | |
| Kan (10) | China | 28 | 63±11 [30–70] | 28 | 64±12 [30–79] | Serum | qRT-PCR | miR-103 | Unknown | 22 | 15 | 6 | 13 | |
| Meng (11) | Germany | 60 | 56 [23–79] | 32 | 56 [42–70] | Serum | qPCR | miR-200a | Unknown | 40 | 3 | 20 | 29 | |
| miR-200b | 56 | 3 | 4 | 29 | ||||||||||
| miR-200c | 51 | 5 | 9 | 27 | ||||||||||
| Todeschini (12) | Italy | 168 | 61 [36–85]; 57 [34–76] | 65 | 61 [38–73]; 52 [26–67] | Serum | qRT-PCR | miR-1246 | 0.41 | 146 | 15 | 22 | 50 | |
| miR-595 | -0.29 | 79 | 10 | 89 | 55 | |||||||||
| miR-2278 | 0.26 | 136 | 22 | 32 | 43 | |||||||||
| Zheng (13) | China | 134 | 53.75 (10.25) | 70 | Unknown | Plasma | qRT-PCR | miR-205 | Unknown | 40 | 4 | 94 | 66 | |
| Zuberi (14) | India | 70 | ≤45 (57.1); >45 (42.5) | 70 | Unknown | Serum | qPCR | miR-125b | 3.76 | 44 | 16 | 26 | 54 | |
| Hong (15) | China | 96 | 58.6±6.2 | 35 | Unknown | Serum | qRT-PCR | miR-221 | Unknown | 85 | 5 | 11 | 30 | |
| Wang (16) | China | 102 | 59.4±9.2 [45–73] | 102 | 57.6±9.7 [43–72] | Serum | qRT-PCR | miR-210 | 1.4 | 98 | 50 | 4 | 52 | |
| miR-126 | 1.3 | 69 | 5 | 33 | 97 | |||||||||
| Mu (17) | China | 71 | 55.2 [34–67] | 134 | 57 [33–72]; 56.5 [35–76] | Plasma | qRT-PCR | miR-205 | Unknown | 65 | 18 | 6 | 116 | |
| miR-212 | 62 | 12 | 9 | 122 | ||||||||||
| miR-429 | 63 | 22 | 8 | 112 | ||||||||||
| Liu (18) | China | 45 | Unknown | 45 | Unknown | Serum | qRT-PCR | miR-145 | 0.411 | 35 | 4 | 10 | 41 | |
EOC, epithelial ovarian cancer; Real-time Quantitative PCR Detecting System (qPCR); qRT-PCR, real-time fluorescence quantitative reverse transcription PCR.
Table 2. Basic data of the literature on the diagnosis of EOC by ctDNA detection.
| First author | Country | Case group (EOC) | Control group | Sample type | Test method | MicroRNA/ctDNA | Cut off value | TP | FP | FN | TN | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of cases | Age (years) | Number of cases | Age (years) | |||||||||||
| Capizzi (19) | Italy | 22 | 64±8 [46–75] | 50 | 42 [35–62] | Plasma | qPCR | ctDNA content | 14.5 ng/mL | 17 | 2 | 5 | 48 | |
| Kamat (20) | U.S.A | 164 | 62 [28–84] | 124 | 53 [33–80]; 46 [29–81] | Plasma | qPCR | ctDNA content | 4,500 GE/mL | 143 | 16 | 21 | 108 | |
| Vanderstichele (21) | Belgium | 57 | 63 [59–73]; 44 [40–47] | 55 | 48 [25–56]; 58 [36–75] | Plasma | LWC | ctDNA content | Unknown | 44 | 5 | 13 | 50 | |
| Zachariah (22) | Switzerland | 21 | 61 [32–90] | 83 | 53 [21–78]; 58 [22–93]; 34 [25–43] | Plasma | qPCR | ctDNA content | 2,653 GE/mL | 16 | 26 | 5 | 57 | |
| Zhang (23) | China | 24 | 51.96±14.12 | 24 | 42.67±12.28; 37.00±13.07 | Plasma | qPCR | ctDNA content | 8.57 ng/μL | 16 | 5 | 8 | 19 | |
| Fu (24) | China | 93 | 55.7 [34–82] | 125 | Unknown | Plasma | SYBR; Green I | ctDNA content | 44 μg/L | 61 | 12 | 32 | 113 | |
| Shao (7) | China | 67 | 57.4 [39–85] | 51 | 55.7 [42–63]; 51.6 [36–68] | Serum | bDNA | ctDNA content | 344.4 μg/L | 56 | 8 | 11 | 43 | |
| Hu (25) | China | 61 | 54.11±1.03 | 39 | 51.00±1.46 | Plasma | FDM | ctDNA content | 0.72 ng/μL | 34 | 2 | 27 | 37 | |
qPCR, real-time fluorescence quantitative PCR; LWC, low coverage whole genome sequencing; SYBR Green I, spot fluorescence staining method; bDNA, branch DNA technology; FDM, fluorescence dye method.
Literature quality evaluation and bias risk evaluation were included
During the evaluation process, it was found in the case selection section that there were 2 consecutive articles included in the case (8,18); one literature included ovarian cancer patients with non-EOC tissue type (21); all literatures were case-control studies; all of the cases included in the original literature were confirmed by histopathological examination. Therefore, the 19 articles were evaluated as “low inapplicability consideration”. Among the diagnostic tests to be evaluated, seven articles had no cut-off value, which were evaluated as “high inapplicability consideration” in the consideration of applicability. Three articles in the gold standard did not clearly describe the histopathological types of the EOC cases, so the bias risk assessment was “unclear”; one article included ovarian cancer patients with non-EOC, which was evaluated as “high inapplicability consideration” in the consideration of applicability. We set the time range ≤3 years as the appropriate time interval, and there were no lost visits or missing cases in all of the original studies. The bias risk assessment and applicability judgment were carried out using the quadas-2 tool, and the results are shown in Figure 2.
Figure 2.
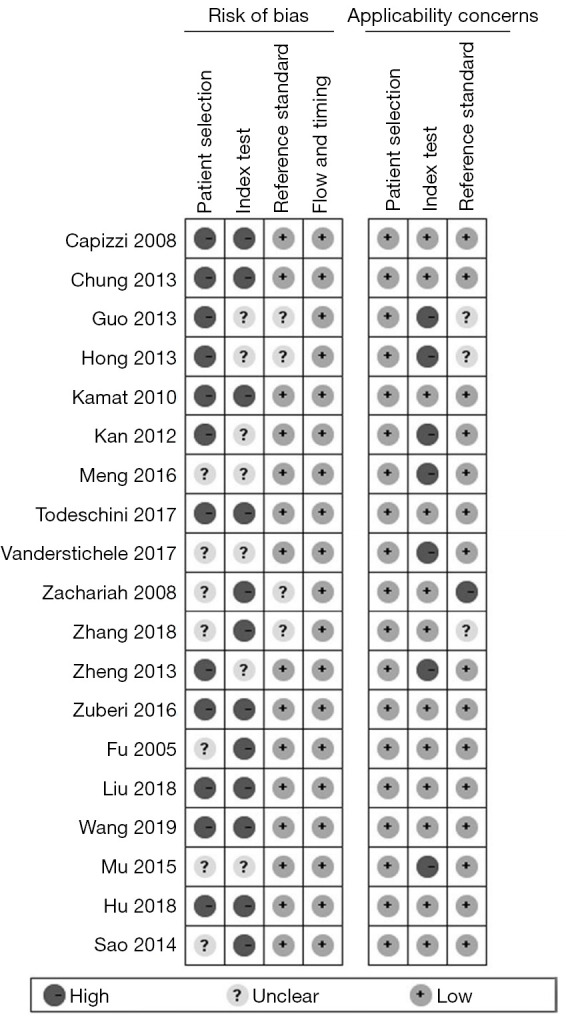
Quality evaluation of the included literature.
Meta-analysis results
The Spearman correlation coefficients of microRNA and ctDNA in the diagnosis of EOC were 0.406 (P=0.068) and 0.095 (P=0.823), respectively, suggesting that there was no threshold effect in the quantitative evaluation, and thus, the random effects model was used for meta-analysis. The combined analysis results of each diagnostic test compared with the gold standard were as follows. Firstly, the meta-analysis results of microRNA in the diagnosis of EOC showed that the combined sensitivity was 0.74 (95% CI: 0.72–0.76), the combined specificity was 0.82 (95% CI: 0.80–0.84), and the combined positive likelihood ratio was 4.62 (95% CI: 3.35–6.38), the combined negative likelihood ratio was 0.24 (95% CI: 0.17–0.35), the combined DOR was 21.62 (95% CI: 13.57–34.44), and the AUC was 0.894.
The meta-analysis results of ctDNA in the diagnosis of EOC showed that the combined sensitivity was 0.76 (95% CI: 0.72–0.80), the combined specificity was 0.86 (95% CI: 0.83–0.89), the combined positive likelihood ratio was 5.81 (95% CI: 3.60–9.38), the combined negative likelihood ratio was 0.29 (95% CI: 0.21–0.41), the combined DOR was 22.61 (95% CI: 13.27–38.52), and the AUC was 0.884. The area under the SROC curve of EOC diagnosed by microRNA and ctDNA was the same (as shown in Figure 3).
Figure 3.
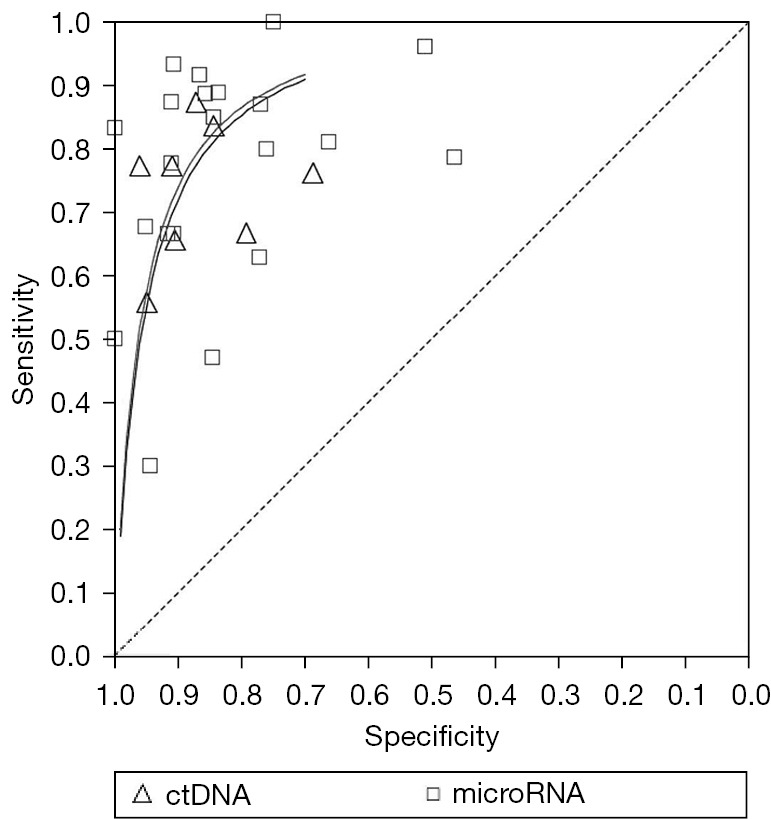
SROC curve of EOC diagnosed by microRNA and ctDNA. SROC, summary receiver operating characteristic; EOC, epithelial ovarian cancer; ctDNA, circulating tumor RNA.
RDOR results of indirect comparison between microRNA and ctDNA in diagnosing EOC
Taking histopathological (his) examination as the common diagnostic standard, the RDOR value of microRNA vs. ctDNA was 0.95 (95% CI: 0.42–2.14) (confidence interval includes 1), and the RDOR value of ctDNA vs. microRNA was 1.05 (95% CI: 0.47–2.37) (confidence interval includes 1). This suggested that there was no significant difference between microRNA and ctDNA in the diagnostic accuracy of EOC (see Figure 4).
Figure 4.
Indirect comparison of microRNA and ctDNA in diagnosing EOC. ctDNA, circulating tumor RNA; EOC, epithelial ovarian cancer.
Heterogeneity test
The results of the heterogeneity test caused by the non-threshold effect showed that the corresponding Cochran-Q =14.06 (P=0.0501>0.05) and I2=50% in eight articles included in ctDNA diagnosis of EOC. There was no statistical heterogeneity among studies based on the P value. The corresponding Cochran-Q =74.88 (P=0.0000) and I2=73.7% in 11 literatures included in microRNA diagnosis of EOC.
Meta regression analysis was conducted according to the following factors to determine the source of heterogeneity: region (divided into “China Group” and “foreign group”); detection method (divided into “qRT-PCR group” and “qPCR group”); cut off value (divided into “cut off value group” and “no cut off value group”); case type (divided into “EOC tissue type clear group” and “EOC tissue type unknown group”); and sample type (divided into “serum group” and “plasma group”). Meta regression analysis found no causes of heterogeneity; however, the heterogeneity decreased after excluding the two studies with “plasma” sample type.
Sensitivity analysis and publication bias
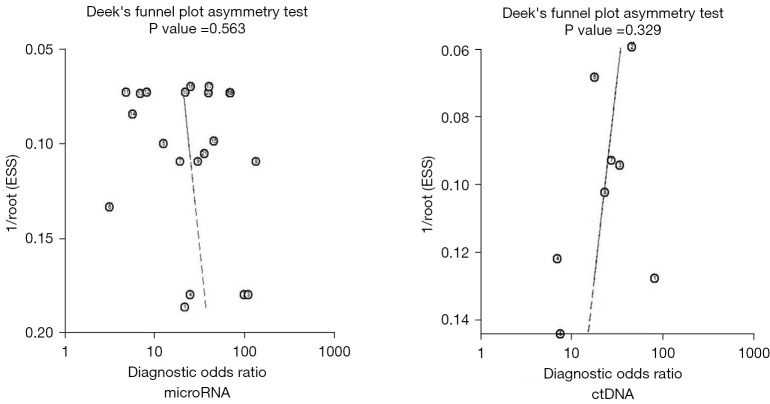
Articles with significantly different results or those with large sample sizes included in the microRNA and ctDNA diagnosis of EOC were removed, and meta-analysis was carried out again. The results of the two diagnostic tests showed no significant changes, indicating that the research of the two diagnostic tests was stable and the results were reliable. The literature included in the two diagnostic tests was tested by linear regression method to check whether there is publication bias, and the Deek's funnel chart was drawn. The slope coefficient results of 21 microRNAs included in 11 articles including microRNA was P=0.563, without obvious publication bias. The slope coefficient of eight literatures including ctDNA was P=0.329, without obvious publication bias (see Figure 5).
Figure 5.
Funnel chart of EOC diagnosed by microRNA and ctDNA. EOC, epithelial ovarian cancer; ctDNA, circulating tumor RNA.
Discussion
In recent years, molecular biological tumor markers such as microRNA and ctDNA have been increasingly used in the diagnosis of malignant tumors (4). Whether the diagnostic efficacy of molecular biological tumor markers for ovarian cancer is better than that of traditional serum tumor markers (CA125 and HE4) has become a research hotspot (23). In this paper, a systematic meta-analysis of the 19 included articles (7-25) was carried out to comprehensively evaluate the diagnostic test that uses histopathological examination as a reference standard to study the diagnostic accuracy of EOC. The diagnostic effect of microRNA and ctDNA on EOC was evaluated by calculating the combined diagnostic effect, fitting the SROC curve, calculating the RDOR results of indirect comparison, and drawing the adjusted indirect comparison forest map. The heterogeneity among the diagnostic tests included in the diagnosis of EOC by microRNA was explored by meta regression analysis. Finally, the reliability of this study was evaluated by sensitivity analysis and detection of publication bias.
The results showed that the sensitivity, specificity, combined DOR, and AUC of microRNA in the diagnosis of EOC were 0.74, 0.82, 21.62, and 0.894. Meta regression analysis did not find the cause of heterogeneity, but the difference between sample types increased the heterogeneity between various studies. For ctDNA diagnosis, the sensitivity, specificity, combined DOR, and AUC of EOC were 0.76, 0.86, 22.61, and 0.884, and the diagnostic efficiency was slightly lower than that of microRNA. We used Revman5.3 software to present the SROC curves of the two diagnostic tests in the same graph. The results showed that the AUC of microRNA for EOC almost overlapped with the AUC of ctDNA, suggesting that the diagnostic accuracy of these two markers was similar. However, this chart can only be used for intuitive descriptive analysis without quantitative comparison. Therefore, we took histopathological examination as the common diagnostic standard and conducted an indirect comparison between microRNA and ctDNA by calculating the RDOR value between them. The RDOR value adjustment indirect comparison forest chart showed that the difference between the two is not statistically significant.
At present, there are considerable difficulties associated with the use of molecular tumor markers for pan-cancer early screening (14). For example, it is impossible to locate the organs of the detected positive results because most of the mutations of proto-oncogenes and tumor suppressor genes are not tissue-specific, and are related to the occurrence and development of a variety of cancers. In addition, a large number of non-pathogenic gene mutations are also carried in normal humans, and with aging, the number of mutations will also increase. Cancer screening technology needs to be able to select cancer-related gene mutations from non-pathogenic somatic gene mutations. The most fundamental need is a baseline of mutation data from an asymptomatic population, which can be used as a comparison standard to obtain accurate identification and localization of cancer-related mutations (26). Therefore, more relevant studies are needed for in-depth analysis in order to improve on traditional serum tumor markers, and encourage widespread clinical use in tumor diagnosis and treatment.
ctDNA related biopsy has the advantages of noninvasive, high sensitivity and specificity, real-time and comprehensive information. It has broad clinical application prospects in early diagnosis, condition monitoring, prognosis evaluation, drug resistance monitoring and medication guidance of ovarian cancer. However, the application of ctDNA in clinic has the following limitations: (I) lack of standardized ctDNA extraction and detection technology; (II) there are deficiencies in the clinical research of liquid biopsy in ovarian cancer (27). Nevertheless, the research and application prospect of ctDNA in ovarian cancer is still worthy of our exploration and discovery, which will take the most solid step for mankind to finally overcome cancer.
There are some limitations in this study that should be noted. Firstly, the 19 included studies did not utilize the blinding method for diagnostic test detection, and the sample sizes of some studies were small. Furthermore, the strength of indirect comparison evidence is lower than that of direct comparison. At the same time, due to the accuracy of the diagnostic test and the imperfect statistical method of mesh meta-analysis, the results of this paper failed to provide the similarity results and overall heterogeneity between the included studies. Therefore, more reasonable and reliable conclusions also need to rely on the accuracy of diagnostic tests, the development and maturity of statistical methods of mesh meta-analysis and the development of related software.
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-2609
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-2609). The authors have no conflicts of interest to declare.
(English Language Editor: A. Kassem)
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013;63:11-30. 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 3.Köbel M, Kalloger SE, Boyd N, et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med 2008;5:e232. 10.1371/journal.pmed.0050232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi PW, Ng SW. The Functions of MicroRNA-200 Family in Ovarian Cancer: Beyond Epithelial-Mesenchymal Transition. Int J Mol Sci 2017;18:1207. 10.3390/ijms18061207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer 2014;14:623-31. 10.1038/nrc3820 [DOI] [PubMed] [Google Scholar]
- 6.Gold B, Cankovic M, Furtado LV, et al. Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. J Mol Diagn 2015;17:209-24. 10.1016/j.jmoldx.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao X, Ji M, Chen X, et al. Study on the value of free DNA in auxiliary diagnosis of ovarian cancer. Journal of Clinical Laboratory 2014;11:821-4. [Google Scholar]
- 8.Chung YW, Bae HS, Song JY, et al. Detection of microRNA as novel biomarkers of epithelial ovarian cancer from the serum of ovarian cancer patients. Int J Gynecol Cancer 2013;23:673-9. 10.1097/IGC.0b013e31828c166d [DOI] [PubMed] [Google Scholar]
- 9.Guo F, Tian J, Lin Y, et al. Serum microRNA-92 expression in patients with ovarian epithelial carcinoma. J Int Med Res 2013;41:1456-61. 10.1177/0300060513487652 [DOI] [PubMed] [Google Scholar]
- 10.Kan CW, Hahn MA, Gard GB, et al. Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer 2012;12:627. 10.1186/1471-2407-12-627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng X, Müller V, Milde-Langosch K, et al. Circulating Cell-Free miR-373, miR-200a, miR-200b and miR-200c in Patients with Epithelial Ovarian Cancer. Adv Exp Med Biol 2016;924:3-8. 10.1007/978-3-319-42044-8_1 [DOI] [PubMed] [Google Scholar]
- 12.Todeschini P, Salviato E, Paracchini L, et al. Circulating miRNA landscape identifies miR-1246 as promising diagnostic biomarker in high-grade serous ovarian carcinoma: A validation across two independent cohorts. Cancer Lett 2017;388:320-7. 10.1016/j.canlet.2016.12.017 [DOI] [PubMed] [Google Scholar]
- 13.Zheng H, Zhang L, Zhao Y, et al. Plasma miRNAs as diagnostic and prognostic biomarkers for ovarian cancer. PLoS One 2013;8:e77853. 10.1371/journal.pone.0077853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuberi M, Khan I, Mir R, et al. Utility of Serum miR-125b as a Diagnostic and Prognostic Indicator and Its Alliance with a Panel of Tumor Suppressor Genes in Epithelial Ovarian Cancer. PLoS One 2016;11:e0153902. 10.1371/journal.pone.0153902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong F, Li Y, Xu Y, et al. Prognostic significance of serum microRNA-221 expression in human epithelial ovarian cancer. J Int Med Res 2013;41:64-71. 10.1177/0300060513475759 [DOI] [PubMed] [Google Scholar]
- 16.Wang B, Xu Z, Zhao H, et al. Relationship between serum mir-210, miR-126 and clinicopathological features and prognosis in patients with ovarian cancer. Chinese Journal of Family Planning 2019;2:221-5, 229. [Google Scholar]
- 17.Mu P, Li L, Jia H, et al. Analysis and significance of plasma mir-205, miR-212 and mir-429 in patients with ovarian cancer? J Clin Oncol 2015;1:30-5. [Google Scholar]
- 18.Liu P, Zou C. Expression and clinical significance of microrna-145 in epithelial ovarian cancer and cell lines. Journal of Nanjing Medical University (Natural Science Edition) 2018;11:1528-32. [Google Scholar]
- 19.Capizzi E, Gabusi E, Grigioni AD, et al. Quantification of free plasma DNA before and after chemotherapy in patients with advanced epithelial ovarian cancer. Diagn Mol Pathol 2008;17:34-8. 10.1097/PDM.0b013e3181359e1f [DOI] [PubMed] [Google Scholar]
- 20.Kamat AA, Baldwin M, Urbauer D, et al. Plasma cell-free DNA in ovarian cancer: an independent prognostic biomarker. Cancer 2010;116:1918-25. 10.1002/cncr.24997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanderstichele A, Busschaert P, Smeets D, et al. Chromosomal Instability in Cell-Free DNA as a Highly Specific Biomarker for Detection of Ovarian Cancer in Women with Adnexal Masses. Clin Cancer Res 2017;23:2223-31. 10.1158/1078-0432.CCR-16-1078 [DOI] [PubMed] [Google Scholar]
- 22.Zachariah RR, Schmid S, Buerki N, et al. Levels of circulating cell-free nuclear and mitochondrial DNA in benign and malignant ovarian tumors. Obstet Gynecol 2008;112:843-50. 10.1097/AOG.0b013e3181867bc0 [DOI] [PubMed] [Google Scholar]
- 23.Zhang R, Pu W, Zhang S, et al. Clinical value of ALU concentration and integrity index for the early diagnosis of ovarian cancer: A retrospective cohort trial. PLoS One 2018;13:e0191756. 10.1371/journal.pone.0191756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu S, Tu H, Zhang G, et al. Quantitative study of circulating DNA in patients with epithelial ovarian cancer. Chinese Journal of Practical Gynecology and Obstetrics 2005;11:659-62. [Google Scholar]
- 25.Hu Q, Jin Y, Li Y, et al. Study on the correlation between plasma free DNA concentration and the diagnosis and clinical characteristics of ovarian cancer. Journal of Practical Obstetrics and Gynecology 2018; 5:357-60. [Google Scholar]
- 26.Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. Cancer Res 2000;60:6116-33. [PubMed] [Google Scholar]
- 27.Li S, Shang G. Current status and challenges of clinical application of circulating tumor DNA. Journal of Multidisciplinary Cancer Management (Electronic Version) 2021;7:72-8. [Google Scholar]