Abstract
Background
Gastric cancer (GC) is the most common malignant tumor of the digestive system, and its mortality rate ranks first among malignant tumors. However, the pathogenesis of GC has not yet been fully elucidated. This study found that microRNA (miRNA)-339 is abnormally expressed in GC tissues. However, the role and molecular mechanism of miRNA-339 in the occurrence and development of GC are still unclear.
Methods
Fluorescence quantitative polymerase chain reaction (qPCR) was used to detect the expression level of miRNA-339 in GC tissues and adjacent tissues and analyze the correlation with the clinicopathological characteristics of GC patients. Cell counting kit-8 (CCK-8) and Transwell experiments detected the effect of overexpression of miRNA-339 on the proliferation, invasion, and migration of GC cells. The luciferase reporter gene detected the downstream target molecules regulated by miRNA-339, and western blot was employed to detect the effect of overexpression of miRNA-339 on the expression of ZNF689.
Results
The results of fluorescence qPCR showed that miRNA-339 was less expressed in GC tissues compared with adjacent tissues, and it was correlated with the patient's clinical tumor, node, metastasis (TNM) grade and lymph node metastasis. Cell function experiments showed that overexpression of miRNA-339 can significantly inhibit the proliferation, invasion, and migration of GC cells. The luciferase reporter gene showed that miRNA-339 can bind to the 3'-UTR region of ZNF689, and overexpression of miRNA-339 can significantly inhibit the expression of ZNF689 in GC cells. Overexpression of ZNF689 can significantly block the ability of overexpression of miRNA-339 to inhibit the proliferation and migration of GC cells.
Conclusions
miRNA-339 inhibits the proliferation and invasion of GC cells through targeted regulation of the expression of ZNF689. In addition, the expression level of miRNA-339 can be used as a biomarker for the prognosis of GC.
Keywords: MicroRNA 339 (miRNA-339), ZNF689, gastric cancer (GC), proliferation, invasion
Introduction
Gastric cancer (GC) is the most common malignant tumor of the digestive system, with the highest mortality rate ranks among malignant tumors (1). Studies have shown that the onset of early GC is hidden, and most patients have reached an advanced stage by the time of diagnosis, with tumor cells already having metastasized, which seriously affects the treatment effect (2). The occurrence and development of GC is a process involving the dysregulation of multiple genes, that is, the activation of proto-oncogenes or the inactivation of tumor suppressor genes (3). Therefore, in-depth study of the molecular mechanisms of GC proliferation and invasion, searching for key molecular therapeutic targets, has important theoretical and practical significance for the clinical treatment of GC in order to develop new targeted drugs. MicroRNA (miRNA) is a single-stranded small molecule RNA composed of about 19–25 nucleotides and non-coding protein, which regulates many biological processes such as cell differentiation, proliferation, apoptosis, carcinogenesis, and hormone secretion (4,5). About one-third of human genes are regulated by miRNAs to varying degrees. These miRNAs may participate in the occurrence and development of tumors by regulating the expression of related genes (6,7). The two arms of the precursor hsa-miR-339 were processed respectively to generate two miRNAs, which were named as miR-339-3p and miR-339-5p, respectively. The miRNA-339 is closely related to the occurrence and development of tumors, and has become a research hotspot in the field of tumors in recent years (8-11). Recent studies have found that miRNA-339-5p plays an important role in inhibiting the invasion and migration of pancreatic cancer cells (8). Overexpression of miRNA-339 significantly inhibits the metastasis of hepatocellular carcinoma (12). In addition, the differential expression of miRNA-339 between patients with liver cirrhosis and liver cancer can be used as a new non-invasive prognostic marker (13). Therefore, miRNA-339 may be a potential target for tumor treatment. However, so far, the specific molecular mechanism of how miRNA-339 regulates key downstream proteins is unclear.
This study preliminarily analyzed the expression of miRNA-339 in GC tissues and its relationship with the clinical pathological characteristics of patients, and explored the effect of miRNA-339 on the proliferation and migration of esophageal cancer cells and related molecular mechanisms. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-994).
Methods
Collection of clinical samples
After the meeting was approved and the patient or family members had provided written informed consent, 46 pairs of GC tissue and para-cancerous tissue were collected between April 2019 and October 2020 from patients in our hospital who had not received any treatment prior to surgery. The specific steps were as follows: The resected cancerous tissues and adjacent tissues were collected after the operation, washed with phosphate buffered saline (PBS) buffer, and put in liquid nitrogen within 30 min for preservation. At the same time, relevant patient clinical data were collected. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by First Affiliated Hospital of Wannan Medical College, Yijishan Hospital (YJ-0219-24).
Cell culture
The GC cell lines (SGC-7901, MKN-45, BGC-823, AGS) and normal gastric fibroblasts (GES-1) used in this study were purchased from the Shanghai Institute of Biological Sciences (Shanghai, China). Roswell Park Memorial Institute (RPMI)-1640 or Dulbecco’s modified eagle medium (DMEM) (Gibco, Waltham, MA, USA) containing 10% fetal bovine serum (FBS) were used in an incubator at 37 °C, 5% CO2, and 95% humidity. The complete medium included 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin. The cells were replaced with medium every 2 days, and subcultured at 1:2–1:3 according to the cell growth status. Cells in the logarithmic growth phase were used for follow-up experimental research and mRNA extraction.
Reverse transcription polymerase chain reaction experiment
TRIzol method was used to extract total RNA in tissues and cells, then NanoDrop 2000 (Thermo Fisher, Waltham, MA, USA) was used to determine the concentration, and these were stored at −80 °C for later use. The reaction was performed on a fluorescent quantitative PCR (qPCR) machine, and U6 was used as the reference gene at the same time. The relative quantitative determination of each gene was carried out using the double standard curve method. The relative expression of each gene was calculated by the 2−ΔΔCt method, where ΔCt = target gene Ct value-internal reference gene Ct value, ΔΔCt = transfection group ΔCt-control group ΔCt.
miRNA-339: forward 5'-ACACTCCAGCTGCGGTCCCTGTCCTCCAGGAG-3'; reverse 5'-TGGTGTCGTGGAGTCG-3'; U6: forward 5'-CTCGCTTCGGCAGCACA-3', reverse 5'-AACGCTTCACGAATTTGCGT-3'.
Transfection experiment
Control mimics and miRNA-339 mimics used for cell transfection were directly purchased from Shanghai Shenggong Bioengineering Co., Ltd. (Shanghai, China). At 24 h before transfection, logarithmic growth phase SGC-7901 and BGC-823 cells were harvested, they were then trypsinized, and complete medium was added to resuspend them. A pipette was used to mix them in order to make a cell suspension. Cells (5×105/well) were inoculated into a 6-well plate to ensure that the cells were evenly distributed in the wells. At 2 h before transfection, the original medium was removed and replaced with a fresh basal medium without antibiotics and serum. After mixing miRNA-339 and Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), they were added to the corresponding culture wells, shaken gently to mix, and placed in an incubator for culture. After 4–6 h, the transfection mixture was removed and replaced with fresh DMEM. The culture plate was then placed in an incubator for 48 h.
Cell Counting Kit-8 detection of cell viability
After 48 h transfection, the cells were digested with trypsin and counted, seeded in a 96-well plate (3×103 cells/well), and then the 96-well plate was placed in a 37 °C, 5% CO2 incubator. At 24, 48, 72, and 96 h, microplate reader (ELIASA) was used to detect the absorbance value (OD value) at 490 nm and a graph of cell growth was drawn.
Invasion and migration experiments
At 24 h after transfection of different cell groups, a single cell suspension was prepared with serum-free DMEM, the cell concentration was adjusted to 5×105 cells/mL, and then 200 µL of cell suspension was added to the upper chamber of each Transwell chamber. A total of 500 µL of DMEM containing 10% serum was added to the lower chamber, and the chamber was placed into a 24-well plate. For invasion experiments, Matrigel glue (Sigma-Aldrich, St. Louis, MO, USA) was pre-laid in the Transwell chamber; the 24-well plate was placed in an incubator and incubated at 37 °C and 5% CO2 for 48 h. Then, the well was removed, washed with PBS 3 times, and the cells in the upper chamber were wiped off carefully with a cotton swab, fixed with 95% ethanol for 10 min, stained with 4 g/L crystal violet, and the movement of the cells to the outer layer of the microporous membrane was observed under an inverted microscope. The number of cells in 10 fields under the double field was randomly counted, the average number of cells in each field was calculated, and the experiment was repeated 3 times.
Luciferase reporter gene
The 3'-UTR of ZNF689 containing the miR-339-binding sites and its mutant was cloned into pGL3-control luciferase reporter vector (Promega Corporation, Fitchburg, WI, USA). The luciferase vector contained pGL3-ZNF689 wild-type (Wt) and mutant (Mut) plasmids in the ZNF689 promoter region, which were transfected into GC cells with Lipofectamine 2000, and then the dual luciferase reporter system was used to detect the initiation of ZNF689 activity. The luciferase activity was calculated according to the standard curve of luciferin activity.
Western blot
The transfected cells were collected, protein lysate (pre-added protease and phosphatase mixed inhibitor) was added, the cells were placed on an ice bath for lysis, centrifuged at 4 °C to obtain the supernatant, bicinchoninic (BCA) protein quantification kit was used for quantification, and prepared and separated by conventional methods gel and concentrated gel, according to 80–60 µg per lane. A sample was added to the gel hole, followed by the conduction of electrophoresis at constant pressure of 40 V (concentrated gel)/100 V (separation gel) until bromophenol blue reached the edge of the gel. Then, the cells were blocked in 8% skimmed milk powder at room temperature 2 h, incubated with the primary antibody diluted in 4% skimmed milk powder at 4 °C overnight, incubated with the secondary antibody diluted in 4% skimmed milk powder at room temperature for 4 hours, and washed with Tris-buffered saline with Tween 20 (TBST). Liquids A and B were mixed in enhanced chemiluminescence (ECL) with equal volume, they were added dropwise to the nitrocellulose membrane, and the medical X-ray film was photographed.
Statistical analysis
The software SPSS 19.0 (IBM Corp., Armonk, NY, USA) was used to analyze the data, and the measurement data were expressed as (mean ± standard deviation (SD)). Normally distributed data were compared using Student’s t-test, and non-normally distributed data were analyzed by analysis of variance (ANOVA). The chi-square test was used for counting data. A P value <0.05 was considered statistically significant.
Results
miRNA-339 is lowly expressed in GC tissues and is correlated with patient clinicopathological characteristics
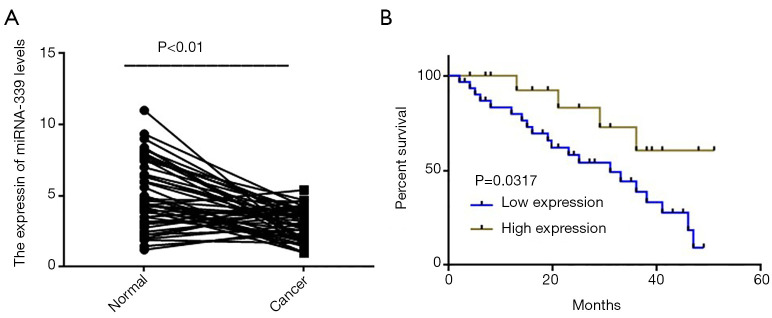
In order to explore the expression of miRNA-339 in GC tissues, we firstly used fluorescence qPCR to detect the expression of miRNA-339 in 46 GC patients and paired adjacent tissue samples. The results showed that compared with adjacent tissues, miRNA-339 was more lowly expressed in GC tissues (Figure 1A). According to the expression level of miRNA-339, esophageal cancer tissues were divided into a miRNA-339 low expression group (n=31) and miRNA-339 high expression group (n=15), and the relationship between the expression level of miRNA-339 and the clinicopathological characteristics of the patient was analyzed. The results showed that the low expression of miRNA-339 is correlated with lymph node metastasis and tumor, node, metastasis (TNM) stage (Table 1). The Kaplan-Meier estimator was used to further analyze the relationship with patient survival, and the results showed that patients with low miRNA-339 expression had a shorter survival time (Figure 1B). The above results showed that the expression level of miRNA-339 can be used as a marker for the diagnosis and prognosis of GC.
Figure 1.
The low expression of miRNA-339 in GC tissues is related to patient survival. (A) Fluorescence qPCR was used to detect the expression of miRNA-339 in 46 cases of GC tissues and adjacent tissues; (B) Kaplan-Meier estimation was used for further analysis of the relationship between the expression level of miRNA-339 and the survival time of GC patients. GC, gastric cancer; qPCR, quantitative polymerase chain reaction.
Table 1. The association of miRNA-339 expression levels with clinical pathology features in GC.
| Characteristics | Number (n=46) | miRNA-339 | P value | |
|---|---|---|---|---|
| Low (n=31) | High (n=15) | |||
| Age (years) | 0.204 | |||
| ≤60 | 26 | 20 | 6 | |
| >60 | 20 | 11 | 9 | |
| Gender | 0.539 | |||
| Male | 24 | 15 | 9 | |
| Female | 22 | 16 | 6 | |
| Tumor size (cm) | 0.058 | |||
| ≤3 cm | 15 | 10 | 5 | |
| >3 cm | 31 | 21 | 10 | |
| Lymph node metastasis | 0.008* | |||
| Yes | 29 | 24 | 5 | |
| No | 17 | 7 | 10 | |
| Differentiation | 0.195 | |||
| Well and moderate | 16 | 13 | 3 | |
| Poor | 30 | 18 | 12 | |
| TNM stage | 0.048* | |||
| I and II | 17 | 8 | 9 | |
| III and IV | 29 | 23 | 6 | |
The chi-square test was used for the association analysis and *P<0.05 was considered statistically significant. GC, gastric cancer; TNM, tumor, node, metastasis.
Overexpression of miRNA-339 can inhibit the proliferation of GC cells
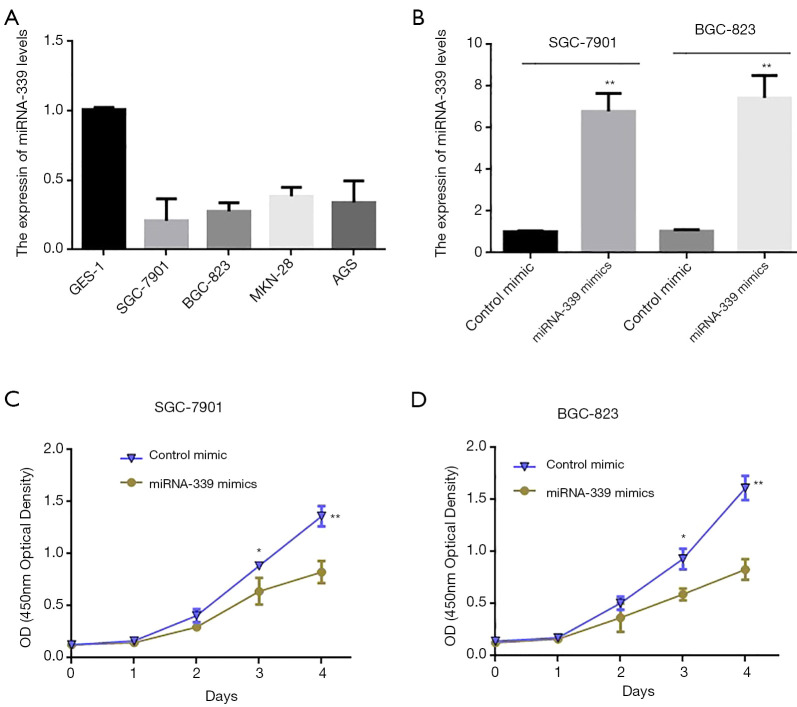
In order to further study the function of miRNA-339 in the occurrence and development of GC, fluorescence qPCR was first used to detect GC cell lines (SGC-7901, MKN-45, BGC-823, AGS) and normal human gastric epithelial cells (GES-1). The results showed that the expression level of miRNA-339 in GC cells was significantly lower than that of normal human gastric epithelial cells (GES-1). Among them, the expression level of miRNA-339 in SGC-7901 and MGC-823 cells was significantly reduced. As their expression level was the lowest (Figure 2A), GC cell lines SGC-7901 and MGC-823 were selected for subsequent functional experiments and molecular mechanism research. First, miRNA-339 mimics were synthesized, then SGC-7901 and MGC-823 cells were transfected to detect the overexpression efficiency of miRNA-339 in the cells. The test results showed that compared with the control mimics, transfection of miRNA-339 can significantly increase the expression level of miRNA-339 in GC cells (Figure 2B). Then, Cell Counting Kit-8 (CCK-8) was used to detect the effect of overexpression of miRNA-339 on the proliferation of GC cells SGC-7901 and MGC-803. The results showed that compared with the blank control group (miRNA-NC), overexpression of miRNA-339 could significantly inhibit the proliferation capacity of GC cells (Figure 2C,D). The above results indicated that overexpression of miRNA-339 can significantly inhibit the proliferation of GC cells.
Figure 2.
Overexpression of miRNA-339 can significantly inhibit the proliferation of GC cells. (A) Fluorescence qPCR was used to detect the expression of miRNA-339 in GC cell lines and normal gastric fibroblasts (GES-1); (B) fluorescence qPCR was used to detect the effect of transfection of miRNA-339 mimics on the expression of miRNA-339 in GC cells; (C,D) CCK-8 was used to detect the effect of transfection of miRNA-339 mimics on the proliferation of GC cells. *P<0.05, **P<0.01. GC, gastric cancer; miRNA, microRNA; CCK-8, cell counting Kit-8; qPCR, quantitative polymerase chain reaction.
Overexpression of miRNA-339 can inhibit the invasion and migration of GC cells
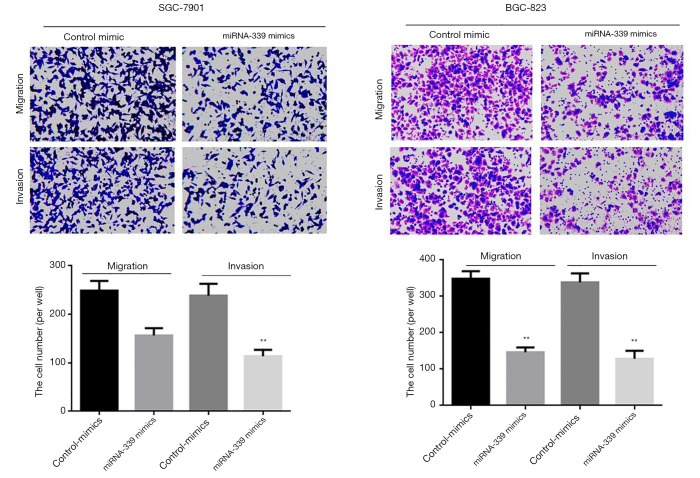
The above analysis shows that the low expression of miRNA-339 is correlated with lymph node metastasis and TNM stage. Therefore, the Transwell experiment was used to further study the effect of miRNA-339 on the invasion and migration of GC cells. First, SGC-7901 and MGC-803 cells were transfected with miRNA-339 mimics, and the cells were inoculated into a Transwell chamber. After 48 h, the cell invasion and migration ability was checked. The results showed that compared with the blank control group (Control mimics), transfection of miRNA-339 mimics can significantly inhibit the invasion and migration of GC cells (Figure 3).
Figure 3.
Transwell experiment detects the effect of overexpression of miRNA-339 on the invasion and migration of GC cells (100×, Crystalline violet staining). *P<0.05, **P<0.01. GC, gastric cancer.
miRNA-339 targets and regulates ZNF689
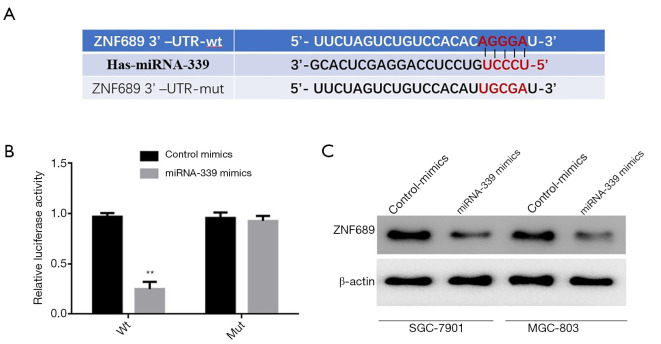
In order to further study the molecular mechanism of miRNA-339 functioning in GC cells, firstly, the binding site of miRNA-339 within the 3'-untranslated region (UTR) of the ZNF689 gene was found by using the bioinformatics online prediction software (TargetScan: http://www.targetscan.org/vert_71/) (Figure 4A). According to the sequence of the 3'-UTR region of the ZNF689 gene, a vector of normal sequence (ZNF689-3'-UTR-wt) and a vector containing mutant bases (ZNF689-3'-UTR-mut) were constructed, respectively, the blank control group (Control mimics) and miRNA-339 mimics were co-transfected into cells, and the luciferase activity was detected. The results showed that miRNA-339 mimics and the normal sequence vector (ZNF689-3'-UTR-wt) can significantly inhibit the cells after transfection and luciferase activity has no significant effect on the mutant (ZNF689-3'-UTR-mut) (Figure 4B). Further, western blot detection showed that compared with the blank control group (Control mimics), the expression level of ZNF689 protein could be significantly inhibited after transfection of miRNA-339 mimics (Figure 4C). The above results showed that miRNA-339 can target the expression of ZNF689 in GC cells.
Figure 4.
miRNA-339 targets and regulates the expression of ZNF689, (A) prediction of the binding site of miRNA-339 and ZNF689 gene 3'-UTR region, (B) luciferase reporter gene detection of miRNA-339 and ZNF689 gene 3'-UTR region binding, (C) western blot to detect the effect of overexpression of miRNA-339 on the expression level of ZNF689. **P<0.01. 3'-UTR, 3'-untranslated region.
miRNA-339 targets and regulates ZNF689 to inhibit the proliferation and invasion of GC cells
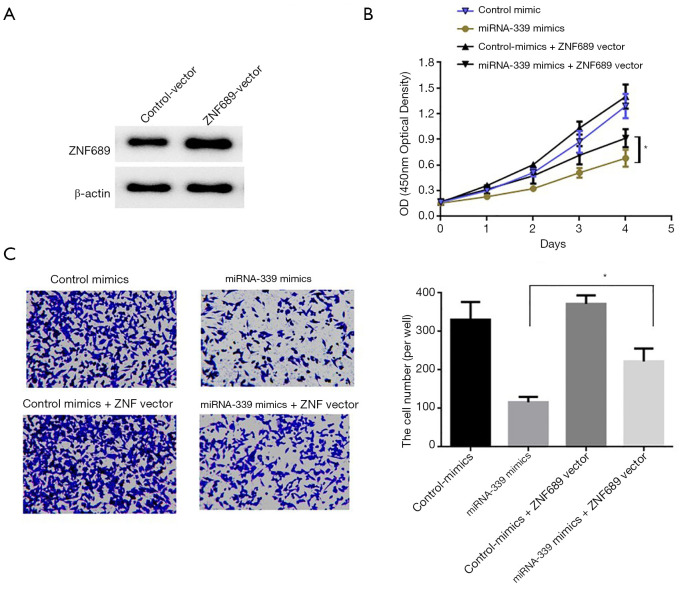
In order to further confirm that miRNA-339 inhibits the invasion ability of GC cells by targeting and regulating the expression of ZNF689, we first constructed an overexpression vector of ZNF689, and then transfected SGC-7901 cells. Western blot was used to detect the overexpression effect of ZNF689. The results showed that compared with the control vector, the expression of ZNF689 in the cells was significantly increased after transfection of the ZNF689 overexpression plasmid (ZNF689-vector) (Figure 5A). In order to further confirm the biological function of miRNA-339 through ZNF689, CCK-8 and Transwell migration test results showed that overexpression of ZNF689 can significantly block the inhibitory ability of overexpression of miRNA-339 on the proliferation and migration of GC cells (Figure 5B,C). The above experimental results indicated that miRNA-339 targets and regulates ZNF689 to inhibit the proliferation and invasion of GC cells.
Figure 5.
Overexpression of ZNF689 can block the inhibitory ability of miRNA-339 on the proliferation and invasion of GC cells. (A) Western blot detection of ZNF689 expression in GC cells after transfection of ZNF689 overexpression vector, (B) CCK-8 detection of the effect of overexpression of ZNF689 in GC cells overexpressing miRNA-339 on cell proliferation, (C) transwell tested the effect of overexpression of ZNF689 in Gc cells overexpressing miRNA-339 on cell migration (100×, Crystalline violet staining). *P<0.05. GC, gastric cancer; CCK-8, cell counting kit-8.
Discussion
As one of the malignant gastrointestinal tumors, GC has a high incidence, morbidity, and mortality rate, making it a serious threat to human health (14). So far, the cause of GC is still unclear, but clinically it is believed that the occurrence of GC is closely related to living and eating habits, genetic susceptibility, and gene expression disorders. Among these causative factors, gene expression disorders are the most important in the occurrence of GC, and 1 such disorder has been the focus of current research (15,16). This study showed that miRNA-339 is under-expressed in GC tissues and is closely related to lymph node metastasis and tumor TNM stage in GC patients. Cell function studies have shown that overexpression of miRNA-339 can significantly inhibit the proliferation, invasion, and migration of GC cells. Further molecular mechanism studies have shown that miRNA-339 regulates the expression of the ZNF689 gene by targeting, and overexpression of ZNF689 can significantly block the inhibitory effect of overexpression of miRNA-339 on the proliferation and migration of GC cells.
The miRNA is a small non-coding RNA that can regulate about 30% of human genes after transcription, and may act as an oncogene or tumor suppressor gene in malignant tumors including GC (17). Studies have shown that abnormal expression of miRNA plays an important role in the early stage of GC and can be used as a candidate biomarker for the early diagnosis of GC (18,19). For example, it was revealed that endogenous miR-558 directly targets the heparanase (HPSE) promoter to attenuate the Smad4-mediated suppression of HPSE expression, thereby promoting the progression of GC (20). In addition, miRNA-194 is abnormally up-regulated in GC. Overexpression of miRNA-194 can promote cell proliferation and migration, while inhibition of miRNA-194 prevents these processes. Molecular mechanism studies have shown that miRNA-194 inhibitors promote the cytoplasmic localization of β-catenin, leading to inhibition of Wnt signal transduction. These findings indicate that miRNA-194, as an oncogene, promotes the proliferation and migration of GC cells by activating Wnt signal transduction (21). In this study, it was found that miRNA-339 is lowly expressed in GC tissues and is closely related to lymph node metastasis and tumor TNM stage in GC patients.
The survival period of patients with low expression of miRNA-339 is significantly shortened, and further cell function experiments showed that overexpression of miRNA-339 can significantly inhibit the proliferation and migration of GC cells. Therefore, it is inferred that miRNA-339 is a tumor suppressor gene in GC, and the low expression of miRNA-339 in GC tissue can significantly promote the occurrence and development of early GC. However, the molecular mechanism of how miRNA-339 exerts its biological functions has remained unclear.
A large number of studies have shown that as non-coding RNA, miRNAs cannot encode proteins but participate in biological functions by regulating the expression of target genes (22,23). For example, miR-146a-5p directly binds to the 3'-UTR of breast cancer cells to down-regulate the expression of interleukin-1 receptor-associated kinase 1 (IRAK1). Restoring the expression of IRAK1 can reverse miR-146a-5p-induced breast cancer. Through the inhibition of cell proliferation and invasion, therefore, there is potential for miR-146a-5p as a new therapeutic target for the treatment of breast cancer (24). In GC, miR-567 is down-regulated in gastric tissue and GC cells. Overexpression of miR-567 significantly inhibits the proliferation and invasion of GC cells. Mechanism studies have shown that miR-567 directly targets PIK3AP1 to induce PI3K/AKT/inactivation of the c-Myc pathway and inhibit the proliferation of GC (25). Therefore, this study used biological information prediction and luciferase report gene experiments to show that miRNA-339 can specifically bind to the 3'-UTR region of ZNF689. Further studies have shown that overexpression of miRNA-339 in GC cells can significantly inhibit the expression of ZNF689. Overexpression of ZNF689 can significantly block the inhibitory effect of overexpression of miRNA-339 on the proliferation and migration of GC cells. Recent studies have shown that ZNF689 is a C2H2 type zinc finger protein (zinc finger proteins, ZNFs), and the abnormal expression of this gene is involved in the occurrence and development of a variety of tumors (26). The expression of ZNF689 is significantly increased in liver cancer tissues, and the positive expression of ZNF689 protein in liver cancer tissues is significantly related to tumor size ≥10 cm, tumor envelope infiltration, and microvascular infiltration, and may be a new predictor of the prognosis of liver cancer patients (27). In addition, knocking out ZNF689 can inhibit the proliferation and migration of liver cancer cells. Further study of its mechanism found that ZNF689 may regulate the epithelial to mesynchymal transition (EMT) process of liver cancer through the Wnt-β-catenin snail signaling pathway (28). However, how ZNF689 regulates the occurrence and development of GC is still unclear. In following research, we will continue to analyze in-depth in vivo and in vitro experiments that show that ZNF689 regulates the proliferation and invasion of GC by affecting those genes.
In summary, this study first clarified that miRNA-339 is under-expressed in GC tissues and is closely related to lymph node metastasis and tumor TNM stage in GC patients. Cell function studies showed that overexpression of miRNA-339 can significantly inhibit the proliferation of GC cells and their invasion and migration capabilities. Further molecular mechanism studies have shown that miRNA-339 regulates expression of the ZNF689 gene by targeting, and overexpression of ZNF689 can significantly block the inhibitory effect of overexpression of miRNA-339 on the proliferation and migration of GC cells. The miRNA miRNA-339 may be a new target for early diagnosis, treatment, or prognosis of patients with GC.
Acknowledgments
Funding: This work was supported by the Anhui Provincial Department of Education Natural Science Key Project (KJ2020A0597).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by First Affiliated Hospital of Wannan Medical College, Yijishan Hospital (YJ-0219-24). The patient or family members had provided written informed consent.
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-994
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-21-994
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-994). The authors have no conflicts of interest to declare.
(English Language Editor: J. Jones)
References
- 1.Marghalani AM, Bin Salman TO, Faqeeh FJ, et al. Gastric carcinoma: Insights into risk factors, methods of diagnosis, possible lines of management, and the role of primary care. J Family Med Prim Care 2020;9:2659-63. 10.4103/jfmpc.jfmpc_527_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machlowska J, Pucułek M, Sitarz M, et al. State of the art for gastric signet ring cell carcinoma: from classification, prognosis, and genomic characteristics to specified treatments. Cancer Manag Res 2019;11:2151-61. 10.2147/CMAR.S188622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Röcken C. Molecular classification of gastric cancer. Expert Rev Mol Diagn 2017;17:293-301. 10.1080/14737159.2017.1286985 [DOI] [PubMed] [Google Scholar]
- 4.Wang JK, Wang Z, Li G. MicroRNA-125 in immunity and cancer. Cancer Lett 2019;454:134-45. 10.1016/j.canlet.2019.04.015 [DOI] [PubMed] [Google Scholar]
- 5.Shen Z, Gu X, Mao W, et al. Characterization of microRNA expression profiles by deep sequencing of small RNA libraries in leukemia patients from Naxi ethnic. Transl Cancer Res 2019;8:160-9. 10.21037/tcr.2019.01.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Z, Sun W, Guo Z, et al. Mechanisms of lncRNA/microRNA interactions in angiogenesis. Life Sci 2020;254:116900. 10.1016/j.lfs.2019.116900 [DOI] [PubMed] [Google Scholar]
- 7.Wang HL, Wang HR, Liang Y, et al. Hsa_circ_0006571 promotes spinal metastasis through sponging microRNA-138 to regulate sirtuin 1 expression in lung adenocarcinoma. Transl Lung Cancer Res 2020;9:2411-27. 10.21037/tlcr-20-1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Z, Zhao S, Wang L, et al. miRNA-339-5p Plays an Important Role in Invasion and Migration of Pancreatic Cancer Cells. Med Sci Monit 2019;25:7509-17. 10.12659/MSM.917038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C, Huang Y, Zhang J, et al. MiRNA-339-5p suppresses the malignant development of gastric cancer via targeting ALKBH1. Exp Mol Pathol 2020;115:104449. 10.1016/j.yexmp.2020.104449 [DOI] [PubMed] [Google Scholar]
- 10.Wang JY, Li H, Ma CM, et al. Acupuncture may exert its therapeutic effect through microRNA-339/Sirt2/NFκB/FOXO1 axis. BioMed Res Int 2015;2015:249013. 10.1155/2015/249013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber CE, Luo C, Hotz-Wagenblatt A, et al. miR-339-3p Is a Tumor Suppressor in Melanoma. Cancer Res 2016;76:3562-71. 10.1158/0008-5472.CAN-15-2932 [DOI] [PubMed] [Google Scholar]
- 12.Zeng H, Zheng J, Wen S, et al. MicroRNA-339 inhibits human hepatocellular carcinoma proliferation and invasion via targeting ZNF689. Drug Des Devel Ther 2019;13:435-45. 10.2147/DDDT.S186352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riazalhosseini B, Mohamed R, Apalasamy YD, et al. Circulating microRNA as a marker for predicting liver disease progression in patients with chronic hepatitis B. Rev Soc Bras Med Trop 2017;50:161-6. 10.1590/0037-8682-0416-2016 [DOI] [PubMed] [Google Scholar]
- 14.Sukri A, Hanafiah A, Mohamad Zin N, et al. Epidemiology and role of Helicobacter pylori virulence factors in gastric cancer carcinogenesis. APMIS 2020;128:150-61. 10.1111/apm.13034 [DOI] [PubMed] [Google Scholar]
- 15.Machlowska J, Baj J, Sitarz M, et al. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int J Mol Sci 2020;21:4012. 10.3390/ijms21114012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molaei F, Forghanifard MM, Fahim Y, et al. Molecular Signaling in Tumorigenesis of Gastric Cancer. Iran Biomed J 2018;22:217-30. 10.29252/ibj.22.4.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu M, Zhu S, Xiong S, et al. MicroRNAs and the PTEN/PI3K/Akt pathway in gastric cancer (Review). Oncol Rep 2019;41:1439-54. 10.3892/or.2019.6962 [DOI] [PubMed] [Google Scholar]
- 18.Hwang J, Min BH, Jang J, et al. MicroRNA Expression Profiles in Gastric Carcinogenesis. Sci Rep 2018;8:14393. 10.1038/s41598-018-32782-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Link A, Kupcinskas J. MicroRNAs as non-invasive diagnostic biomarkers for gastric cancer: Current insights and future perspectives. World J Gastroenterol 2018;24:3313-29. 10.3748/wjg.v24.i30.3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng L, Jiao W, Song H, et al. miRNA-558 promotes gastric cancer progression through attenuating Smad4-mediated repression of heparanase expression. Cell Death Dis 2016;7:e2382. 10.1038/cddis.2016.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng Y, Zhang X, Ma Q, et al. MiRNA-194 activates the Wnt/β-catenin signaling pathway in gastric cancer by targeting the negative Wnt regulator, SUFU. Cancer Lett 2017;385:117-27. 10.1016/j.canlet.2016.10.035 [DOI] [PubMed] [Google Scholar]
- 22.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 23.Vishnoi A, Rani S. MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol Biol 2017;1509:1-10. 10.1007/978-1-4939-6524-3_1 [DOI] [PubMed] [Google Scholar]
- 24.Long JP, Dong LF, Chen FF, et al. miR-146a-5p targets interleukin-1 receptor-associated kinase 1 to inhibit the growth, migration, and invasion of breast cancer cells. Oncol Lett 2019;17:1573-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F, Li K, Yao X, et al. A miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop regulates tumour growth and chemoresistance in gastric cancer. EBioMedicine 2019;44:311-21. 10.1016/j.ebiom.2019.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye Q, Liu J, Xie K. Zinc finger proteins and regulation of the hallmarks of cancer. Histol Histopathol 2019;34:1097-109. [DOI] [PubMed] [Google Scholar]
- 27.Yi PS, Wu B, Deng DW, et al. Positive expression of ZNF689 indicates poor prognosis of hepatocellular carcinoma. Oncol Lett 2018;16:5122-30. 10.3892/ol.2018.9295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shigematsu S, Fukuda S, Nakayama H, et al. ZNF689 suppresses apoptosis of hepatocellular carcinoma cells through the down-regulation of Bcl-2 family members. Exp Cell Res 2011;317:1851-9. 10.1016/j.yexcr.2011.05.012 [DOI] [PubMed] [Google Scholar]