Abstract
Background
PLIN5 is abnormally expressed in many forms of tumors, but its activity and methylation status in human ovarian cancer (OC) have yet to be elucidated.
Methods
RNA sequencing data (RNA-seq) were downloaded from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) database. Differentially expressed genes (DEGs) were identified, and then PLIN5 gene was selected for further study. Expression and methylation levels of PLIN5 were detected by qPCR, western blot, immunohistochemical, and MSP analysis. Moreover, colony formation, transwell, and cell apoptosis assays were employed to explore the abilities of cell proliferation, migration, invasion, and apoptosis, respectively. Furthermore, PLIN5’s function on tumorigenesis was determined by in vivo experiments.
Results
We found that PLIN5 was downregulated in OC tissues by using qPCR, western blot, and immunohistochemical analyses, and MSP also exhibited that PLIN5 was hypermethylated in OC tissues. The expression level of PLIN5 could be restored after treatment with 5-Aza-dC. Furthermore, we found that demethylated PLIN5 could suppress cell proliferation, migration, and invasion of OC, and increase cell apoptosis. Moreover, xenograft experiments showed that demethylated PLIN5 could suppress tumor growth.
Conclusions
Our findings suggest that the expression level of PLIN5 is regulated by methylation, and in OC, PLIN5 can act as a tumor suppressor.
Keywords: Ovarian cancer (OC), PLIN5, methylation, 5-Aza
Introduction
Ovarian cancer (OC), a leading type of malignant tumors in gynecological, and its incidence is increasing globally (1). Despite the progress in surgery, chemotherapy, and radiotherapy, the clinical prognosis of OC remains unsatisfactory, and most OC patients ultimately die of recurrence and metastasis (2). Hence, it is necessary to reveal the potential molecular mechanisms of OC occurrence and metastasis to identify new epigenetic biomarkers for OC therapy.
Both epigenetic and genetic aberrant changes are seen as essential to carcinogenesis (3). DNA methylation is the most characterized epigenetic modification. The methylation of CpG island leads to the inactivation of tumor suppressor gene transcription, which has become an important part of cancer epigenetic research (4). Therefore, investigation on the changes in DNA methylation in cancer can help to better elucidate the mechanisms of tumor development.
In this study, we used the TCGA and GTEx datasets to screen out PLIN5 as a hub gene and as a new molecular target in OC. Perilipin 5 (PLIN5) is a vital member of the perilipin family, and is especially prominent in oxidative tissues, including in heart, brown adipose, skeletal muscle, and liver tissues (5). PLIN5 is considered to mediate mitochondrial function and to control the concentration and release of fatty acids to sustain lipid droplet homeostasis (6). Moreover, we explored the expression and degree of methylation of PLIN5 in OC tissues and analyzed its effects on the proliferation and metastasis of OC cells.
Methods
Gene expression data
RNA sequencing data (RNA-seq) of the OC samples was downloaded from TCGA (7), and RNA-seq of the normal ovarian specimen was collected from the GTEx project (8). The edgeR package was employed to screen out the DEGs with |logFC| >1 and false discovery rate (FDR) <0.05 (9). In all transcriptomic data, we used only protein-related genes. Those protein genes with logFC >1 and FDR <0.05 were deemed as upregulated, and those genes with logFC <−1 and FDR <0.05 were deemed as downregulated.
Function enrichment analyses
Gene Ontology (GO) (10) and KEGG pathway (11) enrichment analyses were applied by the DAVID (12). Besides, GO terms comprised the biological process (BP), cellular component (CC), and molecular function (MF). FDR <0.05 were considered as significant.
Clinical samples and cell culture
This study examined all patients with OC who underwent surgery Affiliated Hospital of Nantong University. And none of the patients received any treatment before the operation, and all patients had given their informed written consent. Fresh tissue samples were immediately stored at −80 °C. Moreover, the study protocol has been approved by the Affiliated Hospital of Nantong University.
The SKOV3, HO8910, OV90, and HOSE cell lines were cultured at 37 °C and 5% CO2 in DMEM or RPMI-1640 medium (Gibco) with 10% fetal bovine serum (FBS) (Invitrogen). Transfection was applied by using liposome Lipofectamine 3000 (Invitrogen). Transfected cells were harvested and used for further experimental research.
Quantitative real-time polymerase chain reaction (qPCR)
Total RNA was extracted by TRIzol (Invitrogen). Then, cDNA was reversely transcribed with a SuperScript First-Strand Synthesis System (Invitrogen). qPCR reactions were applied by using the SYBR Green PCR Master Mix (Roche). Primers: PLIN5 forward, 5'-TGCCCATGACGGAGGAAGA-3', PLIN5 reverse, 5'-GAGCCGAGGCGCACAAA-3'. The relative expression levels were normalized by the Ct value of GAPDH using a 2−ΔΔCt method.
Western blot analysis
The lysates were extracted, and total protein was isolated by 10% SDS/PAGE gels and transferred to nitrocellulose membranes. The membranes were blocked in 5% fat-free milk for 1 h at room temperature, and then incubated with the primary antibodies overnight at 4 °C: rabbit polyclonal anti-PLIN5 (Abcam; 1:1,000; cat. no. ab222811), and mouse monoclonal anti-β-actin (1:2,500). After washing three times with Tris-buffered saline (TBS) containing 0.1% Tween-20, the membrane was incubated secondary antibody, anti-rabbit IgG conjugated IRDye800 (1:5,000, Rockland Gilbertsville, CA) for 2 h at room temperature. At last, the protein bands were tested using the ECL Plus kit (ZSbio).
Immunohistochemical staining
Paraffin-embedded tissues were analyzed by using tissue microarray (TMA) analysis. Tissue sections were dewaxed and rehydrated in graded ethanol. Then, antigen retrieval was achieved by boiling sections in EDTA buffer (pH 6.0) for 3 min. The endogenous peroxidase activity was quenched with 3% hydrogen peroxide for 30 min, and the TMA slides were stained overnight with a rabbit polyclonal anti-PLIN5 antibody (Abcam) at 4 °C The goat anti-rabbit secondary antibody (Sigma) combined with horseradish peroxidase was incubated at 37 °C for 30 min. Then, slides were treated with horseradish peroxidase and 3,3-diaminobenzidine chromogen solution, and then counterstained with hematoxylin. The semiquantitative scoring system was based on both the staining intensity and the percentage of cells at that intensity. According to the pathological score, the staining intensity was divided into 0 (−, no staining), 1 (+, weak staining), 2 (++, moderate staining), or 3 (+++, intense staining). The percentage of stained cells was evaluated and multiplied by the intensity score to generate an intensity percentage score. Finally, the staining score was the sum of the four intensity percentage scores, ranging from 0 (no staining) to 300 (100% of cells with +++ staining intensity).
Methylation-specific PCR (MSP)
Genomic DNA was extracted by using a DNeasy tissue kit (Qiagen). The sodium bisulfite reaction was applied to 5 µg genomic DNA using the EpiTect Bisulfite Kit (Qiagen). The primers of MSP were as follows: (M): forward, 5'-GATGATAGGTGGTTAAATTTGGTTC-3' and reverse, 5'-TTAAACTAACCCAATACCCTACGAT-3'; and unmethylated-specific primers (U): forward, 5'-ATGATAGGTGGTTAAATTTGGTTTG-3' and reverse, 5'-CCTTAAACTAACCCAATACCCTACAA-3'.
Demethylation with 5-aza-2'-deoxycytidine (5-Aza-dC) treatment
Cells were seeded into six-well plates at a concentration of 1×106 cells per well and cultured at room temperature for 24 h. Next, cells were processed by using 1 µM of 5-Aza-dC (13) (Sigma) for 72 h. And the culture medium was changed every day.
Colony formation assay
Cells were seeded into six-well plates in complete medium and cultured for 2 weeks. Cells were fixed with 100% methanol for 10 min and stained with 0.5% crystal violet for 20 min at room temperature, and the number of colonies (>50 cells) was calculated under an optical microscope.
Cell apoptosis
Briefly, after 72 h of transfection, cells were harvested and resuspended in the Annexin-binding buffer at a concentration of 1×106 cells/mL, then stained in the dark with Annexin V-FITC and PI for 15 min at room temperature, and then analyzed with the FACSCalibur flow cytometer.
Transwell assay
For the migration assays, cells (1×105 cells/well) were added to the upper Transwell chambers (Corning, 8-µm pore size). Cells were cultured in serum-free medium, and 10% FBS serum medium was used as the chemical attractant in the lower chambers. For the invasion assays, cells were plated on the upper side of the chambers with Matrigel (BD Biosciences) and incubated in the medium. A medium containing 10% FBS was injected into the lower chamber to attract cells. After incubation in a humidified atmosphere with 5% CO2 at room temperature for 24 h, cells that did not migrate or invade to the pores with cotton wool were removed. Cells on the lower membrane were fixed with 100% methanol for 10 min and stained with 0.2% crystal violet for 20 min at room temperature. Finally, images of the inserts were taken under an inverted microscope (Olympus), and the migrated and invaded cells were calculated in five randomly fields.
Xenograft experiments
Firstly, BALB/c nude mice were applied for tumorigenesis experiments. Then, the mice were subcutaneously injected or inoculated with 1×106 cells/100 µL per side, 7 per group). Tumor length (L) and width (W) were assessed weekly. The calculation formula of tumor volume was as follows: tumor volume = width2 × length/2. After about 4 weeks, the mice were sacrificed and the peritoneal metastatic nodules were measured.
Statistical analysis
All experiments were performed in triplicate. SPSS 21.0 statistical software (SPSS, IL, USA) was performed for the statistical analysis. A paired t-test was employed to analyze the statistical significance of the differences between the two groups. A one-way analysis of variance (ANOVA) test was employed to analyze the statistical significance of the differences between the three groups. P<0.05 was determined as statistically significant.
Results
Identification of DEGs in OC and enrichment analyses
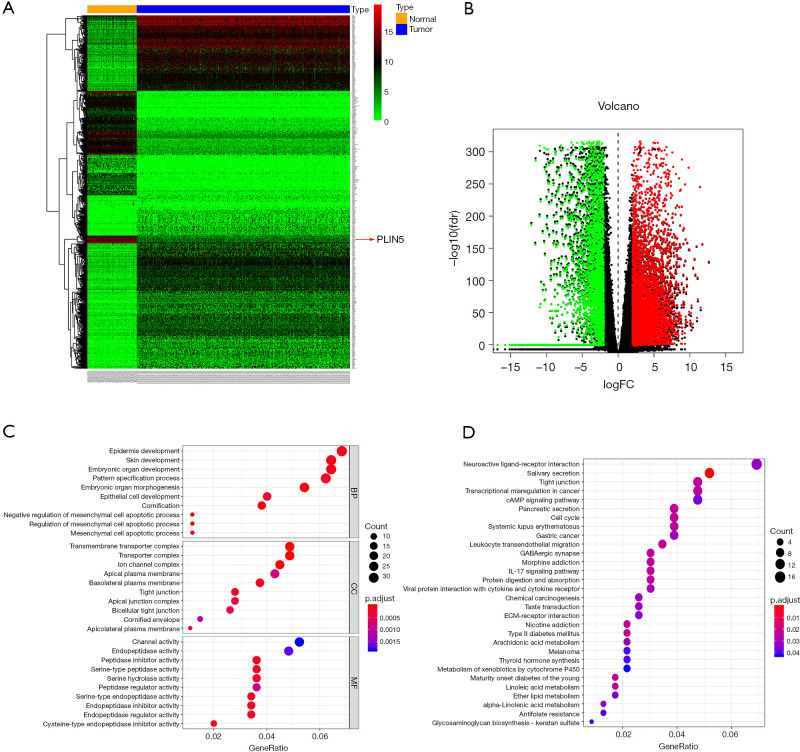
To identify the expression pattern of genes, we screened DEGs from the RNA-Seq data of the OC cases in TCGA and normal cases from the GTEx database. The heatmaps exhibited the upregulated and downregulated DEGs in datasets (Figure 1A). Also, the volcano plot showed 634 DEGs, including 446 upregulated genes and 188 downregulated genes (Figure 1B). To further investigate the biological effects of these DEGs in OC, we completed an enrichment analysis. The results of GO analysis revealed that these DEGs were most dramatically enriched in cornification (BP), transmembrane transporter complex (CC), and peptidase inhibitor activity (MF). Moreover, KEGG pathway analysis presented that these DEGs were primarily enriched in salivary secretion, pancreatic secretion, among other processes (Figure 1C,D).
Figure 1.
Hierarchical cluster analysis and functional enrichment analysis. (A,B) Heatmaps and volcano plot of DEGs between OC and normal tissues in TCGA and GTEx datasets; (C,D) GO and KEGG pathway analysis of the DEGs in OC. DEGs, differentially expressed genes; OC, ovarian cancer; TCGA, The Cancer Genome Atlas; GTEx, Genotype-Tissue Expression; GO, Gene Ontology.
Expression and methylation levels of PLIN5 in OC tissues and cells
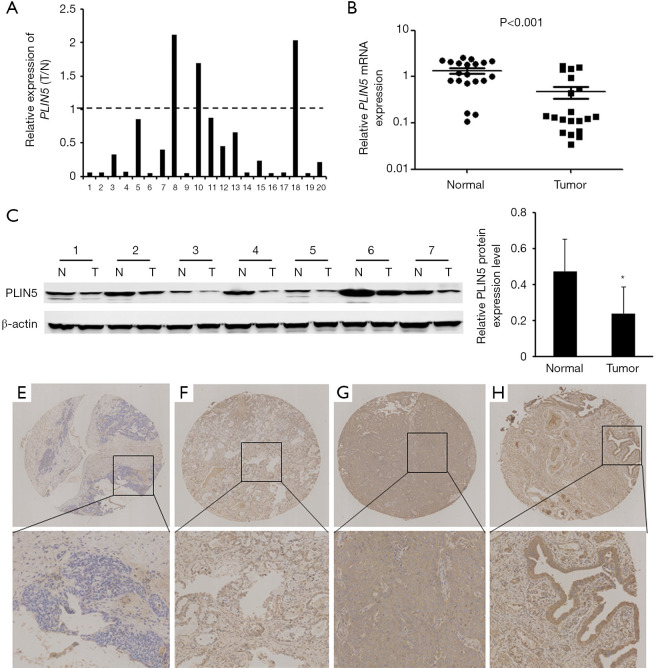
The levels of PLIN5 expression was examined with qPCR analysis, which indicated that PLIN5 expression were markedly lower in OC tissues compared to the normal tissues (Figure 2A,B). The results were confirmed to examine the PLIN5 protein level by western blot in 20 random pairs of OC and matched adjacent normal tissues. Then, the representative results of the western blot in 7 cases were exhibited in Figure 2C. The above results demonstrated that the PLIN5 protein level was reduced in the tumor samples compared with matched adjacent normal tissues (Figure 2D; P<0.05). These above data indicated that PLIN5 was downregulated in OC tissues. Furthermore, TMA-based immunohistochemical analysis was performed in 57 OC tissues and 12 normal ovarian tissues. The representative results were shown in Figure 2E,F,G,H. The positive staining was mainly localized in the cytoplasm of tumor cells. PLIN5 showed strongly positive staining in 12 normal ovarian tissues, and 57 OC tissues were positively stained for PLIN5, 32 cases showed low expression of PLIN5, and 25 cases showed high expressions of PLIN5. Moreover, the correlation between the expression of PLIN5 and the clinicopathologic factors of these OC patients are summarized in Table 1. The data showed that low PLIN5 expression was observably related to distant metastasis (P=0.010).
Figure 2.
Expression levels of PLIN5 in ovarian cancer. (A) qPCR analysis indicated that PLIN5 expression was downregulated in 20 OC tissues. (B) The downregulation of PLIN5 was statistically significant compared to normal tissues. (C) Western blot analysis of representative paired samples of OC (T) and their matched adjacent normal tissues (N). (D) Relative average PLIN5 protein expression levels were significantly downregulated in OC tissues compared with the corresponding adjacent normal tissues (*, P<0.05). Representative patterns of PLIN5 expression in ovarian tissues were present by immunohistochemical analysis. (E) OC samples weakly positively stained with a staining index marked as +. (F) OC samples moderately positively stained with a staining index marked as ++. (G) OC samples strongly positively stained with a staining index marked as +++. (H) Normal ovarian samples strongly positively stained with a staining index marked as +++. Original magnification, ×40 or ×200. OC, ovarian cancer.
Table 1. Correlation of clinicopathologic characteristics with PLIN5 expression.
| Characteristics | Total (n=57) | PLIN5 expression | P value | |
|---|---|---|---|---|
| Low (n=32) | High (n=25) | |||
| Age (years) | 0.366 | |||
| ≤50 | 22 | 14 | 8 | |
| >50 | 35 | 18 | 17 | |
| Tumor size (cm) | 0.492 | |||
| ≤5 | 20 | 10 | 10 | |
| >5 | 37 | 22 | 15 | |
| Location | 0.845 | |||
| Right | 13 | 8 | 5 | |
| Left | 12 | 6 | 6 | |
| Bilateral | 32 | 18 | 14 | |
| Histology | 0.130 | |||
| Serous adenocarcinoma | 40 | 19 | 21 | |
| Clear cell carcinoma | 8 | 6 | 2 | |
| Mucinous adenocarcinoma | 9 | 7 | 2 | |
| FIGO stage | 0.376 | |||
| I | 8 | 6 | 2 | |
| II | 13 | 8 | 5 | |
| III | 32 | 15 | 17 | |
| IV | 4 | 3 | 1 | |
| Lymph node metastasis | 0.446 | |||
| Yes | 5 | 2 | 3 | |
| No | 52 | 30 | 22 | |
| Distant metastasis | 0.010* | |||
| M0 | 14 | 12 | 2 | |
| M1 | 43 | 20 | 23 | |
*, P<0.05 was considered significant.
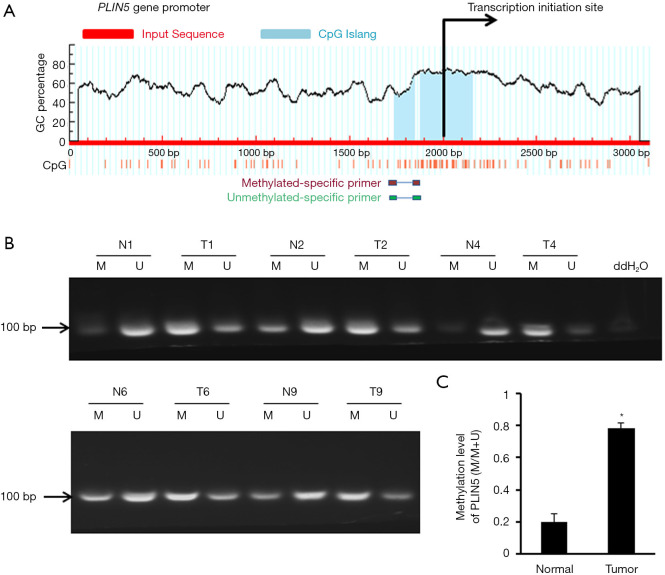
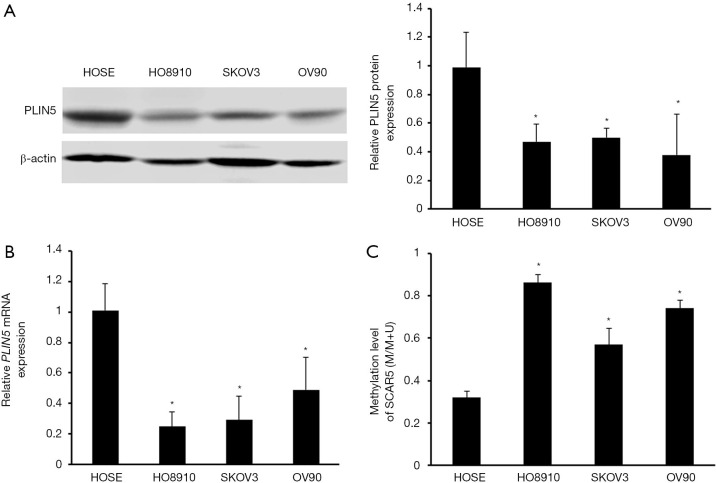
Previous studies have shown that the methylation status of the CpG islands may closely correlated with the regulation of gene expression (14). Therefore, we used MethPrimer software to search for the status of CpG islands in the promoter region of PLIN5 and found significant CpG islands (Figure 3A). MSP analysis was employed to detect the methylation status of PLIN5 in OC and normal tissues. And the methylation levels of PLIN5 in cancer tissues were distinctly increased compared with normal tissues (Figure 3B,C). Next, PLIN5 expression in SKOV3, HO8910, and OV90 cell lines was measured by western blot and qPCR analyses. The results exhibited that the protein and mRNA levels of PLIN5 were decreased in OC cell lines, compared with HOSE cell lines (Figure 4A,B). Furthermore, the methylation levels of PLIN5 in HO8910, SKOV3, and OV90 were elevated compared with that of the HOSE cell line (Figure 4C, P<0.05).
Figure 3.
PLIN5 expression is regulated by promoter region methylation in ovarian cancer. (A) MethPrimer analysis showed that the scheme of the location of the CpG islands is located in the region of the PLIN5 gene promoter; (B) representative MSP results indicated the PLIN5 degree of methylation in normal tissues and tumor tissues; (C) compared with that of the normal tissues, the PLIN5 methylation degree in tumor tissues was increased. *, P<0.05.
Figure 4.
Expression and methylation status of PLIN5 in ovarian cancer cells. (A) The protein expression levels of PLIN5 were detected by western blot in OC cell lines. Western blot analysis indicated the PLIN5 protein expression levels in SKOV3, OV90, and HO8910 cell lines compared with the HOSE cell line. *, P<0.05. (B) qPCR analysis indicated the PLIN5 mRNA expression levels in SKOV3, OV90, and HO8910 cell lines compared with the HOSE cell line. *, P<0.05. (C) The methylation levels of PLIN5 in HO8910, SKOV3, and OV90 were dramatically increased compared with that of the HOSE cell line. *, P<0.05. OC, ovarian cancer.
Restored PLIN5 expression inhibited the progression of OC cells
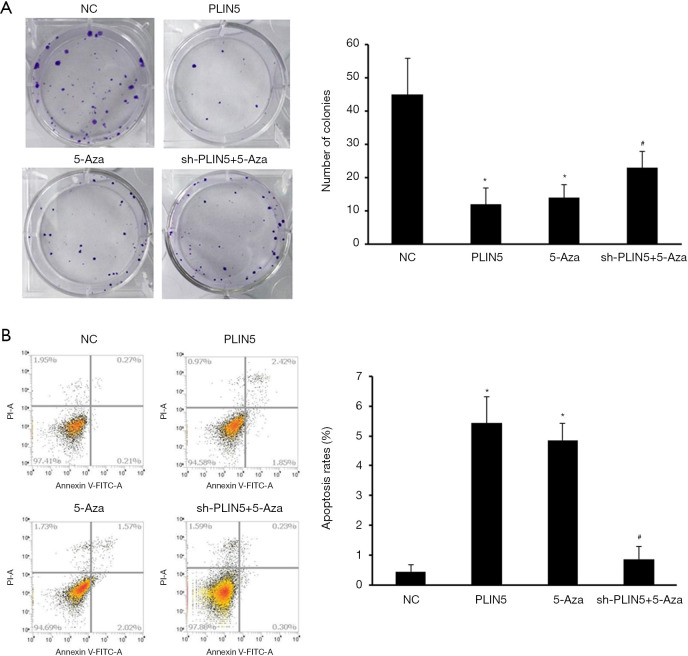
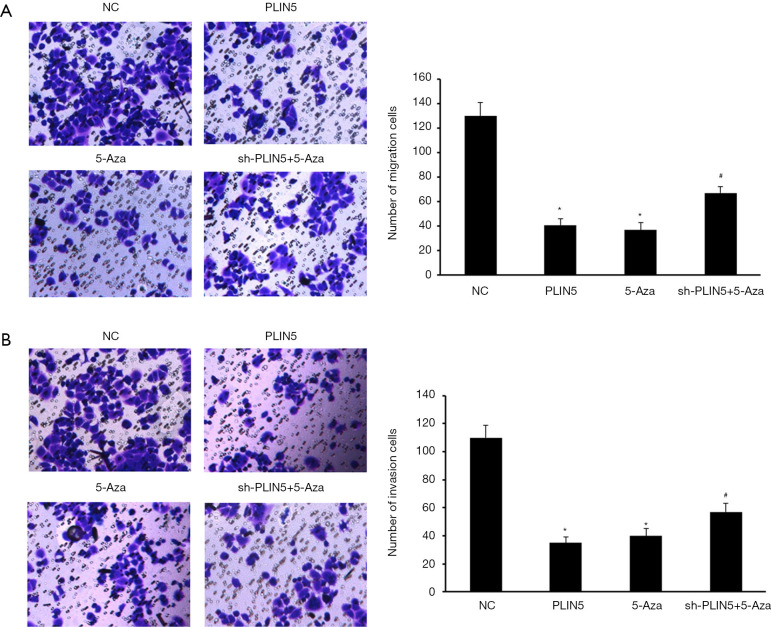
Firstly, the number of clones was remarkably lower in the PLIN5 overexpression group and the 5-Aza group than in the NC and sh-PLIN5 + 5-Aza groups by using colony formation assay (Figure 5A). The apoptosis rate of the OV90 cells transfected with PLIN5 or 5-Aza was dramatically increased compared with the NC and sh-PLIN5 + 5-Aza groups (Figure 5B, P<0.05). In addition, transwell assays exhibited that the ability of cell migration and invasion decreased significantly in the PLIN5 and 5-Aza groups relative to the NC and sh-PLIN5 + 5-Aza groups (Figure 6A,B).
Figure 5.
Restored expression of PLIN5 inhibits cell proliferation. (A) The number of clones was decreased in the PLIN5-overexpressing group and the 5-Aza group compared with the NC group. *, P<0.05, compared with the NC group; #, P<0.05, compared with the 5-Aza group. Original magnification, ×100. (B) The apoptosis rate of SKOV3 cells transfected with PLIN5 or 5-Aza dramatically increased compared with the NC group. *, P<0.05, compared with the NC group; #, P<0.05, compared with the 5-Aza group.
Figure 6.
Restored expression of PLIN5 inhibits cell migration and invasion. (A,B) Transwell assays revealed that the abilities of cell migration and invasion were remarkedly reduced in the PLIN5 and 5-Aza groups compared to the control group. *, P<0.05, compared with the NC group; #, P<0.05, compared with the 5-Aza group. Representative photos of stained cells are shown with the original magnification of ×100.
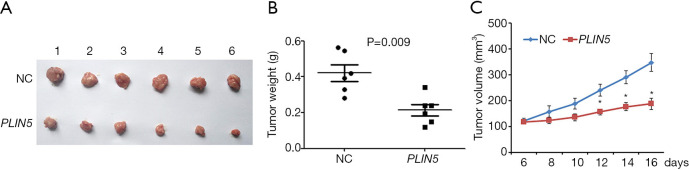
To further assess the effects of PLIN5 on the proliferation of OC cells in vivo, we observed tumor weight and volume after transfected OV90 cells with PLIN5 overexpression plasmid. The results showed that tumor weight and volume in the PLIN5 group were lower than those in the NC group (Figure 7, P<0.05). All the above results suggested that PLIN5 may serve as a tumor suppressor gene, which could suppress cell proliferation, migration, and invasion of OC while promoting apoptosis.
Figure 7.
Overexpression of PLIN5 in vivo inhibits tumor growth. (A,B,C) Tumor xenograft results showed that tumor weight and volume were reduced in the PLIN5 group compared with the control group. *, P<0.05.
Discussion
Nowadays, the poor prognosis of OC patients is attributed primarily to the challenging diagnosis and likelihood of early metastasis. These two factors highlight the importance of revealing the molecular mechanisms of an OC metastasis. To improve the prognosis of OC patients, new biomarkers and therapeutic targets are urgently needed (15). In the current study, the DEGs in OC patients were analyzed by using data from the TCGA and GTEx databases, and we found that PLIN5 was significantly downregulated in the tumor samples. Moreover, we identified a significant CpG island in the promoter region of PLIN5. Therefore, PLIN5 was selected for further study.
PLIN5 is reported to be specifically expressed in fatty acid oxidized tissues, such as adipose, heart, liver, and muscle tissue (16). The preliminary research in PLIN5 focused on its function in the heart because of its high oxidative capacity, and most of these studies have pointed out the protective effect of PLIN5 against hepatic lipotoxicity in the liver (17). Interestingly, PLIN5 overexpression was found to enhance cellular lipid droplet accumulation, while knockdown of PLIN5 promoted fatty acid oxidation metabolism (18). Another recent study has reported that PLIN5 exists in renal cell carcinoma, rhabdomyosarcoma, liposarcoma (16), and hepatocellular carcinoma (19). Thus, we believe that PLIN5 could regulate reactive oxygen species (ROS) levels, and lipid content and lipolysis of cells, which could affect the development of a tumor. Furthermore, the down-regulation of PLIN5 may induce tumorigenesis (20). In this study, to further verify the PLIN5 expression level and methylation degree, western blot, qPCR, immunohistochemical and MSP analysis were employed and showed that PLIN5 was hypermethylated and PLIN5 expression was reduced in OC tissues.
Our study also explored the biological functions of the PLIN5 gene in the genesis and development of OC. The results of the experiments revealed that PLIN5 overexpression could dramatically suppress cell proliferation, migration, and invasion, and promote apoptosis in vitro. Additionally, it is suggested that the changes in DNA methylation may be of significance to the diagnosis, prognosis, and treatment of OC (21). Studies have shown that DNA hypermethylation silences the genes needed for tumor development (22). Moreover, DNA methylation is a reversible epigenetic modification. 5-Aza, a DNA methyltransferase inhibitor, can reverse the hypermethylation in the promoter region, re-express tumor-related genes, and inhibit tumor cell growth (23). Therefore, we added the 5-Aza + sh-PLIN5 treatment group to further exhibit a significant effect on cell proliferation, apoptosis, migration, and invasion of OC after DNA methylation inhibition was significantly improved compared with the PLIN5 overexpression and 5-Aza treatment groups.
Conclusions
To conclude, we found that the promoter region methylation regulates the expression of PLIN5 in OC. Also, we showed that PLIN5 expression level was downregulated in OC, and methylation of PLIN5 is associated with OC metastasis. At last, it is suggested that the PLIN5 may be a potential therapeutic target for OC.
Acknowledgments
Funding: This work was supported by grants from the Nantong Science and Technology Project (MS32017009).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of the Affiliated Hospital of Nantong University (2013-066). Informed consent was provided by all participants. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1221). The authors have no conflicts of interest to declare.
References
- 1.Jayson GC. Ovarian cancer. Lancet 2014;384:1376-88. 10.1016/S0140-6736(13)62146-7 [DOI] [PubMed] [Google Scholar]
- 2.Karnezis AN, Cho KR, Gilks CB, et al. The disparate origins of ovarian cancers: pathogenesis and prevention strategies. Nat Rev Cancer 2017;17:65-74. 10.1038/nrc.2016.113 [DOI] [PubMed] [Google Scholar]
- 3.Kelly AD, Issa JPJ. The promise of epigenetic therapy: reprogramming the cancer epigenome. Curr Opin Genet Dev 2017;42:68-77. 10.1016/j.gde.2017.03.015 [DOI] [PubMed] [Google Scholar]
- 4.Carrió E, Suelves M. DNA methylation dynamics in muscle development and disease. Front Aging Neurosci 2015;7:19. 10.3389/fnagi.2015.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalen KT, Dahl T, Holter E, et al. LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochim Biophys Acta 2007;1771:210-27. 10.1016/j.bbalip.2006.11.011 [DOI] [PubMed] [Google Scholar]
- 6.Mason RR, Watt MJ. Unraveling the roles of PLIN5: linking cell biology to physiology. Trends Endocrinol Metab 2015;26:144-52. 10.1016/j.tem.2015.01.005 [DOI] [PubMed] [Google Scholar]
- 7.Zhu Y, Qiu P, Ji Y. TCGA-Assembler: open-source software for retrieving and processing TCGA data. Nat Methods 2014;11:599-600. 10.1038/nmeth.2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.GTEx Consortium . The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580-5. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun J, Nishiyama T, Shimizu K, et al. TCC: an R package for comparing tag count data with r obust normalization strategies. BMC Bioinformatics 2013;14:219. 10.1186/1471-2105-14-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W, Liu J, Rajapakse JC. Gene Ontology Enrichment Improves Performances of Functional Similarity of Genes. Sci Rep 2018;14;8:12100. [DOI] [PMC free article] [PubMed]
- 11.Altman T, Travers M, Kothari A, et al. A systematic comparison of the MetaCyc and KEGG pathway databases. BMC Bioinformatics 2013;14:112. 10.1186/1471-2105-14-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis G, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, visualization, and Integrated Discovery. Genome Biol 2003;4:P3. 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- 13.Xing XQ, Li B, Xu SL, et al. 5-Aza-2'-deoxycytidine, a DNA methylation inhibitor, attenuates hypoxic pulmonary hypertension via demethylation of the PTEN promoter. Eur J Pharmacol 2019;855:227-34. 10.1016/j.ejphar.2019.05.021 [DOI] [PubMed] [Google Scholar]
- 14.Gao D, Herman JG, Guo M. The clinical value of aberrant epigenetic changes of DNA damage repair genes in human cancer. Oncotarget 2016;7:37331-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van den Brand D, Mertens V, Massuger LFAG, et al. siRNA in ovarian cancer - Delivery strategies and targets for therapy. J Control Release 2018;283:45-58. 10.1016/j.jconrel.2018.05.012 [DOI] [PubMed] [Google Scholar]
- 16.Hashani M, Witzel HR, Pawella LM, et al. Widespread expression of perilipin 5 in normal human tissues and in diseases is restricted to distinct lipid droplet subpopulations. Cell Tissue Res 2018;374:121-36. 10.1007/s00441-018-2845-7 [DOI] [PubMed] [Google Scholar]
- 17.Wang C, Zhao Y, Gao X, et al. Perilipin 5 improves hepatic lipotoxicity by inhibiting lipolysis. Hepatology 2015;61:870-82. 10.1002/hep.27409 [DOI] [PubMed] [Google Scholar]
- 18.Li H, Song Y, Zhang LJ, et al. LSDP5 Enhances Triglyceride Storage in Hepatocytes by Influencing Lipolysis and Fatty Acid β-Oxidation of Lipid Droplets. PLoS One 2012;7:e36712. 10.1371/journal.pone.0036712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Y, Zhang X, Zhang L, et al. Perilipin5 protects against lipotoxicity and alleviates endoplasmic reticulum stress in pancreatic β-cells. Nutr Metab (Lond) 2019;16:50. 10.1186/s12986-019-0375-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan Y, Jin Y, Wang Q, et al. Perilipin 5 Protects against Cellular Oxidative Stress by Enhancing Mitochondrial Function in HepG2 Cells. Cells 2019. doi: . 10.3390/cells8101241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen P, Huhtinen K, Kaipio K, et al. Identification of Prognostic Groups in High-Grade Serous Ovarian Cancer Treated with Platinum-Taxane Chemotherapy. Cancer Res 2015;75:2987-98. 10.1158/0008-5472.CAN-14-3242 [DOI] [PubMed] [Google Scholar]
- 22.Lee ST, Wiemels JL. Genome-wide CpG island methylation and intergenic demethylation propensities vary among different tumor sites. Nucleic Acids Res 2016;44:1105-17. 10.1093/nar/gkv1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seelan RS, Mukhopadhyay P, Pisano MM, Greene RM. Effects of 5-Aza-2'-deoxycytidine (decitabine) on gene expression. Drug Metab Rev 2018;50:193-207. 10.1080/03602532.2018.1437446 [DOI] [PubMed] [Google Scholar]