Abstract
Background
Non-small cell lung cancer (NSCLC) is a huge threat to sufferers’ life and overall health. Long non-coding RNA (lncRNA) distal-less homeobox 6 antisense RNA 1 (DLX6-AS1) has been revealed to function as a carcinogenesis factor in some cancers. This research aimed to scrutinize the role and mechanism underlying DLX6-AS1 in NSCLC tumorigenesis and progression.
Methods
The levels of DLX6-AS1, microRNA-16-5p (miR-16-5p), and BMI1 mRNA were estimated via reverse transcription-quantitative PCR (RT-qPCR) assay. The protein levels were disclosed by western blot assay. Cell proliferative potential was estimated by colony formation and Cell Counting Kit-8 (CCK-8) assays. Cell migration was estimated by Transwell and wound healing assay. A Transwell assay was executed to estimate cell invasion. The relationships of DLX6-AS1, miR-16-5p, and BMI1 were forecasted by bioinformatics analysis, and confirmed by luciferase reporter assay and RNA immunoprecipitation (RIP) assay. A xenograft mice model was employed to to inspect the function of DLX6-AS1 knockdown on NSCLC tumorigenesis in vivo.
Results
DLX6-AS1 was overexpressed in NSCLC tissues and cells, and was inextricably linked with the poor prognosis of NSCLC patients. Depletion of DLX6-AS1 oppressed cell proliferation, migration, invasion, epithelial-mesenchymal transition (EMT) but promoted apoptosis in NSCLC. MiR-16-5p is a target of DLX6-AS1 and directly targets BMI1. Moreover, the anti-tumor impacts of miR-16-5p were overturned by overexpression of DLX6-AS1 or BMI1 in NSCLC cells. Additionally, DLX6-AS1 silencing inhibited tumor growth of NSCLC in vivo.
Conclusions
In conclusion, lncRNA DLX6-AS1 downregulation suppressed the tumorigenesis and progression of NSCLC via miR-16-5p/BMI1 axis in vitro and in vivo, elucidating the vital roles and downstream targets of DLX6-AS1 in NSCLC.
Keywords: Long non-coding RNA distal-less homeobox 6 antisense RNA 1 (lncRNA DLX6-AS1), microRNA-16-5p (miR-16-5p), BMI1, non-small cell lung cancer (NSCLC)
Introduction
Lung cancer has been categorized into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) by WHO (1), which is the most frequent malignancy and the major contributors of cancer deaths, accounting for about 11.6% of the total cancer cases and 18.4% of all deaths globally in 2018 (2). NSCLC, the most common type of lung cancer, accounts for more than 85% of all lung cancer cases (3,4). NSCLC is currently classified according to its pathological characteristics, with squamous cell carcinoma and adenocarcinoma as the two major NSCLC subtypes (5). The 60-month survival rate for all stages of lung cancer patients in the USA is approximately 18% and declines to 5% for those with metastatic cancer. Unfortunately, NSCLC patients are often not diagnosed until the advanced stages (3,6). Therefore, a better understanding of the molecular pathogenesis pertinent to the tumorigenesis and progression of NSCLC might contribute to discovering biomarkers and more efficacious therapeutic strategies.
Non-coding RNAs, including long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), are a class of transcripts have no or limited protein-coding potential (7,8). LncRNA have been suggested to exert an important role in multiple pathophysiological processes, such as chromatin modification, gene transcription, splicing, translation, cell cycle regulation and cell differentiation (9,10). Numerous biological processes precisely regulated the carcinogenesis and progression of cancers, which often involve silencing tumour-suppressor genes or activating oncogenes (11). The aberrant versions of lncRNAs have been confirmed in a sort of tumors, indcluding NSCLC (12). In addition, several lncRNAs were disclosed to function as onco-lncRNAs and tumour-suppressor lncRNAs in NSCLC (13-16). Therefore, an in-depth knowledge of the regulatory mechanism underlying abnormally expressed lncRNA is of great importance for the the discovery of new and promising therapeutic targets for NSCLC.
Several research has disclosed the carcinogenic effect of lncRNA distal-less homeobox 6 antisense RNA 1 (DLX6-AS1) in several forms of cancers. For instance, DLX6-AS1 silence attenuated hepatocellular cancer (HCC) cell proliferative, migratory, and invasive capacities and impeded HCC tumor growth through modulating the microRNA-203/matrix metalloproteinase-2 pathway (17). Additionally, DLX6-AS1 deficiency resulted in reduced cell proliferative ability and increased apoptotic cell rate in vitro together with the inhibition of tumor growth in vivo via the regulation of microRNA-26a/phosphatase and tensin homolog axis in renal cell cancer (18). Moreover, DLX6-AS1 expression was prominently elevated in lung adenocarcinoma (a type of NSCLC) tissues in contrast with adjacent normal tissues, and associated the differentiation and TNM stage of lung adenocarcinoma (19), indicating that DLX6-AS1 might be implicated in the tumorigenesis and progression of NSCLC.
MicroRNA-16 (miR-16), located at chromosome 13q14, is a tumor-inhibiting factor in multiple cancers (20,21). For example, miR-16-5p was found to repress proliferation, metastasis and epithelial-mesenchymal transition (EMT) and induce apoptosis by targeting SRY-box 5 in pituitary tumor cells (22). In addition, miR-16-5p overexpression resulted in decreased proliferation and invasive potential of colorectal cancer (CRC) cells, but elevated cell apoptosis, and reduced CRC tumor growth in vivo by targeting KRAS (23). Moreover, multivariate logistic regression analysis revealed that miR-16-5p could function as an independent diagnostic and prognostic factor in NSCLC (24,25). Additionally, previous documents reported that miR-16 could inhibit the development and progression of NSCLC (26-28).
Bioinformatic analysis disclosed the binding fragments of miR-16-5p in DLX6-AS1. However, whether DLX6-AS1 mediate the tumorigenesis and progression of NSCLC by miR-16-5p is still unknown. In current research, we aimed to scrutinize the effects of DLX6-AS1 in regulating NSCLC progression, including proliferation, migration, invasion, EMT, and in vivo tumor growth. And the regulatory mechanism of DLX6-AS1 in regulating NSCLC progression was examined. Our results first disclosed that DLX6-AS1 controls the tumorigenesis and progression of NSCLC by miR-16-5p/BMI1 axis, which deepens our comprehension on the pathogenesis of NSCLC and provides theoretical support for using DLX6-AS1 as a therapeutic target for NSCLC.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-1240).
Methods
Ethical statement
All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Haikou People’s Hospital and informed consent was taken from all the patients.
All animal experiments were fulfilled in an SPF feeding environment following the National Institutes of Health guidelines and approved by the Institutional Animal Care and Use Committee of Haikou People’s Hospital affiliated with Central South University. A protocol was prepared before the study without registration.
Tissue collection and cell culture
NSCLC samples and paired normal tissues were resected from 33 primary NSCLC patients at Haikou People’s Hospital from 2011 to 2016 and stored at −80 °C. Patients who had received chemotherapy and biological/targeted therapy for NSCLC were excluded from the research. And the patients who were newly diagnosed with histologically confirmed NSCLC were included.
A549 and H1299 cells (two NSCLC cell lines) were procured from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Human bronchial epithelial cells (16HBE) were purchased from Biovector Science Lab, Inc. (Beijing, China). A549 cells were cultivated in a DMEM medium (Thermo Fisher Scientific, Rockford, IL, USA) complemented with 10% FBS (Thermo Fisher Scientific). H1299 cells were cultivated in RPMI-1640 medium (Thermo Fisher Scientific) with 10% FBS (Thermo Fisher Scientific). 16HBE cells were maintained in keratinocyte serum-free medium (Thermo Fisher Scientific) completed with bovine pituitary extract (0.05 mg/mL, Sigma-Aldrich, St. Louis, MO, USA), hydrocortisone (500 ng/mL, Sigma-Aldrich), human epidermal growth factor (5 ng/mL, Sigma-Aldrich) and insulin (0.005 mg/mL, Sigma-Aldrich). All cells were maintained in an incubator containing humidified air and 5% CO2 at 37 °C.
Reagent and cell transfection
Small interference RNA against DLX6-AS1 (si-DLX6-AS1), miR-16-5p mimic, miR-16-5p inhibitor (anti-miR-16-5p) and matching controls (si-NC, miR-NC and anti-miR-NC), DLX6-AS1 or BMI1 overexpression plasmid pcDNA-DLX6-AS1 or pcDNA-BMI1 (BMI1) and their negative control (pcDNA) was ordered from GenePharma Co., Ltd. (Shanghai, China). Cell transfection was conducted using Lipofectamine 2000 reagent (Thermo Fisher Scientific).
Western blot assay
Proteins were isolated from NSCLC xenograft tumors and cells via RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific) comprising protease inhibitor (Roche, Basle, Switzerland) and quantified using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Afterwards, protein samples (30 µg/sample) was separated using SDS-PAGE and transferred onto polyvinylidene difluoride membranes. After blocking with 5% skim milk, the membranes were hatched overnight at 4 °C using primary antibodies. The following antibodies were used: anti-E-cadherin (1:10,000; ab40772), anti-N-cadherin (1:1,000; ab18203), anti-BMI1 (1:10,000; ab126783), and anti-β-actin (1:5,000; ab179467). Next, horseradish peroxidase (HRP)-conjugated Goat Anti-Rabbit IgG H&L (HRP) (1:20,000; ab205718) was used to bind with matching primary antibody. All antibodies were procured from Abcam Inc. (Cambridge, UK). Subsequently, the blots were visualized using Clarity Western ECL Substrate (Bio-Rad Laboratories, Hercules, CA, USA) and quantified using Quantity One Software (Bio-Rad) via densitometry analysis.
Reverse transcription-quantitative PCR (RT-qPCR) assay
Total RNA was derived from NSCLC tissues and cells using TRIzol reagent (Thermo Fisher Scientific). Before reverse transfection, RNA was hatched with RNase-free DNase I (Thermo Fisher Scientific) to eliminate DNA contamination. The miR-16-5p level was then examined using TaqMan MicroRNA Assay kit (Thermo Fisher Scientific) with U6 snRNA as the internal control. For the analysis of BMI1, β-actin, and DLX6-AS1 expression, cDNA was cloned using high-capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) and quantified using SYBRTM Green PCR Master Mix (Thermo Fisher Scientific). The primer sequences were presented as follows: 5'-GGGTAGCAGCACGTAAATA-3' (forward) and 5'-CAGTGCGTGTCGTGGAGT-3' (reverse) for miR-16-5p; 5'-CTCGCTTCGGCAGCACA-3' (forward) and 5'-AACGCTTCACGAATTTGCGT-3' (reverse) for U6 small nuclear RNA (U6); 5'-GGTGGGGAACCTCCAAACAT-3'(forward) and 5'-CAGGCATGTAGCCTGCAGAT-3' (reverse) for DLX6-AS1; 5'-TTCCACTCTGCCTTCAGCGG-3' (forward) and 5'-CCACATCCCCATTGTCTCGC-3' (reverse) for BMI1; 5'-GGATTCCTATGTGGGCGACGA-3' (forward) and 5'-GCGTACAGGGATAGCACAGC-3' (reverse) for β-actin. β-actin was used for the normalization of BMI mRNA and DLX6-AS1.
RNA immunoprecipitation (RIP) assay
The enrichment level of DLX6-AS1 in IgG or AGO2 immunoprecipitation (IP) complex was examined using EZ-Magna RIP kit (Millipore, Billerica, MA, USA). Cells were gathered at 48 hours after transfection and lysed in RIP lysis buffer supplemented with protease and ribonuclease inhibitors (Sigma-Aldrich). After centrifugation, cell supernatants were hatched with magnetic beads coupled with IgG or AGO2 antibody (Millipore). Subsequently, RNA in the IP complex was purified. Finally, the DLX6-AS1 level was evaluated by RT-qPCR assay.
Luciferase reporter assay
The sequences of DLX6-AS1 and BMI1 3'UTR harboring wide-type (WT) or mutant-type (MUT) miR-16-5p binding fragments were introduced into psiCHECK-2 luciferase reporter vector (Hanbio Biotechnology Co., Ltd., Shanghai, China), and named DLX6-AS1WT, DLX6-AS1 MUT, BMI1 3'UTR-WT, or BMI1 3'UTR-MUT reporter. These reporters together with miR-16-5p mimic or miR-NC were introduced into A549 and H1299 cells for 48 h, luciferase activities were discovered using the dual-luciferase reporter assay kit (Promega, Madison, WI, USA).
Cell Counting Kit-8 (CCK-8) assay
Cell proliferative capacity was estimated through CCK-8 assay using CCK-8 kit (MedChemExpress, Monmouth Junction, NJ, USA). First, cells transfected with corresponding miRNA mimic, miRNA inhibitor, or overexpression plasmid were plated into 96-well plates. At 0, 24, 48, 72 hours post-transfection, each well was incubated with 10 µL of CCK-8 solution for 2 hours. Next, the optical density (OD) values were determined at 450 nm.
Colony formation assay
Transfected cells were inoculated into 6-well plates for about 2 weeks. Cells were then settled with methanol for 15 minutes and treated with 1% crystal violet staining solution (Sigma-Aldrich) for 20 minutes. Next, the number of positive cell colonies (>30 cells) was calculated.
Wound healing assay
NSCLC cells were inoculated into 6-well plates for 24 hours in a complete medium. A plastic tip was used to wound the cell monolayers. After being washed with PBS three times, the cells were incubated for 24 hours. An inverted microscope determined the average distance of migrating cells at designated time points.
Transwell assay
Transwell assay was conducted using Transwell chamber (8 µm pore size, Corning, Corning, NY, USA) in the absence (for migration assay) or presence (for invasion assay) of matrigel (Corning). Briefly, cells in serum-free medium were placed into the superior chamber, and the medium with 20% FBS was filled into the lower chamber. Forty-eight hours later, cells on the upper membranes were erased by cotton swabs, whereas cells that migrated or invaded into lower membranes were settled with methanol, spotted with 0.1% crystal violet, and assessed in 10 random fields with a microscope.
Mouse experiment
Lentiviruses harboring DLX6-AS1 knockdown sequences (sh-DLX6-AS1) and its negative control sh-NC were ordered from Hanbio Biotechnology Co., Ltd. (Shanghai, China). Twelve male BALB/c athymic nude mice (6 weeks old, 18–22 g) were procured from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China) and randomly divided into sh-NC and sh-DLX6-AS1 groups with 6 mice in each group. For the xenograft growth assay, the mice were subcutaneously inoculated with H1299 cells infected with sh-NC or sh-DLX6-AS1 lentiviruses. Tumor volume was then checked every 7 days for 35 days and calculated using the formula of volume = 0.5 × length × width2. On day 35, mice were euthanized by CO2 inhalation, and xenograft tumors were dissected, weighed, and stored for the following RT-qPCR and western blot assay. It is noted that mice reaching humane endpoints including severe weight loss (>15%), inability to obtain food or water, severe lethargy or any other signs of distress necessitate euthanasia, and no mice reached any humane endpoints during the study.
Immunohistochemical staining
All xenograft tumor samples were deparaffinized, rehydrated using graded alcohol, and washed with PBS. Tissue biopsies were blocked with 3% BSA at room temperature for 1 hour and probed with anti-BMI1 antibodies diluted 1:500 at 4 °C overnight. The corresponding secondary antibody was used for 1 hour at 37 °C. Rabbit-specific HRP/DAB (ABC) IHC Detection Kit (Abcam) was then used, and image data were acquired by an inverted fluorescent microscope.
Statistical analysis
The data of three repetitions were expressed as means ± standard deviations using Graphad Prism 6 software. The differences between paired samples were estimated through Student’s t-test. And one-way analysis of variance (ANOVA) along with Tukey post-hoc test was employed for multiple comparisons. Statistical significance was presented as P<0.05.
Results
DLX6-AS1 was over expressed in NSCLC tissues and cells and connected with poor overall survival of NSCLC patients
Firstly, the RT-qPCR assay disclosed that the DLX6-AS1 level was markedly elevated in 33 cases of NSCLC tissues as opposed to adjacent normal tissues (Figure 1A). Moreover, patients were categorized as the low (n=14) and high DLX6-AS1 expression group (n=19), with the mean value of DLX6-AS1 expression in NSCLC tissues as the cutoff point (Figure 1B). Furthermore, Kaplan-Meier analysis revealed that patients with low DLX6-AS1 expression had better overall survival, while those with high levels had poor outcomes (Figure 1B). Moreover, an obvious upregulation of DLX6-AS1 level was observed in two NSCLC cells (A549 and H1299) (Figure 1C).
Figure 1.
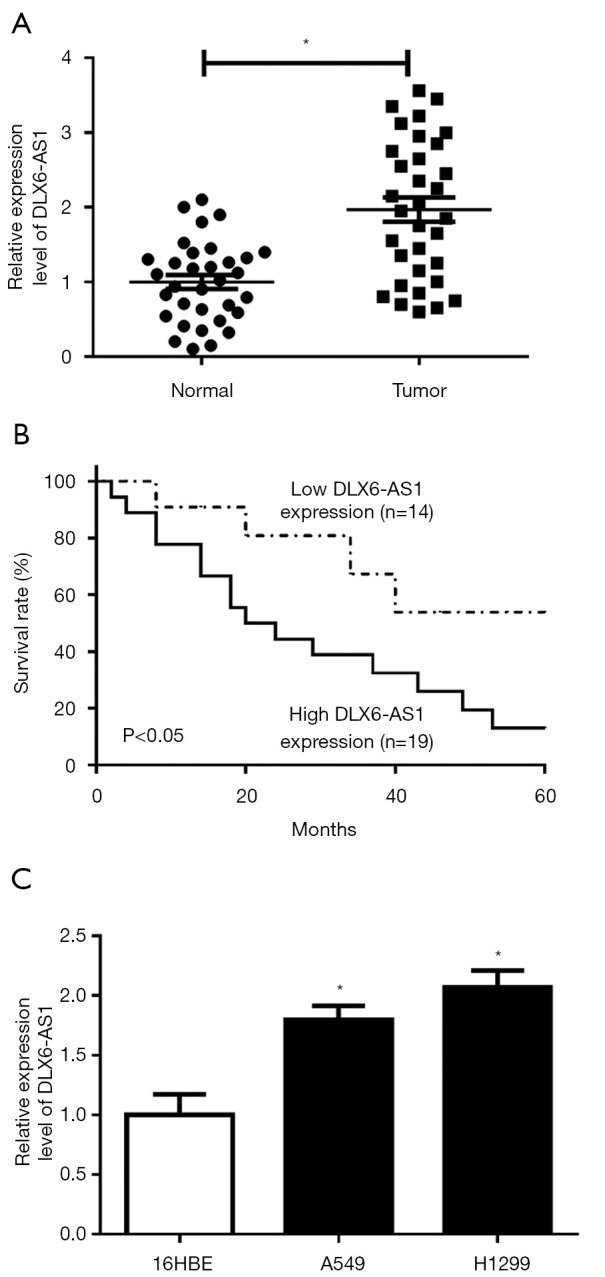
DLX6-AS1 was overexpressed in NSCLC tissues and cells and associated with a lower overall survival rate of NSCLC patients. (A) The RT-qPCR assay was conducted to detect the expression of DLX6-AS1 in 33 pairs of NSCLC tissues and adjacent normal tissues. (B) Kaplan-Meier survival analysis of overall survival for the 33 NSCLC patients with DLX6-AS1 expression level in tumor tissues. (C) RT-qPCR detected the expression of DLX6-AS1 in 16HBE, A549, and H1299 cells. *, P<0.05. DLX6-AS1, distal-less homeobox 6 antisense RNA 1; NSCLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative PCR.
DLX6-AS1 downregulation curbed proliferation, migration, invasion, and EMT in NSCLC cells
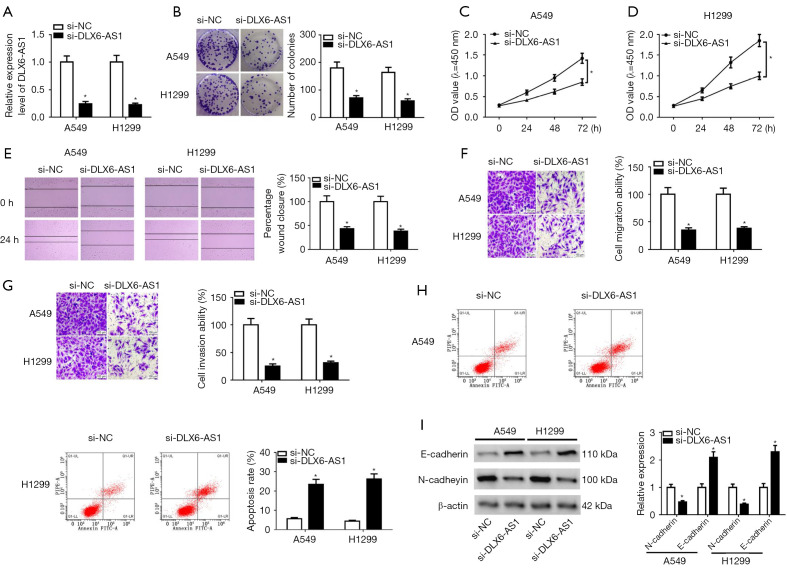
To investigate the function and molecular mechanisms of DLX6-AS1 in NSCLC cells, si-DLX6-AS1 and si-NC were introduced into A549 and H1299 cells. Transfection efficiency analysis disclosed that the introduction of si-DLX6-AS1 markedly reduced DLX6-AS1 level (Figure 2A), meaning that si-DLX6-AS1 could be used for subsequent experiments. Besides, DLX6-AS1 downregulation induced a notable downregulation of cell colony number in A549 and H1299 cells (Figure 2B), suggesting DLX6-AS1 downregulation suppressed NSCLC cell proliferation. Moreover, CCK8 assay indicated that an obvious reduction of cell proliferative ability was observed in DLX6-AS1-depleted NSCLC cells (Figure 2C,2D). Wound healing and Transwell assays uncovered that DLX6-AS1 downregulation expedited cell migration and invasion abilities (Figure 2E-2G). However, flow cytometry showed that the apoptosis cells were increased in cells transfected with si-DLX6-AS1 (Figure 2H). EMT is a dynamic process, which participates in the regulation of tumor initiation, metastasis, and therapeutic resistance (29). For analysis the function of DLX6-AS1 on EMT process, the protein levels of EMT markers were investigated. The results unveiled that DLX6-AS1 knockout resulted in increased E-cadherin expression and the decrease of N-cadherin expression in A549 and H1299 cells (Figure 2I), implying that DLX6-AS1 downregulation hampered cell EMT. These outcomes illustrated that DLX6-AS1 downregulation impeded proliferation, migration, invasion and EMT, but prompted apoptosis in NSCLC cells.
Figure 2.
DLX6-AS1 knockdown inhibited proliferation, migration, invasion, and EMT but induced apoptosis in NSCLC cells. (A-I) A549 and H1299 cells were transfected with si-NC or si-DLX6-AS1. (A) The expression of DLX6-AS1 was detected by RT-qPCR assay after transfection for 48 hours. (B-D) Colony formation assay (B, crystal violet staining) and CCK-8 assay (C,D) were performed to detect the proliferation ability in A549 and H1299 cells after transfection. (E, crystal violet staining) Cell migration was detected by wound healing assay in A549 and H1299 with si-NC or si-DLX6-AS1 transfection. (F,G) A Transwell assay was conducted to measure the cell migration and invasion potential (crystal violet staining, ×200 magnification). (H) Cell apoptosis was detected by flow cytometry in A549 and H1299 after transfection. (I) The protein levels of E-cadherin and N-cadherin were measured using western blot assay at 48 hours post-transfection. *, P<0.05. DLX6-AS1, distal-less homeobox 6 antisense RNA 1; EMT, epithelial-mesenchymal transition; NSCLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative PCR; CCK-8, Cell Counting Kit-8.
DLX6-AS1 suppressed miR-16-5p activity by direct interaction in NSCLC cells
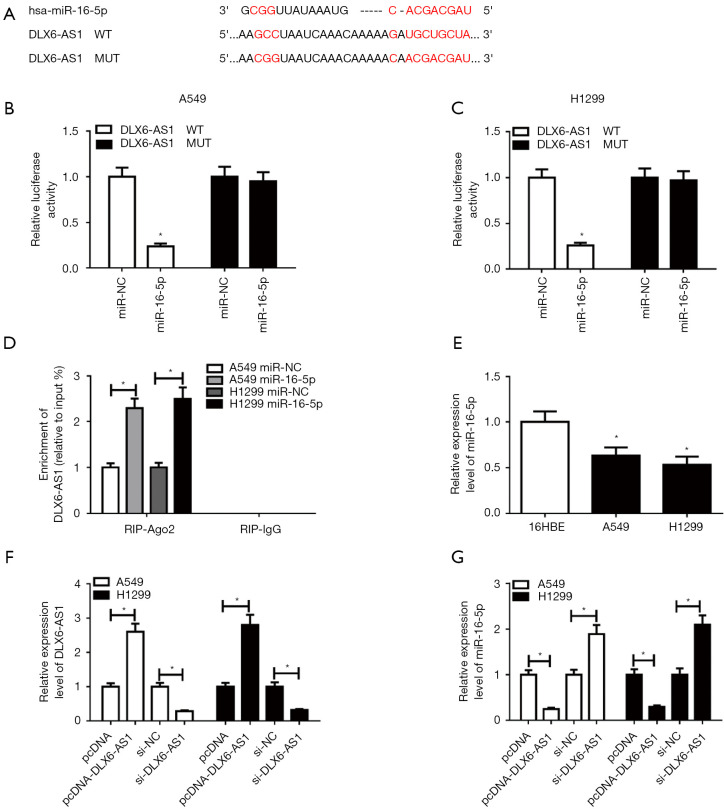
Next, DIANA-LncBase database (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=site/page&view=software) was used to search for miRNAs with binding fragments of DLX6-AS1. Among these candidates, miR-16-5p was picked owing to its crucial role in tumorigenesis and the progression of NSCLC (20-22). To further demonstrate this prediction, DLX6-AS1 WT and DLX6-AS1 MUT reportes (Figure 3A) were generated and then respectively transfected into A549 and H1299 cells with miR-16-5p mimic or miR-NC. The results uncovered that miR-16-5p overexpression markedly diminished the luciferase activity of DLX6-AS1 WT group but had no noticeable impact on luciferase activity of cell transfected with DLX6-AS1 MUT, suggesting that DLX6-AS1 could bind to miR-16-5p through predicted binding sites (Figure 3B,3C). Ago2, a core component of the RNA-induced silencing complex (RISC), functions as a crucial player in the biogenesis and maturation of miRNAs (30). Hence, potential miRNA targets can be obtained in the AGO2 IP complex. RIP assay exhibited that the enrichment of DLX6-AS1 in NSCLC cells with miR-16-5p transfection (Figure 3D), which further demonstrated the interconnection between DLX6-AS1 and miR-16-5p. Moreover, the miR-16-5p level was noticeably downregulated in NSCLC cells in contrast with 16HBE cells (Figure 3E). Transfection efficiency analysis manifested that the transfection of pcDNA-DLX6-AS1 induced a notable increase of DLX6-AS1 level, while si-DLX6-AS1 reduced DLX6-AS1 level in A549 and H1299 cells (Figure 3F). Further analysis unveiled that miR-16-5p level was remarkably decreased in DLX6-AS1 overexpressed A549 and H1299 cells but was elevated in miR-16-5p-knockout cells (Figure 3G). Thus, DLX6-AS1 functions as a sponge for miR-16-5p in NSCLC cells.
Figure 3.
DLX6-AS1 suppressed miR-16-5p expression by direct interaction in NSCLC cells. (A) The complementary binding sites of DLX6-AS1 and miR-16-5p, and mutant sites in DLX6-AS1 MUT reporter. (B,C) DLX6-AS1 WT or DLX6-AS1 MUT reporter was transfected into A549 and H1299 cells along with miR-16-5p mimic or miR-NC, followed by the measurement of luciferase activity at 48 hours upon transfection. (D) A549 and H1299 cells were transfected with miR-16-5p mimic or miR-NC. At 48 hours following transfection, RIP assay and RT-qPCR assay was used to examine the enrichment pattern of DLX6-AS1 in IgG or AGO2 IP complex. (E) The RT-qPCR assay was performed to detect miR-16-5p expression in 16HBE, A549, and H1299 cells. (F,G) A549 and H1299 cells were transfected with pcDNA, pcDNA-DLX6-AS1, si-NC, or si-DLX6-AS1, respectively. DLX6-AS1 and miR-16-5p level were subsequently measured by RT-qPCR assay at 48 hours after transfection. *, P<0.05. DLX6-AS1, distal-less homeobox 6 antisense RNA 1; miR-16-5p, microRNA-16-5p; NSCLC, non-small cell lung cancer; MUT, mutant-type; WT, wide-type; RIP, RNA immunoprecipitation; RT-qPCR, reverse transcription-quantitative PCR.
Anti-miR-16-5p inversed DLX6-AS1 knockdown mediated anti-tumor effects on NSCLC cells
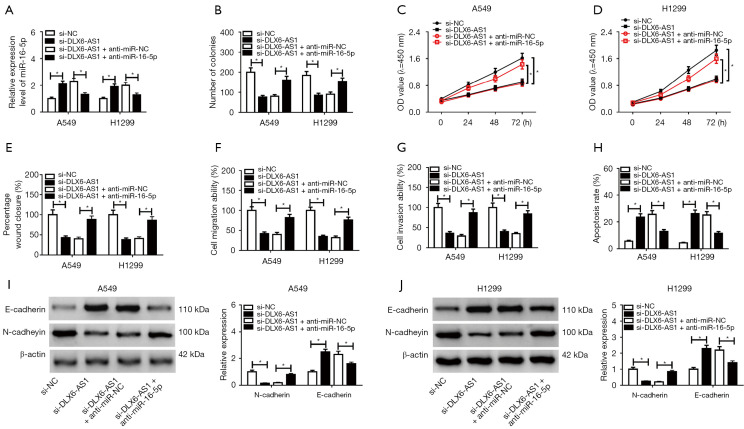
To further confirm whether DLX6-AS1 exerts its role through regulating miR-16-5p, A549 and H1299 cells were introduced with si-NC, si-DLX6-AS1, si-DLX6-AS1 + anti-miR-NC, si-DLX6-AS1 + anti-miR-16-5p. As displayed in Figure 4A, the level of miR-16-5p was elevated in cells with DLX6-AS1 downregulation, but anti-miR-16-5p reversed this up-regulation effect. Subsequent in vitro assay revealed that knockdown of miR-16-5p revoked the functions of si-DLX6-AS1 on cell proliferation, migration, invasion, apoptosis, and EMT in A549 and H1299 cells (Figure 4B-4J). In short, DLX6-AS1 regulates the progression of NSCLC cells through miR-16-5p.
Figure 4.
Anti-miR-16-5p inversed DLX6-AS1 knockdown mediated inhibitory effects on proliferation, migration, invasion, EMT, and promotion effects on apoptosis of NSCLC cells. (A-J) A549 and H1299 cells transfected with si-NC, si-DLX6-AS1, si-DLX6-AS1 + anti-miR-NC or si-DLX6-AS1 + anti-miR-16-5p respectively. (A) RT-qPCR was performed to detect the expression of miR-16-5p in A549 and H1299 cells after transfection for 48 hours. (B-D) Colony formation assay (B) and CCK-8 assay (C,D) were performed to detect the proliferation ability in A549 and H1299 cells after transfection. (E) Cell migration was detected by wound healing assay in A549 and H1299 with si-NC or si-DLX6-AS1 transfection. (F,G) A Transwell assay was conducted to measure the cell migration and invasion potential. (H) Cell apoptosis was detected by flow cytometry in A549 and H1299 after transfection. (I,J) E-cadherin and N-cadherin protein levels were measured using western blot assay at 48 hours post-transfection in A549 (I) and H1299 (J). *, P<0.05. miR-16-5p, microRNA-16-5p; DLX6-AS1, distal-less homeobox 6 antisense RNA 1; EMT, epithelial-mesenchymal transition; NSCLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative PCR; CCK-8, Cell Counting Kit-8.
BMI1 was a target of miR-16-5p
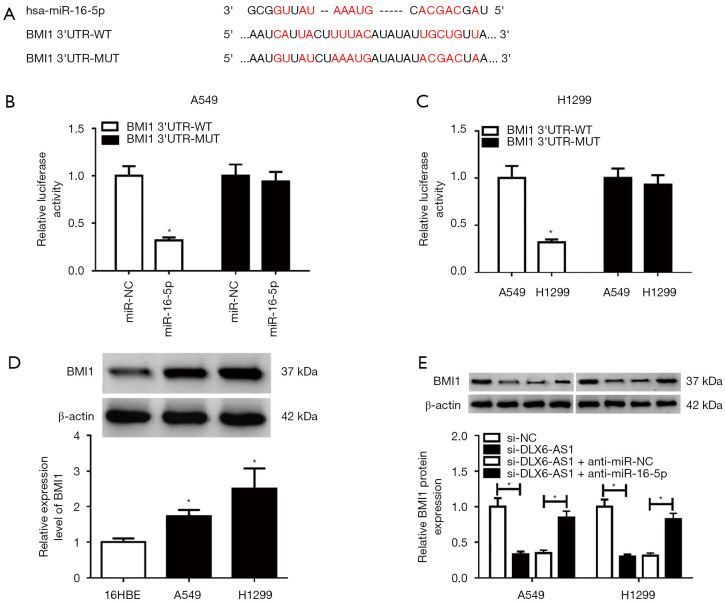
Next, bioinformatics analysis by miRTarBase online website (http://mirtarbase.mbc.nctu.edu.tw/php/search.php) showed the existence of binding fragments between miR-16-5p and BMI1 3'UTR (Figure 5A). Furthermore, the luciferase activity of miR-16-5p mimic and BMI1 3'UTR-WT group in A549 and H1299 cells was decreased (Figure 5B,C), while no obvious change was observed in BMI1 3'UTR-MUT and miR-16-5p mimic co-transfected group (Figure 5B,5C), implying that miR-16-5p could couple with BMI1 3'UTR. In addition, BMI1 was highly expressed in A549 and H1299 cells (Figure 5D). And downregulation of DLX6-AS1 minimized the protein level of BMI1, while it was upregulated in cells cointroduced with si-DLX6-AS1 and anti-miR-16-5p (Figure 5E). Hence, DLX6-AS1 regulates BMI1 by miR-16-5p.
Figure 5.
BMI1 was a target of miR-16-5p. (A) Putative binding sites between miR-16-5p and BMI1 3'UTR via the miRTarBase website, together with the mutant sites in BMI1 3'UTR-MUT reporter. (B,C) Forty-eight hours later, luciferase activity was detected in A549 and H1299 cells cotransfected with BMI1 3'UTR-WT or BMI1 3'UTR-MUT reporter and miR-16-5p mimic or miR-NC. (D) BMI1 protein level was examined by western blot assay in 16HBE, A549 and H1299 cells. (E) The level of BMI1 protein was assessed by western blot assay in A549 and H1299 cells transfected with si-NC, si-DLX6-AS1, si-DLX6-AS1 + anti-miR-NC, and si-DLX6-AS1 + anti-miR-16-5p, respectively, at 48 hours post transfection. *, P<0.05. miR-16-5p, microRNA-16-5p; DLX6-AS1, distal-less homeobox 6 antisense RNA 1; MUT, mutant-type; WT, wide-type.
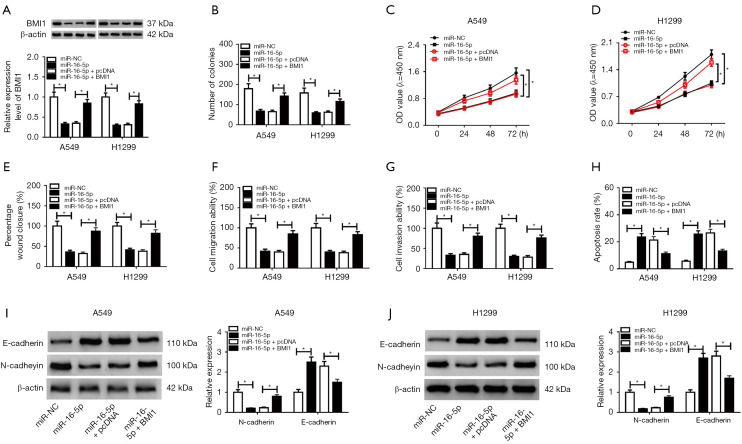
MiR-16-5p partly inverted the promotion impacts of BMI1 on NSCLC cell progression
To further investigate the function of BMI1 in DLX6-AS1/miR-16-5p-mediated NSCLC cell progression, A549 and H1299 cells were introduced with miR-NC miR-16-5p, miR-16-5p + pcDNA, and miR-16-5p + BMI1, respectively. The protein and mRNA levels of BMI1 were lessened in cells with miR-16-5p transfection (Figure 6A), while BMI1 inversed this decreasing effect. Furthermore, in vitro assay revealed that BMI1 overexpression invalidated the prohibition effects of miR-16-5p on cell proliferation, migration, invasion, and EMT, and the promotion effect on apoptosis (Figure 6B-6J). Above all, DLX6-AS1 functions as a sponge for miR-16-5p to upregulate BMI1 in NSCLC cells.
Figure 6.
MiR-16-5p partly reversed the promotion effects of BMI1 on proliferation, migration, invasion, and EMT, and inhibition effects on apoptosis of NSCLC cells. (A-J) A549 and H1299 cells were transfected with miR-NC, miR-16-5p, miR-16-5p + pcDNA, and miR-16-5p + BMI1, respectively. (A) The protein and mRNA levels of BMI1 in A549 and H1299 cells after transfection. (B-D) Cell proliferative capacity was estimated by colony formation assay (B) and CCK-8 assay (C,D). (E) Cell migration ability was detected by wound healing assay. (F,G) Cell migration and invasion abilities were detected by Transwell assay at 48 hours upon transfection. (H) Flow cytometry was performed to analyze cell apoptosis in A549 and H1299 cells after transfection. (I,J) Protein levels of E-cadherin and N-cadherin were measured by western blot assay at 48 hours post-transfection. *, P<0.05. miR-16-5p, microRNA-16-5p; DLX6-AS1, distal-less homeobox 6 antisense RNA 1; CCK-8, Cell Counting Kit-8.
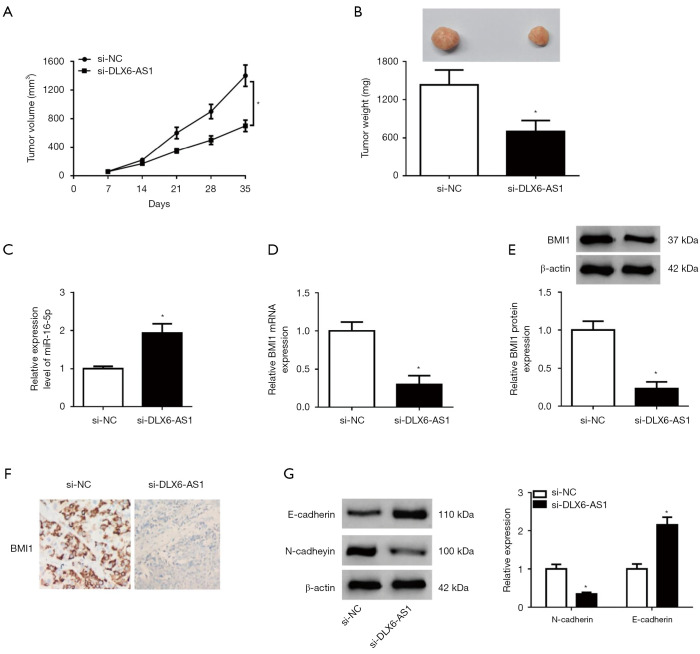
DLX6-AS1 silencing constrained the growth of NSCLC xenografts through upregulating miR-16-5p and downregulating BMI1
Next, in vivo experiments disclosed that DLX6-AS1 silencing constrained the growth of NSCLC xenografts (Figure 7A,7B). Moreover, DLX6-AS1 knockdown promoted miR-16-5p expression and repressed BMI mRNA and protein expression in xenograft tumors of NSCLC (Figure 7C-7E). Meanwhile, IHC staining for BMI1 indicated that BMI1-positive cells were significantly declined in tumor tissues with DLX6-AS1 silencing (Figure 7F). Additionally, E-cadherin expression was markedly elevated and N-cadherin expression was strikingly attenuated in xenograft tumors of NSCLC following the depletion of DLX6-AS1 (Figure 7G). Thus, DLX6-AS1 knockdown suppressed the tumorigenesis and progression of NSCLC xenografts via miR-16-5p/BMI1 axis.
Figure 7.
DLX6-AS1 knockdown suppressed the growth of NSCLC xenografts through upregulating miR-16-5p and downregulating BMI1. (A-F) H1299 cells infected with sh-NC or sh-DLX6-AS1 lentiviruses were subcutaneously injected into the flanks of mice in sh-NC or sh-DLX6-AS1 group, respectively. (A) Tumor volume was measured every 7 days for total 35 days. (B) Xenograft tumors were resected and weighed on day 35 post-injection. (C,D) The RT-qPCR assay was performed to detect the expression of miR-16-5p (C) and BMI1 mRNA (D) in NSCLC xenograft tumor samples. (E) The protein level of BMI1 in NSCLC xenograft tumors was measured by western blot assay. (F) The expression of BMI1 in xenograft tumors of NSCLC was detected by the immunohistochemical staining method (×200 magnification). (G) Protein levels of E-cadherin and N-cadherin were determined by western blot assay in xenograft tumors of NSCLC. *, P<0.05. DLX6-AS1, distal-less homeobox 6 antisense RNA 1; NSCLC, non-small cell lung cancer; miR-16-5p, microRNA-16-5p; RT-qPCR, reverse transcription-quantitative PCR.
Discussion
NSCLC is a major type of lung cancer, which was classified into three types: adenocarcinoma, squamous cell carcinoma and large cell (31). Even though great advance has been made in the treatment of NSCLC, the 5-year survival rate for NSCLC remains low (23%) (32). Thus, it is still needed to explore the pathogenesis of NSCLC to find potential therapeutic targets.
LncRNAs that are larger than 200 nucleotide (nt) and miRNAs approximately 20 nt in length have been reported to be potential oncogenic factors or tumor suppressors in several cancers, including NSCLC (33,34). Moreover, miRNAs can exert their function by regulating protein-coding genes expression via sequence complementarity (35). Additionally, there is progressive evidence that lncRNAs exert as sponges of miRNAs and consequently downregulate the expression of miRNA and increase the target mRNA levels (36,37). Our research demonstrated that lncRNA DLX6-AS1 promoted NSCLC tumorigenesis and progression through miR-16-5p/BMI1 axis.
Our results revealed that DLX6-AS1 level was remarkably upregulated in NSCLC tissues and cells, and were closely linked with the poor prognosis of NSCLC patients. In addition, the downregulation of DLX6-AS1 restrained NSCLC cell proliferation, migration, invasion, and EMT, and triggered cell apoptosis in vitro, as well as impeded tumorigenesis in NSCLC xenograft models. Similarly, a recent study demonstrated that downregltion of DLX6-AS1 blocked pancreatic cancer development by regulating miR-181b/Zinc finger E-box-binding homeobox 2 axes in (38). Moreover, DLX6-AS1 knockdown impaired pancreatic cancer cell EMT in vitro and in vivo (38). Additionally, Li et al. demonstrated that DLX6-AS1 overexpression facilitated glioma cell proliferation in vitro and expedited glioma cell xenograft growth in vivo through sequestering miR-197-5p from E2F1 (39).
Bioinformatics analysis demonstrated that miR-16-5p is a target of DLX6-AS1, which was further confirmed by luciferase reporter assay and RIP assay. miR-16-5p was reported to be lower expressed in serum samples of NSCLC patients (24). Moreover, Ke et al. disclosed that the abnormal expression of miR-16-5p caused a notable downregulation of cell proliferative, migratory, and invasive capacities through inhibiting hepatoma-derived growth factor in NSCLC (26). Additionally, miR-16 overexpression constrained cell proliferation, but expedited cell apoptosis and blocked cell cycle process by targeting wip1 in NSCLC (27). Our study demonstrated that DLX6-AS1 functioned its tumor promoter role in NSCLC by sponging miR-16-5p.
BMI1 was forecasted to be a target of miR-16-5p. Moreover, BMI1 depletion attenuated cell proliferative, migratory, and invasive capabilities, and impeded cell EMT in NSCLC. The suppression impacts of miR-16-5p mimic on NSCLC cell development were abrogated by miR-16-5p mimic. DLX6-AS1 functioned as a endogenous sponge of miR-16-5p to insulate it from its target BMI1, leading to an increasing BMI1 level. Additionally, DLX6-AS1 knockdown inhibited NSCLC tumorigenesis through controlling miR-16-5p/BMI1 axis in vivo.
BMI1, a core regulatory component of polycomb repressive complex-1, has been established as an oncogenic factor in cancer initiation and progression (40,41). Additionally, multiple studies showed that BMI1 could promote the tumorigenesis and development of NSCLC (42-44). For instance, the depletion of BMI1 controlled cell proliferation and hampered cell cycle progression in NSCLC (44). In addition, BMI silecing diminished the migratory and metastasis abilities of NSCLC cells (45). Moreover, BMI1 expression was connected to tumor size, poor differentiation, distant metastasis, and poor prognosis in patients with NSCLC (46-48).
There are still some deficiencies in the current research work. We have confimed the effect of DLX6-AS1 on EMT process in vitro, however, whether DLX6-AS1 regulated the metastasis of NSCLC in vitro is still known. In vivo animal and clinical research are indispensable in the future. Besides, as a part of the epigenetic machinery, miRNAs could be epigenetically modified by DNA methylation and histone modifications (49). Several research has suggested that the methylation alterations of miRNA methylation might be a potential target of the early diagnosis and treatment of cancer (50,51). In current research, we have disclosed that miR-16-5p is lower expressed in NSCLC. However, whether miR-16-5p methylation is altered in NSCLC cases is still unknown. Thus, it is efficient to scrutinize the methylation level of miR-16-5p and its correlation with the carcinogenesis and progression of NSCLC.
Conclusions
Our combined data revealed that DLX6-AS1 facilitated NSCLC cell proliferation, migration, invasion, EMT in vitro, and promoted NSCLC tumor growth in vivo by downregulating miR-16-5p to upregulate BMI1 expression, providing us important direction for NSCLC treatment strategies. However, the downstream or downstream regulatory pathways and interactive networks of DLX6-AS1/miR-16-5p/BMI1 axis still need further investigation. Moreover, it is necessary to deeply understand the effects of the DLX6-AS1/miR-16-5p/BMI1 axis on lung cancer development and progression, including SCLC and NSCLC.
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Haikou People’s Hospital and informed consent was taken from all the patients. All animal experiments were fulfilled in an SPF feeding environment following the National Institutes of Health guidelines and approved by the Institutional Animal Care and Use Committee of Haikou People’s Hospital affiliated with Central South University. A protocol was prepared before the study without registration.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-1240
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-21-1240
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-1240). The authors have no conflicts of interest to declare.
(English Language Editors: G. Stone and J. Chapnick)
References
- 1.Lemjabbar-Alaoui H, Hassan OU, Yang YW, et al. Lung cancer: Biology and treatment options. Biochim Biophys Acta 2015;1856:189-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Ettinger DS, Akerley W, Borghaei H, et al. Non-small cell lung cancer. J Natl Compr Canc Netw 2012;10:1236-71. 10.6004/jnccn.2012.0130 [DOI] [PubMed] [Google Scholar]
- 4.Ettinger DS, Wood DE, Akerley W, et al. Non-small cell lung cancer, version 1.2015. J Natl Compr Canc Netw 2014;12:1738-61. 10.6004/jnccn.2014.0176 [DOI] [PubMed] [Google Scholar]
- 5.Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014;14:535-46. 10.1038/nrc3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanoue LT, Tanner NT, Gould MK, et al. Lung cancer screening. Am J Respir Crit Care Med 2015;191:19-33. 10.1164/rccm.201410-1777CI [DOI] [PubMed] [Google Scholar]
- 7.Beermann J, Piccoli MT, Viereck J, et al. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev 2016;96:1297-325. 10.1152/physrev.00041.2015 [DOI] [PubMed] [Google Scholar]
- 8.Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J 2017;474:4219-51. 10.1042/BCJ20170079 [DOI] [PubMed] [Google Scholar]
- 9.Statello L, Guo CJ, Chen LL, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 2021;22:96-118. 10.1038/s41580-020-00315-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661-7. 10.1038/onc.2017.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Wang R, Zhang K, et al. Long non-coding RNAs in non-small cell lung cancer as biomarkers and therapeutic targets. J Cell Mol Med 2014;18:2425-36. 10.1111/jcmm.12431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin T, Fu Y, Zhang X, et al. A seven-long noncoding RNA signature predicts overall survival for patients with early stage non-small cell lung cancer. Aging (Albany NY) 2018;10:2356-66. 10.18632/aging.101550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Chen X, Lei T, et al. Integrative Analysis of NSCLC Identifies LINC01234 as an Oncogenic lncRNA that Interacts with HNRNPA2B1 and Regulates miR-106b Biogenesis. Mol Ther 2020;28:1479-93. 10.1016/j.ymthe.2020.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao L, Zhang X, Shi Y, et al. LncRNA SNHG14 contributes to the progression of NSCLC through miR-206/G6PD pathway. Thorac Cancer 2020;11:1202-10. 10.1111/1759-7714.13374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang G, Wang Q, Zhang X, et al. LncRNA FENDRR suppresses the progression of NSCLC via regulating miR-761/TIMP2 axis. Biomed Pharmacother 2019;118:109309. 10.1016/j.biopha.2019.109309 [DOI] [PubMed] [Google Scholar]
- 16.Yang CB, Xiao SW, Cheng SM, et al. LncRNA FAS-AS1 inhibits the progression of non-small cell lung cancer through regulating miR-19a-5p. Eur Rev Med Pharmacol Sci 2020;24:3775-85. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, He X, Jin T, et al. Long non-coding RNA DLX6-AS1 aggravates hepatocellular carcinoma carcinogenesis by modulating miR-203a/MMP-2 pathway. Biomed Pharmacother 2017;96:884-91. 10.1016/j.biopha.2017.10.056 [DOI] [PubMed] [Google Scholar]
- 18.Zeng X, Hu Z, Ke X, et al. Long noncoding RNA DLX6-AS1 promotes renal cell carcinoma progression via miR-26a/PTEN axis. Cell Cycle 2017;16:2212-9. 10.1080/15384101.2017.1361072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Li P, Zhao W, et al. Expression of long non-coding RNA DLX6-AS1 in lung adenocarcinoma. Cancer Cell Int 2015;15:48. 10.1186/s12935-015-0201-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang E, Liu R, Chu Y. miRNA-15a/16: as tumor suppressors and more. Future Oncol 2015;11:2351-63. 10.2217/fon.15.101 [DOI] [PubMed] [Google Scholar]
- 21.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ 2010;17:215-20. 10.1038/cdd.2009.69 [DOI] [PubMed] [Google Scholar]
- 22.Renjie W, Haiqian L. MiR-132, miR-15a and miR-16 synergistically inhibit pituitary tumor cell proliferation, invasion and migration by targeting Sox5. Cancer Lett 2015;356:568-78. 10.1016/j.canlet.2014.10.003 [DOI] [PubMed] [Google Scholar]
- 23.You C, Liang H, Sun W, et al. Deregulation of the miR-16-KRAS axis promotes colorectal cancer. Sci Rep 2016;6:37459. 10.1038/srep37459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan L, Qi H, Teng J, et al. Identification of serum miRNAs by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumour Biol 2016;37:7777-84. 10.1007/s13277-015-4608-3 [DOI] [PubMed] [Google Scholar]
- 25.Navarro A, Diaz T, Gallardo E, et al. Prognostic implications of miR-16 expression levels in resected non-small-cell lung cancer. J Surg Oncol 2011;103:411-5. 10.1002/jso.21847 [DOI] [PubMed] [Google Scholar]
- 26.Ke Y, Zhao W, Xiong J, et al. Downregulation of miR-16 promotes growth and motility by targeting HDGF in non-small cell lung cancer cells. FEBS Lett 2013;587:3153-7. 10.1016/j.febslet.2013.08.010 [DOI] [PubMed] [Google Scholar]
- 27.Gu Y, Wang XD, Lu JJ, et al. Effect of mir-16 on proliferation and apoptosis in human A549 lung adenocarcinoma cells. Int J Clin Exp Med 2015;8:3227-33. [PMC free article] [PubMed] [Google Scholar]
- 28.Bandi N, Zbinden S, Gugger M, et al. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res 2009;69:5553-9. 10.1158/0008-5472.CAN-08-4277 [DOI] [PubMed] [Google Scholar]
- 29.Li C. New Functions of Long Noncoding RNAs during EMT and Tumor Progression. Cancer Res 2019;79:3536-8. 10.1158/0008-5472.CAN-19-1205 [DOI] [PubMed] [Google Scholar]
- 30.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014;15:509-24. 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- 31.Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. 10.1097/JTO.0000000000000630 [DOI] [PubMed] [Google Scholar]
- 32.Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019;69:363-85. 10.3322/caac.21565 [DOI] [PubMed] [Google Scholar]
- 33.Florczuk M, Szpechcinski A, Chorostowska-Wynimko J. miRNAs as Biomarkers and Therapeutic Targets in Non-Small Cell Lung Cancer: Current Perspectives. Target Oncol 2017;12:179-200. 10.1007/s11523-017-0478-5 [DOI] [PubMed] [Google Scholar]
- 34.Wei MM, Zhou GB. Long Non-coding RNAs and Their Roles in Non-small-cell Lung Cancer. Genomics Proteomics Bioinformatics 2016;14:280-8. 10.1016/j.gpb.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev 2015;87:3-14. 10.1016/j.addr.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sui J, Li YH, Zhang YQ, et al. Integrated analysis of long non-coding RNA-associated ceRNA network reveals potential lncRNA biomarkers in human lung adenocarcinoma. Int J Oncol 2016;49:2023-36. 10.3892/ijo.2016.3716 [DOI] [PubMed] [Google Scholar]
- 37.Yang S, Ning Q, Zhang G, et al. Construction of differential mRNA-lncRNA crosstalk networks based on ceRNA hypothesis uncover key roles of lncRNAs implicated in esophageal squamous cell carcinoma. Oncotarget 2016;7:85728-40. 10.18632/oncotarget.13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An Y, Chen XM, Yang Y, et al. LncRNA DLX6-AS1 promoted cancer cell proliferation and invasion by attenuating the endogenous function of miR-181b in pancreatic cancer. Cancer Cell Int 2018;18:143. 10.1186/s12935-018-0643-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Zhang H, Wu X. Long noncoding RNA DLX6-AS1 accelerates the glioma carcinogenesis by competing endogenous sponging miR-197-5p to relieve E2F1. Gene 2019;686:1-7. 10.1016/j.gene.2018.10.065 [DOI] [PubMed] [Google Scholar]
- 40.Jiang L, Li J, Song L. Bmi-1, stem cells and cancer. Acta Biochim Biophys Sin (Shanghai) 2009;41:527-34. 10.1093/abbs/gmp040 [DOI] [PubMed] [Google Scholar]
- 41.Cao L, Bombard J, Cintron K, et al. BMI1 as a novel target for drug discovery in cancer. J Cell Biochem 2011;112:2729-41. 10.1002/jcb.23234 [DOI] [PubMed] [Google Scholar]
- 42.Dovey JS, Zacharek SJ, Kim CF, et al. Bmi1 is critical for lung tumorigenesis and bronchioalveolar stem cell expansion. Proc Natl Acad Sci U S A 2008;105:11857-62. 10.1073/pnas.0803574105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen T, Xu C, Chen J, et al. MicroRNA-203 inhibits cellular proliferation and invasion by targeting Bmi1 in non-small cell lung cancer. Oncol Lett 2015;9:2639-46. 10.3892/ol.2015.3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu Q, Su B, Liu D, et al. Antisense RNA-mediated suppression of Bmi-1 gene expression inhibits the proliferation of lung cancer cell line A549. Oligonucleotides 2007;17:327-35. 10.1089/oli.2007.0070 [DOI] [PubMed] [Google Scholar]
- 45.Meng X, Wang Y, Zheng X, et al. shRNA-mediated knockdown of Bmi-1 inhibit lung adenocarcinoma cell migration and metastasis. Lung Cancer 2012;77:24-30. 10.1016/j.lungcan.2012.02.015 [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, Tian T, Sun W, et al. Bmi-1 overexpression as an efficient prognostic marker in patients with nonsmall cell lung cancer. Medicine (Baltimore) 2017;96:e7346. 10.1097/MD.0000000000007346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vrzalikova K, Skarda J, Ehrmann J, et al. Prognostic value of Bmi-1 oncoprotein expression in NSCLC patients: a tissue microarray study. J Cancer Res Clin Oncol 2008;134:1037-42. 10.1007/s00432-008-0361-y [DOI] [PubMed] [Google Scholar]
- 48.Zhang X, Sun J, Wang H, et al. IGF-1R and Bmi-1 expressions in lung adenocarcinoma and their clinicopathologic and prognostic significance. Tumour Biol 2014;35:739-45. 10.1007/s13277-013-1100-9 [DOI] [PubMed] [Google Scholar]
- 49.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011;21:381-95. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konno M, Koseki J, Asai A, et al. Distinct methylation levels of mature microRNAs in gastrointestinal cancers. Nat Commun 2019;10:3888. 10.1038/s41467-019-11826-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oltra SS, Peña-Chilet M, Vidal-Tomas V, et al. Methylation deregulation of miRNA promoters identifies miR124-2 as a survival biomarker in Breast Cancer in very young women. Sci Rep 2018;8:14373. 10.1038/s41598-018-32393-3 [DOI] [PMC free article] [PubMed] [Google Scholar]