Abstract
Objective:
The aim of this study was to compare IL-1β levels in gingival crevicular fluid (GCF) from healthy and periodontitis sites of IL-1B(3954)-Single Nucleotide Polymorphism (SNP) positive and IL-1B(3954)-SNP negative periodontitis subjects in association with their bacterial profiles.
Background:
Susceptibility to periodontitis has been associated with several risk factors, including allelic variants at multiple gene loci. Variations in the IL-1 gene cluster have been linked with increased risk for periodontitis. IL-1B(3954)-SNP has been previously associated with increased levels of IL-1β in GCF or periodontal tissues in chronic periodontitis patients, as well as higher levels of specific periodontal pathogens. There is insufficient evidence to conclude if IL-1B gene polymorphisms affect the susceptibility to periodontitis by ultimately modulating the levels of IL-1β in GCF, the subgingival microbial profile or both.
Materials and Methods:
GCF, subgingival plaque, and buccal epithelial cells were collected from 32 individuals with periodontitis. GCF IL-1β levels were measured by an enzyme-linked immunosorbent assay (ELISA). Bacterial plaque samples were analyzed for 11 periodontal pathogens using polymerase chain reaction (PCR) analysis with specific primers for the 16SrRNA gene of each bacterium. IL-1B(3954)-SNP status was determined by identifying the carriers of the polymorphic T allele.
Results:
A significant association was shown between IL-1B(3954)-SNP and IL-1β GCF levels (amount and concentration). The concomitant presence of two or three red complex bacterial species was associated with increased IL-1β GCF levels in periodontitis sites (site-level analysis). The concurrent presence of all three red complex periodontal pathogens and IL-1B(3954)-SNP was associated with the highest IL-1β GCF levels in periodontitis sites.
Conclusions:
Our results indicate an independent association of both IL-1B(3954)-SNP and red complex bacterial species with increased IL-1β levels in GCF of periodontitis sites. A better understanding of the interaction between genetics, bacteria, and inflammation is essential to develop more effective diagnostic, prognostic, and therapeutic tools for periodontitis.
Keywords: bacteria, gingival crevicular fluid, interleukin-1 beta, polymorphism, single nucleotide
1 ∣. INTRODUCTION
Periodontitis is the 6th most prevalent chronic disease in the world and the primary cause for tooth loss in adults.1 It is estimated at least 47% of adults aged 30 years and older have periodontitis in the United States.2 Periodontitis is a complex infectious-inflammatory disease of the gingiva and of the supporting structures of the teeth with several etiologic and contributory factors.3 Although periodontal diseases are initiated and sustained by dental bacterial plaque, patients are not equally susceptible and do not respond similarly to treatment; it is ultimately the host immune response that largely drives the pathological process.4 Thus, periodontal diseases are considered “eco-genetic” diseases, whereby patients become susceptible by genetic or environmental factors5 that trigger a chronic (hyper)-inflammatory response against the microbiome and cause a disruption of immune-bacteria homeostasis in the periodontium.6,7
The regulation of inflammatory mediators by endogenous mechanisms and the balance between pro- and anti-inflammatory mediators determines the severity of tissue destruction.8 Several mechanisms in the pathogenesis of periodontitis associated with either an up-regulation of pro-inflammatory mediators or host immune deficiencies appear to be genetically determined.9 Interleukin-1 (IL-1) is an important pro-inflammatory cytokine largely responsible for initiating the cascade of inflammatory responses against periodontal pathogens and exists in three forms: IL-1α, IL-1β, and interleukin-1 receptor antagonist (IL-1RA).10 Increased levels of IL-1β have been detected in gingival crevicular fluid (GCF) and gingival tissues of individuals with periodontitis showing a strong association with the severity of periodontal tissue destruction.10,11
Heritability has been considered an important predisposing factor for periodontitis with an estimated magnitude of genetic contribution to overall periodontitis susceptibility as 33%-50%.12 Common single nucleotide polymorphisms (SNPs) associated with altered gene activity or protein production affecting the host response to bacteria play an important role in the severity and progression of periodontitis.13 Substantial evidence supports that genetic differences in the degree of the inflammatory response, based on the levels of expression of inflammatory mediators, are associated with increased risk for periodontitis.14 Although no single-gene variant has yet been determined, variations in the IL-1 gene cluster have been linked with increased host susceptibility to several inflammatory diseases including coronary artery disease, Alzheimer's disease, and periodontitis.15-17
In 1997, non-smoking Caucasian subjects with positive IL-1A(−889) and IL-1B(+3953)-SNPs were found to have an increased risk for severe periodontitis.16 Several studies subsequently explored the role of IL-1 gene polymorphisms in periodontitis with mixed results, mainly due to a substantial heterogeneity in the clinical phenotype and ethnicity across the reports.18 Subjects with positive IL-1B(+3953)-SNP demonstrated higher levels of IL-1β in GCF before and after periodontal therapy.19
Variations in susceptibility to periodontitis have been also attributed to differences in the composition of subgingival microbiota among different host genotypes. Individuals with positive IL-1 SNPs had higher levels of “red” and “orange” complex species compared to individuals with negative ones.20 Specific periodontal pathogens may though account for higher IL-1β levels in periodontal tissues21 affecting the local cytokine periodontal milieu. However, there is insufficient evidence to conclude if IL-1B gene polymorphisms affect the susceptibility to periodontitis by ultimately modulating the levels of IL-1β in GCF, the subgingival microbial profile or both.
Hence, the aim of the present study is to compare the levels of IL-1β in GCF collected from healthy and periodontitis sites in IL-1B(3954)-SNP positive and IL-1B(3954)-SNP negative subjects with periodontitis. As an exploratory analysis, we also sought to determine any association of the above primary outcome with the bacterial profiles at the same periodontal sites.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Study population
Individuals (n = 32) in this cross-sectional study were enrolled from the Department of Periodontology, Tufts University School of Dental Medicine from November 2013 until November 2014 in full accordance with the World Medical Association Declaration of Helsinki. Overall inclusion criteria: All subjects were ≥18 years of age, in good systemic health and had ≥20 natural teeth excluding 3rd molars. Specific inclusion criteria: Subjects diagnosed with periodontitis were included in the study confirming bleeding on probing (BOP) ≥20% of probing sites, presence of ≥6 teeth with ≥1 site with probing depth (PD) ≥5 mm, clinical attachment level (CAL) >3 mm and BOP, and radiographic evidence of interproximal bone loss >2 mm.22 Exclusion criteria: acute intraoral infection, diagnosis of diabetes mellitus; HIV/AIDS; rheumatoid arthritis; presence of systemic conditions that require antibiotic coverage for routine periodontal procedures (eg, heart conditions, joint replacements); use of antibiotics or daily use of anti-inflammatory drugs or any type of periodontal treatment including dental prophylaxis in the last 3 months; pregnant or nursing women; and smokers. All participants were informed about the study protocol and signed a consent form previously approved by Tufts Health Sciences Institutional Review Board (IRB #11000) prior to enrollment.
2.2 ∣. Clinical examination
Before collecting any samples, clinical measurements were performed on the same day by the same and only experienced examiner (P.P.) at six sites/tooth (mesiobuccal, buccal, distobuccal, distolingual, lingual, and mesiolingual) at all teeth excluding third molars. The clinical parameters included: (i) Modified O’Leary Plaque Score Index23 (PI) (0 or 1), (ii) BOP (0 or 1), (iii) PD and (iv) CAL. PD and CAL measurements were made to the nearest mm using a North Carolina periodontal probe (PCPUNC 15; Hu-Friedy Manufacturing Company). The same examiner also performed radiographic evaluation of any interproximal bone loss for each subject based on radiographs exposed <12 months prior to enrollment in the study.
2.3 ∣. GCF collection
Two sites on two different teeth in each subject were chosen for bacterial plaque and GCF collection: a healthy site (absence of BOP, PD ≤3 mm, CAL ≤3 mm, and no bone loss) and a periodontitis site (PD ≥5 mm, CAL >3 mm, BOP, and bone loss >2 mm). GCF was collected from the site with the deepest PD among the periodontitis sites, and from one healthy site randomly selected among the healthy ones in a subject's mouth.22 In case of multiple sites with the same PD, the most accessible one was chosen. After removal of supragingival plaque, the area was isolated with cotton rolls and the sampling site was air-dried. Periopaper strips (PerioPaper, Oraflow) were inserted gently into the orifice of the sulcus/pocket, 1-2 mm subgingivally, in order to avoid any mechanical injury, and were left in place for 30 seconds. Strips visibly contaminated by saliva or blood were discarded. GCF volume was determined using Periotron 8000 (Oraflow), which was calibrated by the investigators based on a protocol described before.24 Readings were converted to an actual volume (μl) by reference to the standard (calibration) curve, by using the specific software that accompanies the device. Periopaper strips were placed immediately in de-identified labeled individual Eppendorf tubes and were then stored at −80°C until cytokine analysis.
2.4 ∣. GCF IL-1β levels quantification (ELISA)
IL-1β levels in GCF were determined in duplicate samples by an enzyme-linked immunosorbent assay (ELISA) (Human IL-1 beta/IL-1F2 DuoSet/DY201, R&D systems). The IL-1β detection range was 3.91-250 pg/ml. ELISA assay was based on an antibody sandwich method using microtiter plates, which were coated with the appropriate human cytokine. Reagent Diluent 250 μL (DY999, R&D Systems) was added to each sample. The samples were then shaken and centrifuged in order to purify more proteins of the strip into the Eppendorf tube. A microplate pre-coated with capture antibody was provided and an HRP-labeled antibody (detection antibody, Streptavidin-HRP) was added and bound to the captured analyte. Tetramethylbenzidine (TMB) substrate solution was added to the wells and a blue color developed in proportion to the amount of analyte present in the sample. The optical density (O.D.) of each well was determined within 30 minutes using a microplate reader set to 450 nm and 570 nm. The standard curve was created by reducing the data using a computer software capable of generating a four-parameter logistic (4-PL) curve-fit. GCF analysis was performed blindly to the genetic status.
2.5 ∣. Microbiological assessment
Subgingival plaque samples were collected from the same tooth sites selected for GCF collection immediately after GCF collection, allowing for comparisons on a site-by-site basis.25 Two sterile paper points, one following the other, were placed for 20 seconds down to the bottom of each pocket and then placed in de-identified labeled individual Eppendorf tubes. All samples were shipped for analysis using PCR (OralDNA®Labs).
Bacterial DNA was extracted from the processed sample through a combination of mechanical disruption of the bacterial cell and ion-exchange column purification and tested for eleven bacterial species associated with periodontal disease provided by a commercial test kit (MyPerioPath®, OralDNA®Labs). Universal 16SrRNA and species-specific primers were used to confirm the presence and levels of bacterial DNA in each sample. The bacterial species were tested using asymmetric multiplexed PCR with primers and molecular beacons designed to specific gene regions of each bacterial species.26 Amplification and detection were performed using a Qiagen RotorGene (Qiagen, Germany). Fluorescent emission resulting from molecular beacon hybridization was read at the end of the PCR reaction and compared to the fluorescence of known plasmid standards to provide a semi-quantitative analysis of patient sample concentration for each bacterium. The sensitivity of this assay permitted detection of 103 cells of each given species.
2.6 ∣. Analysis of genetic polymorphisms
Cells from buccal mucosa were obtained from each subject for the IL-1B(3954)-SNP (rs1143634) gene polymorphism assessment using a cytology brush following a previously established protocol.27 The cytology brush was used vigorously on both sites of buccal mucosa (internal surface of cheeks) for at least 30 seconds. The brush was then placed into a transfer tube and shipped for genotype analysis↑. The carriers of the polymorphic T allele (CT and TT genotypes) were combined into the same group that was called IL-1B(3954)-SNP positive, while another group with the homozygous C allele (CC genotype) was named IL-1B(3954)-SNP negative.28
2.7 ∣. Statistical analysis
Demographic data and clinical variables from the two different genotype groups were calculated at the subject-level with counts and percentages for categorical variables and mean (SD) for continuous variables. Pearson's chi-squared or Fisher's exact test was used to test to compare categorical variables, while Student's independent t test or Mann-Whitney U test was used for continuous variables. Bacteria levels and IL-1β amount (pg) and concentration (pg/ul) were compared between the two different genotype groups. Normality of data was assessed using the Kolmogorov-Smirnov test. The Wilcoxon signed-rank test (paired data) was used for comparisons (GCF volume, amount/ concentration of IL-1β in GCF) between healthy and periodontitis sites (overall). For periodontitis sites, a linear regression model with the log-transformed outcome measure was used to assess differences in IL-1β levels between the two genotype groups. For comparisons between genotype groups, a mixed effect linear regression model with random intercept was used for IL-1β levels while a mixed-effect logistic regression model with random intercept was used for bacteria detection.
A mixed-effect Poisson regression model with the subject as the random intercept was used to assess the effect of IL-1B(3954)-SNP genotype on number of red bacteria present. A 2-way ANOVA with Tukey's HSD adjustment for multiple comparisons was used to assess the joint effects of IL-1B(3954) genotype and red complex bacteria presence on IL-1β levels within healthy and periodontitis sites separately. An interaction term between IL-1B(3954) genotype and red complex bacteria was used to assess independence of the dependent variables. McNemar's test was used to assess the association between detection levels of each bacteria strain (%) at the subject-level while chi-square test was used for separate site-level analysis. All analyses were performed using SAS version 9.2 and 9.3 (SAS Institute, Inc., North Carolina). Statistical significance was set at a 2-sided P < .05.
Assuming α=0.05, a sample of 30 subjects would provide 80% power to detect a significant difference in GCF IL-1β levels between IL-1B(3954)-SNP positive and IL-1B(3954)-SNP negative periodontitis subjects considering that the prevalence of the IL-1B(3954)-SNP in this population was 20% (nQuery Advisor, 7.0).19
3 ∣. RESULTS
3.1 ∣. IL-1B(3954)-SNP frequency analysis
Demographic data of study participants are presented in Table 1. No significant differences were found regarding age, gender, and ethnicity distribution between two different genotype groups. The allele frequencies and genotype distributions for IL-1B(3954)-SNP in our study population are presented in Table 2. The prevalence of IL-1B(3954)-SNP positive genotype (CT, TT) was 40.6% of our study population (Table 1). The heterozygous polymorphic genotype (CT) (61.5%) was more frequent among IL-1B(3954)-SNP positive subjects comparing to the homozygous polymorphic genotype (TT) (38.5%). Overall, the C allele was more prevalent than the T allele (Table 2).
TABLE 1.
Demographic data and clinical characteristics
| Variable |
IL-1B(3954)-SNP positive (n = 13) |
IL-1B(3954)-SNP negative (n = 19) |
Total | P-value |
|---|---|---|---|---|
| N subjects (count, %) | 13 (40.6%) | 19 (59.4%) | 32 (100%) | |
| Age (mean, SD) | 47.00 (12.51) | 45.92 (16.12) | 46.56 (14.56) | .841a |
| Gender | 4 females (30.8%) | 9 females (47.4%) | 13 females (40.6%) | .348b |
| 9 males (69.2%) | 10 males (52.6%) | 19 males (59.4%) | ||
| Ethnicity | ||||
| White (Non-Hispanic) | 9 (69.2%) | 8 (42.1%) | 17 (53.1%) | .112c |
| American Black | 3 (23.1%) | 4 (21.1%) | 7 (21.9%) | |
| Asian | 0 (0%) | 6 (31.6%) | 6 (18.8%) | |
| Hispanic, Other | 1 (7.7%) | 1 (5.3%) | 2 (6.3%) | |
| Clinical parameters | ||||
| PD mean (SD) | 3.25 (0.42) | 3.23 (0.58) | 3.24 (0.51) | .43d |
| CAL mean (SD) | 3.74 (0.46) | 3.62 (0.64) | 3.67 (0.57) | .24d |
| BOP mean (%) | 55.00 (14.97) | 57.00 (13.25) | 56.19 (13.77) | .730d |
| PI mean (%) | 71.98 (34.78) | 86.01 (20.14) | 80.31 (27.44) | .241d |
| Tooth loss | 1.69 ± 2.31 | 2.11 ± 1.89 | .436d | |
| # Teeth Periodontitis | 12.08 ± 4.66 | 10.95 ± 3.77 | .289d | |
| % Teeth Periodontitis | 46 ± 17 | 42 ± 13 | .748d |
Note: Values are given as counts (percentages) or as means (standard deviations).
No statistical significant differences were found.
Determined by the independent t test.
Determined by the chi-square test.
Determined by the Fisher exact test.
Determined by the Mann-Whitney test.
TABLE 2.
Frequencies of IL-1B(3954)-SNP
| Genotype or Allele | Total subjects (%) |
Subjects with IL- 1B(3954)-SNP positive |
|---|---|---|
| IL-1B genotype | ||
| CC | 19 (59.37%) | 0% |
| CT | 8 (25%) | 61.5% |
| TT | 5 (15.63%) | 38.5% |
| CT+TT | 13 (40.63%) | |
| Allele | ||
| C | 46 (71.87%) | 8 (30.7%) |
| T | 18 (28.13%) | 18 (69.3%) |
Note: CC: IL-1B(3954)-SNP negative.
CT/TT: IL-1B(3954)-SNP positive.
3.2 ∣. IL-1B(3954)-SNP and clinical parameters
No significant difference was found between subjects with or without IL-1B(3954)-SNP for PD, CAL, BOP, or PI (Table 1). Although IL-1B(3954)-SNP positive subjects showed an increased number of teeth affected with periodontitis (≥ 1 site with PD ≥5 mm, CAL >3 mm and BOP, and radiographic evidence of interproximal bone loss >2 mm) compared to IL-1B(3954)-SNP negative subjects, there was no statistical significant difference between the two groups.
3.3 ∣. IL-1B(3954)-SNP and IL-1β GCF levels
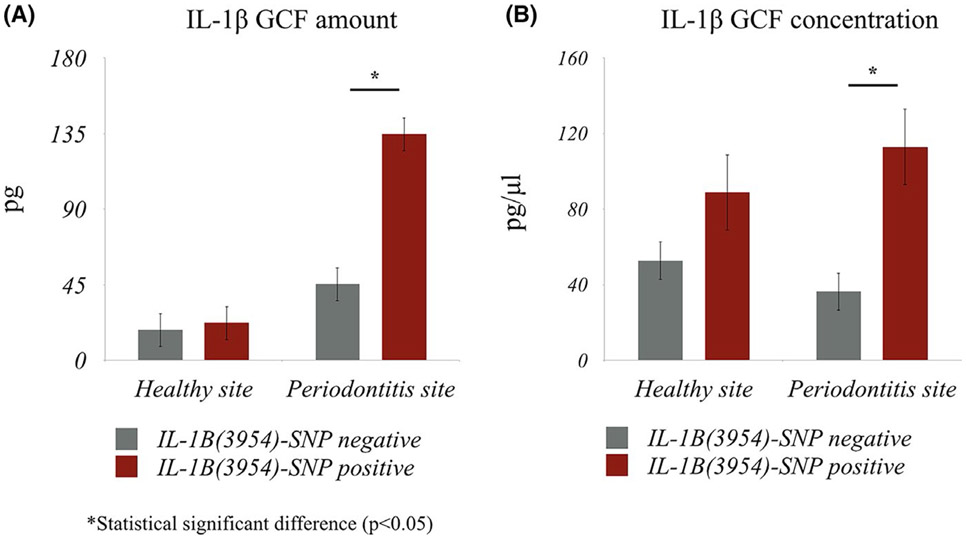
IL-1β was detected in 100% of all samples. Using the Wilcoxon signed-rank test (paired data) significant differences were found for both the amount of IL-1β in GCF and the volume of GCF (P < .001) between healthy and periodontitis sites (overall) (data not shown). Subjects with IL-1B(3954)-SNP positive showed significantly higher IL-1β amount (P < .01) and concentration (P < .01) in GCF in periodontitis sites compared to IL-1B(3954)-SNP negative subjects (site-level analysis) (Linear Regression Model, Figure 1A,B). No significant differences in IL-1β GCF levels were found in healthy sites between two different genotype groups. At the subject-level analysis (combining healthy and periodontitis sites), IL-1B(3954)-SNP positive subjects had significantly higher IL-1β concentration in GCF (Mixed-effect regression model, P = .013, Table 3), while GCF IL-1β amount differences did not reach significance (P = .058, Table 3). Adjusting for age, gender, and ethnicity in our analysis did not affect the significance of the associations (data not shown).
FIGURE 1.
The amount (A) and the concentration (B) of IL-1β in GCF in healthy and periodontitis sites between subjects with IL-1B(3954)-SNP positive and IL-1B(3954)-SNP negative. Statistical significant differences were found between IL-1β amount (P < 0.001) and IL-1β concentration (P < 0.001) in GCF in periodontitis sites between IL-1B(3954)-SNP positive and IL-1B(3954)-SNP negative subjects
TABLE 3.
Association of IL-1B(3954)-SNP and the detection of periodontal pathogens and IL-1β levels in GCF on a site-level and subject-level analysis
| Variable | Site-level analysis (Periodontitis sites) |
Subject-level analysis (Healthy and Periodontitis sites) |
|||||
|---|---|---|---|---|---|---|---|
|
IL-1B(3954)-SNP Negative (n = 19) |
IL-1B(3954)-SNPPositive (n = 13) |
P-value |
IL-1B(3954)-SNP Negative (n = 19 pairs) |
IL-1B(3954)-SNP Positive (n = 13pairs) |
P-value | ||
| IL-1β Amount (pg) | Mean (SD) | 45.47 (33.60) | 134.6 (127.8) | <.01 | 31.81 (30.41) | 78.36 (106.3) | .058 |
| Median [Min-Max] | 47.28 [3.21, 146.49] | 81.69 [32.93, 450.17] | 20.41 [1.17, 146.49] | 42.56 [1.16, 450.17] | |||
| IL-1β Concentration (pg/μl) | Mean (SD) | 36.37 (26.88) | 113.0 (101.6) | <.01a,* | 44.57 (57.89) | 100.9 (104.5) | .013 |
| Median [Min-Max] | 37.82 [2.56, 117.19] | 85.03 [26.34, 360.13] | 31.86 [1.15, 352.41] | 52.03 [6.08, 369.38] | |||
| A.actinomycetemcomitans | Count (%) | 6 (31.58) | 4 (30.77) | 1.00d | 9 (23.68) | 4 (15.38) | .525e |
| F.nucleatum | Count (%) | 6 (31.58) | 9 (69.23) | .036 | 6 (15.79) | 13 (50.00) | <.01 |
| P.gingivalis | Count (%) | 9 (47.37) | 9 (69.23) | .221e | 10(26.32) | 13 (50.00) | .082e |
| T.forsythia | Count (%) | 19 (100) | 13 (100) | 1.00d | 28 (73.38) | 25 (96.15) | .052e |
| T.denticola | Count (%) | 11 (57.89) | 9 (69.23) | .713d | 13 (34.21) | 11 (42.31) | .516e |
Abbreviations: A.a: Aggregatibacter actinomycetemcomitans; F.n: Fusobacterium nucleatum; P.g: Porphyromonas gingivalis; T.d: Treponema Denticola; T.f: Tannerella forsythia.
Linear regression.
Mixed-effect regression model with random intercept.
Chi-square test.
Fisher exact test.
Mixed-effect logistic regression with random intercept.
Statistical significant difference (P < .05).
3.4 ∣. IL-1B(3954)-SNP and periodontal pathogens
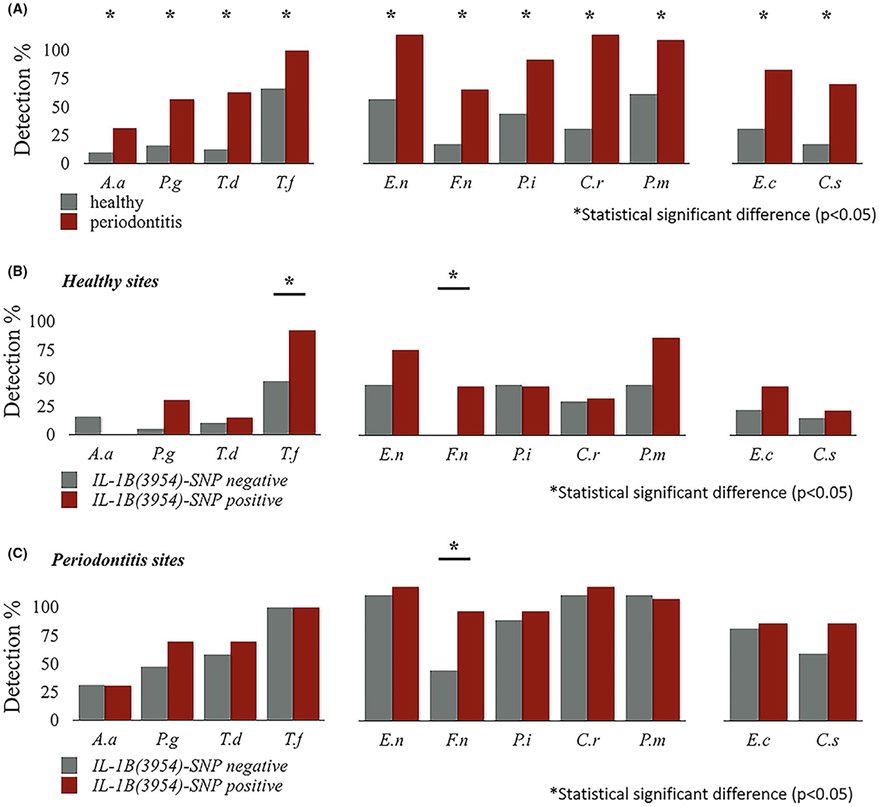
Higher frequencies of detection of all investigated bacterial species were found in periodontitis sites than in healthy ones (Figure 2A, McNemar's Test). Frequency detection of Fusobacterium nudeatum and Tannerella forsythia was significantly higher in healthy sites in IL-1B(3954)-SNP positive compared with IL-1B(3954)-SNP negative subjects (P = .02; P = .01 respectively, Figure 2B). Frequency detection of F.nudeatum was significantly higher in periodontitis sites in IL-1B(3954)-SNP positive compared with IL-1B(3954)-SNP negative subjects (69.23% vs. 31.58%, P = .036, Figure 2C). At the subject-level analysis, a significant association was found between F.nudeatum and IL-1B(3954)-SNP status (Table 3, mixed-effect logistic regression model). IL-1B(3954)-SNP positive was associated with 1.4 times higher (95% CI 0.92-2.12) presence of red complex bacteria than IL-1B(3954)-SNP negative; however, this association was not statistically significant at the 0.05 level of significance (P = .099, mixed-effect Poisson regression model).
FIGURE 2.
The frequency of detection of different types of bacteria in healthy and periodontitis sites between subjects with IL-1B(3954)-SNP positive and IL-1B(3954)-SNP negative. Higher frequencies of detection of all tested periodontal pathogens were found in periodontitis sites than in healthy sites in overall population (all sites) (A). The frequency detection of F.nucleatum and T.forsythia was significantly higher in healthy sites in IL-1B(3954)-SNP positive compared to IL-1B(3954)-SNP negative subjects (P = 0.02; P = 0.01) (B). The frequency detection of F.nucleatum was found to be significantly higher in periodontitis sites in IL-1B(3954)-SNP positive (69.23%) compared to IL-1B(3954)-SNP negative (31.58%) subjects (P = 0.036) (C)
3.5 ∣. Periodontal pathogens and IL-1β GCF levels
The concomitant presence of two or three red complex bacterial species was associated with increased IL-1β GCF levels in periodontitis sites independently of IL-1B(3954)-SNP status. Indeed, subjects with all three red complex bacteria present in periodontitis sites had 6.2 times higher IL-1β GCF concentration compared to subjects with only one red complex pathogen (P < .001, Tukey's HSD adjustment for multiple comparisons) (data not shown).
3.6 ∣. IL-1B(3954)-SNP, red complex periodontal pathogens, and IL-1β GCF levels
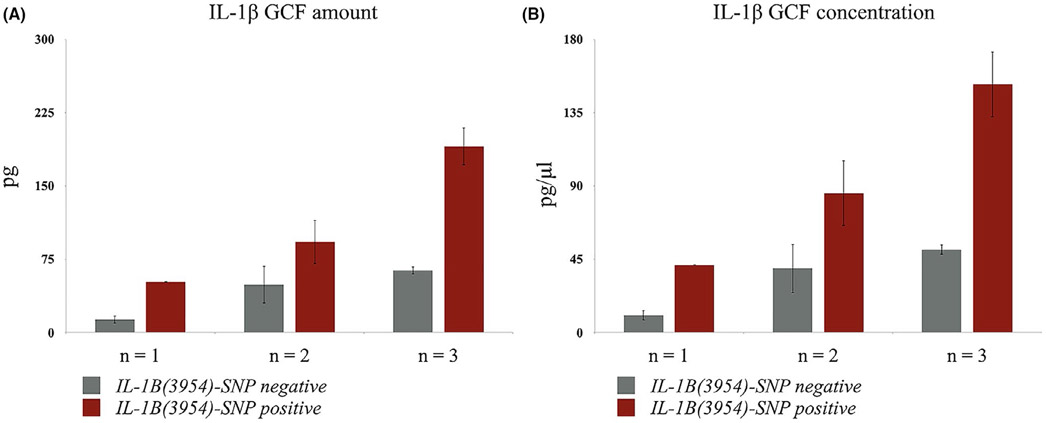
Using a 2-way ANOVA model, both IL-1B(3954)-SNP and number of red complex bacterial species had significant effects on GCF IL-1β levels of periodontitis sites. Analysis presented here is unadjusted for demographic variables. Models adjusted for age, gender, and ethnicity (white vs non-white) were also performed but no differences compared to unadjusted models were found. The interaction term between the two dependent variables in ANOVA analysis was not significant (P = .7714) suggesting the absence of any additive effect between the two variables; thus, data presented here are for the ANOVA model with the interaction term removed. Subjects with both positive IL-1B(3954)-SNP and three red complex bacterial species showed the highest GCF IL-1β levels (amount and concentration) in periodontitis sites (Figure 3). Based on the 2-way ANOVA model with periodontitis sites and IL-1B(3954)-SNP, periodontitis sites with three red complex bacterial species present had 4.7 times higher IL-1β GCF amount compared to periodontitis sites with only one of red complex bacteria species present (P < .01 for both, Figure 3). After controlling for red complex bacteria species, periodontitis sites from subjects with IL-1B(3954)-SNP positive had 2.3 times higher IL-1β GCF amount and 2.4 times higher IL-1β GCF concentration compared to periodontitis sites from subjects with IL-1B(3954)-SNP negative (P < .01 for both, Figure 3).
FIGURE 3.
The effect of IL-1B(3954)-SNP on both GCF amount (A) and concentration (B) of IL-1β and number (n) of red complex bacteria species (P.gingivalis, T.forsythia, T.denticola) in periodontitis sites. Both IL-1B(3954)-SNP and number of red complex bacteria species had significant effects on the IL-1β GCF amount and concentration. After controlling for IL-1B(3954)-SNP, periodontitis sites with three red complex bacteria species present had 4.7 times higher IL-1β GCF amount and 4.6 times higher IL-1β GCF concentration compared to periodontitis sites with one of red complex bacteria species present. After controlling for red complex bacteria species, periodontitis sites from subjects with IL-1B(3954)-SNP positive had 2.3 times higher IL-1β GCF amount and 2.4 times higher IL-1β GCF concentration compared to periodontitis sites from subjects with IL-1B(3954)-SNP negative.
4 ∣. DISCUSSION
Periodontitis is a complex, multi-factorial disease involving repetitive interactions between bacteria, host immune/inflammatory responses and modifying genetic and environmental factors.7,29 Pathologic shifts in the subgingival microbial profile have been observed in periodontitis sites that may have occurred by an over-growth of commensal bacterial or a dysregulated host immune response to oral biofilm, resulting in a “dysbiosis” of the subgingival microbiota and a disruption of tissue homeostasis.30 However, this microbial shift favoring periodontal pathogens also requires a suitable environment and a “susceptible” host to manifest clinically.5 In this study, we show that the concurrent presence of all three red complex periodontal pathogens and IL-1B(3954)-SNP is associated with the highest IL-1β GCF levels in periodontitis sites. Our data implicate both IL-1B(3954)-SNP and red complex species as possible modulators of host immune response in the micro-environment of the periodontal sulcus/pocket favoring the establishment of periodontal disease.
Our data demonstrate significantly higher GCF volume and amount of IL-1β in GCF from periodontitis sites compared with healthy ones within the same individuals. Although the concentration of IL-1β in GCF was not significantly different, this is still an important finding. IL-1β is produced at high amounts in healthy sites of subjects with periodontitis; IL-1β secretion could favor the establishment of a suitable “inflammo-philic” subgingival environment with higher risk for future periodontal tissue breakdown and change into a periodontitis site with deeper PD and bone loss. Increased IL-1β levels in GCF have been associated with periodontitis severity, progression, tooth loss, and disease recurrence.10,19,31 One of the limitations of previous studies is that increased IL-1β levels may have been due to increased PD or the presence of specific bacteria. Therefore, we explored the direct contribution of IL-1B(3954)-SNP to increased GCF levels of IL-1β in subjects with periodontitis with regards to the presence of periodontal pathogens and comparing sites of similar PD. In the present study, considerable attention was given to the clinical selection of periodontitis sites and healthy sites. Our study design allowed us to control several known risk factors for periodontal immune responses by excluding smokers and any systemic disease or use of medications that could interfere with the synthesis of IL-1β or microbial colonization. Although our population was derived from different ethnic groups, no significant differences in age, gender, and ethnicity distribution between different IL-1B(3954)-SNP subjects were found. Furthermore, there were no significant differences in the clinical periodontal variables between IL-1B(3954)-SNP positive and IL-1B(3954)-SNP negative subjects that allowed us to compare our primary study outcomes (IL-1β GCF levels and bacterial profile) in an unadjusted fashion. The prevalence of IL-1B(3954)-SNP in our study population was 40.63% with frequencies of C (71.87%) and T alleles (28.13%) similar to ones reported in other similar studies.32,33 The use of paper strips in our study is suitable for simultaneous determination of microbial and immunological parameters and has shown the highest detected cytokine levels, especially for IL-1β with ELISA.25,34,35
Identifying local and/or systemic factors responsible for overproduction of IL-1β in the periodontium is critical in establishing a risk profile for subjects with periodontitis. While several studies have associated IL-1B gene polymorphisms with an increased risk for periodontitis, few studies have shown the mechanisms behind this association with clinical data.11,16,28,32,33 Most of the studies have been in vitro where IL-1B gene polymorphisms result in increased secretion of IL-1β from several immune cells, including monocytes and polymorphonuclear leukocytes (PMNs).36-38 Here, we investigated the functional role of IL-1B(3954)-SNP in modulating IL-1β synthesis directly in the periodontal pocket's microenvironment by measuring IL-1β levels in GCF adjusting for the presence of periodontal pathogens, a key missing point from previous similar studies. Our study confirms that IL-1B(3954)-SNP is associated with higher IL-1β levels in GCF in healthy and periodontitis sites of subjects with periodontitis.
Our data on the positive effect of IL-1B(3954)-SNP on IL-1β levels are in accordance with previous studies.19,32 However, our results are more directly relevant as we measured actual IL-1β levels in GCF (secretion) of periodontitis subjects, rather than IL-1β gene expression from gingival biopsies as Ferreira et al. studied.32 In contrast to our findings, Engebretson et al. reported that only shallow sites (<4 mm) showed higher IL-1β levels in GCF in positive IL-1B(3954)-SNP periodontitis subjects but no differences in deeper sites (≥4 mm)19 as shown in our study. Moreover, Yücel et al. found no significant differences for IL-1β levels in GCF in periodontitis subjects with different IL-1B(3954)-SNP status; however, GCF samples in this study were collected and pooled from 6 maxillary sites/subject indicating significant heterogeneity in GCF collection compared to our strict collection of GCF from one specific site/subject.33
The investigation of the relationship between host genetic variants and the microbial colonization, named “infectogenomics,” is gaining more interest lately.39 A recent systematic review concluded that there is no evidence yet of any genetic polymorphism association with presence/counts of subgingival bacteria.39 F. nucleatum was the only bacterial species (among 11 we studied) significantly more frequently detected in periodontitis sites in IL-1B(3954)-SNP positive compared with IL-1B(3954)-SNP negative subjects. We also show that the frequency of detection of F. nucleatum and T. forsythia is significantly higher in healthy sites in IL-1B(3954)-SNP positive compared with IL-1B(3954)-SNP negative subjects possibly predisposing progression from gingivitis to periodontitis. In addition, subjects with two or three red complex bacterial species present in periodontitis sites had higher IL-1β GCF concentration compared to subjects with only one red complex pathogen suggesting a crucial pro-inflammatory role for the combination of red complex bacteria.
Periodontitis sites from IL-1B(3954)-SNP positive subjects and three red complex bacterial species present showed the highest IL-1β GCF levels. The interaction term between red complex bacteria and IL-1B(3954)-SNP was not significant suggesting that there is no additive effect and the effects of the two variables are independent of each other. Our findings provide a possible biologic mechanism of how IL-1B(3954)-SNP may increase the risk of periodontitis by favoring both the growth of “inflammo-philic” periodontal pathogens and increased levels of IL-1β independently resulting in the disruption of both microbial and host immune homeostasis in the periodontium. Indeed, inflammation and dysbiosis positively reinforce each other and collectively sustain periodontal disease pathogenesis, promoting progression of periodontitis.40 Furthermore, our findings align with a recent genome-wide association study (GWAS) that showed that biological phenotypes, such as subgingival bacterial colonization and biomarker mediators, had the highest heritability, suggesting that these biologically informed traits are promising targets for the conduct of genomics investigations, according to the goals of personalized oral health.41 Our results indicate an independent association of both IL-1B(3954)-SNP and red complex bacterial species with increased IL-1β levels in GCF of periodontitis sites.
The diversity in genetic, infectious, and immunologic subtypes of periodontal disease argues for personalized therapy. A recent GWAS demonstrated that IL-37 variants with functional roles in decreased expression of IL-37 lead to upregulation of IL-1β levels in GCF contributing to a hyper-inflammatory periodontal environment.42 A better understanding of the interaction between genetics, bacteria, and inflammation is essential to develop more effective diagnostic, prognostic, and therapeutic tools for periodontitis. A personalized approach combining gene biomarkers with conventional risk factors to stratify populations for their needs for prevention has been previously proposed.43 GCF biomarkers combined with the presence of specific pathogens has been recommended for monitoring periodontal disease activity and progression.31 Our findings provide additional evidence that IL-1B(3954)-SNP can modulate the subclinical profile of the periodontal sulcus and should be combined with other bacteria and immune biomarkers to allow early identification of susceptible individuals/sites. One of the strengths of our study was that we evaluated the effect of IL-1B(3954)-SNP on both site-level and subject-level, an approach that was missing from previous similar studies. Recognizing the small sample size in our study as a potential limitation, our results should be interpreted carefully before being generalized in larger populations. Considering that our sample size was properly calculated based on the effect of IL-1B gene polymorphism on IL-1β GCF levels as a primary outcome,18 caution should be applied on extrapolating the results regarding the secondary outcomes. However, proper statistical adjustments for all possible confounding factors were made in our analyses and our study can generate new valid hypotheses for further investigation. Future studies on large populations among different ethnic groups including GWAS with healthy controls, longer observation data, and monitoring treatment outcomes will be needed to validate our findings before clinical application.
ACKNOWLEDGEMENTS
Supported in part by USPHS grant K08DE027119 to Dr. E. Papathanasiou from the National Institute of Dental and Craniofacial Research (NIDCR). This research project was supported by the Office of Advanced Graduate Education and the Department of Periodontology at Tufts University School of Dental Medicine. MyPerioPath® tests and IL-1B(3954)-SNP analysis were supported by OralDNA®Labs, MN. Dr. R. McGlennen is a principal share-holder and an officer of the company, Access Genetics, OralDNA®Labs, MN. Ms. Carrie Brown is a biostatistician at The Emmes Corporation, Rockville, MD. The authors declare that there are no conflicts of interest in this study.
Footnotes
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest in this study.
PRINCIPAL FINDING
Both IL-1B(3954)-SNP and red complex bacterial species were independently associated with increased GCF-IL-1β levels in periodontitis sites.
REFERENCES
- 1.Kassebaum NJ, Bernabe E, Dahiya M, Bhandari B, Murray CJ, Marcenes W. Global burden of severe periodontitis in 1990–2010: a systematic review and meta-regression. J Dent Res. 2014;93:1045–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ, Cdc Periodontal Disease Surveillance workgroup: James Beck GDRP. Prevalence of periodontitis in adults in the United States. J Dent Res. 2012;91:914–920. 2009 and 2010. [DOI] [PubMed] [Google Scholar]
- 3.Slots J Periodontitis: facts, fallacies and the future. Periodontol. 2000;2017(75):7–23. [DOI] [PubMed] [Google Scholar]
- 4.Kinane DF, Stathopoulou PG, Papapanou PN. Periodontal diseases. Nat Rev Dis Primers. 2017;3:17038. [DOI] [PubMed] [Google Scholar]
- 5.MarkBartold P,Van Dyke TE. Host modulation: control ling the inflammation to control the infection. Periodontol. 2000;2017(75):317–329. [DOI] [PubMed] [Google Scholar]
- 6.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–490. [DOI] [PubMed] [Google Scholar]
- 7.Loos BG, Van Dyke TE. The role of inflammation and genetics in periodontal disease. Periodontol. 2000;2020(83):26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kantarci A, Hasturk H, Van Dyke TE. Host-mediated resolution of inflammation in periodontal diseases. Periodontol. 2000;2006(40):144–163. [DOI] [PubMed] [Google Scholar]
- 9.Kinane DF, Hart TC. Genes and gene polymorphisms associated with periodontal disease. Crit Rev Oral Biol Med. 2003;14:430–449. [DOI] [PubMed] [Google Scholar]
- 10.Graves DT, Cochran D. The contribution of interleukin-1 and tumor necrosis factor to periodontal tissue destruction. J Periodontol. 2003;74:391–401. [DOI] [PubMed] [Google Scholar]
- 11.Stashenko P, Fujiyoshi P, Obernesser MS, Prostak L, Haffajee AD, Socransky SS. Levels of interleukin 1 beta in tissue from sites of active periodontal disease. J Clin Periodontol. 1991;18:548–554. [DOI] [PubMed] [Google Scholar]
- 12.Michalowicz BS, Aeppli D, Virag JG, et al. Periodontal findings in adult twins. J Periodontol. 1991;62:293–299. [DOI] [PubMed] [Google Scholar]
- 13.Nibali L, Di lorio A, Tu YK, Vieira AR. Host genetics role in the pathogenesis of periodontal disease and caries. J Clin Periodontol. 2017;44(Suppl 18):S52–S78. [DOI] [PubMed] [Google Scholar]
- 14.Yoshie H, Kobayashi T, Tai H, Galicia JC. The role of genetic polymorphisms in periodontitis. Periodontol. 2000;2007(43):102–132. [DOI] [PubMed] [Google Scholar]
- 15.Karimbux NY, Saraiya VM, Elangovan S, et al. Interleukin-1 gene polymorphisms and chronic periodontitis in adult whites: a systematic review and meta-analysis. J Periodontol. 2012;83:1407–1419. [DOI] [PubMed] [Google Scholar]
- 16.Kornman KS, Crane A, Wang H-Y, et al. The interleukin-1 genotype as a severity factor in adult periodontal disease. J Clin Periodontol. 1997;24:72–77. [DOI] [PubMed] [Google Scholar]
- 17.Dinarello CA. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev. 2018;281:8–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grigoriadou ME, Koutayas SO, Madianos PN, Strub JR. Interleukin-1 as a genetic marker for periodontitis: review of the literature. Quintessence Int. 2010;41:517–525. [PubMed] [Google Scholar]
- 19.Engebretson SP, Lamster IB, Herrera-Abreu M, et al. The influence of interleukin gene polymorphism on expression of interleukin-1beta and tumor necrosis factor-alpha in periodontal tissue and gingival crevicular fluid. J Periodontol. 1999;70:567–573. [DOI] [PubMed] [Google Scholar]
- 20.Socransky SS, Haffajee AD, Smith C, Duff GW. Microbiological parameters associated with IL-1 gene polymorphisms in periodontitis patients. J Clin Periodontol. 2000;27:810–818. [DOI] [PubMed] [Google Scholar]
- 21.Bodet C, Chandad F, Grenier D. Inflammatory responses of a macrophage/epithelial cell co-culture model to mono and mixed infections with Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia. Microbes Infect. 2006;8:27–35. [DOI] [PubMed] [Google Scholar]
- 22.Papathanasiou E, Teles F, Griffin T, et al. Gingival crevicular fluid levels of interferon-gamma, but not interleukin-4 or −33 or thymic stromal lymphopoietin, are increased in inflamed sites in patients with periodontal disease. J Periodontal Res. 2014;49:55–61. [DOI] [PubMed] [Google Scholar]
- 23.O'Leary TJ, Drake RB, Naylor JE. The plaque control record. J Periodontol. 1972;43:38. [DOI] [PubMed] [Google Scholar]
- 24.Chappie IL, Landini G, Griffiths GS, Patel NC, Ward RS. Calibration of the Periotron 8000 and 6000 by polynomial regression. J Periodontal Res. 1999;34:79–86. [DOI] [PubMed] [Google Scholar]
- 25.Teles R, Sakellari D, Teles F, et al. Relationships among gingival crevicular fluid biomarkers, clinical parameters of periodontal disease, and the subgingival microbiota. J Periodontol. 2010;81:89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. [DOI] [PubMed] [Google Scholar]
- 27.Woo JG, Sun G, Haverbusch M, et al. Quality assessment of buccal versus blood genomic DNA using the Affymetrix 500 K GeneChip. BMC Genet. 2007;8:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masamatti SS, Kumar A, Baron TK, Mehta DS, Bhat K. Evaluation of interleukin -1B (+3954) gene polymorphism in patients with chronic and aggressive periodontitis: a genetic association study. Contemp Clin Dent. 2012;3:144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet. 2002;3:391–397. [DOI] [PubMed] [Google Scholar]
- 30.Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J Dent Res. 2012;91:816–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinney JS, Morelli T, Oh M,et al. Crevicular fluid biomarkers and periodontal disease progression. J Clin Periodontol. 2014;41:113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreira SB Jr, Trombone AP, Repeke CE, et al. An interleukin-1beta (IL-1beta) single-nucleotide polymorphism at position 3954 and red complex periodontopathogens independently and additively modulate the levels of IL-1beta in diseased periodontal tissues. Infect Immun. 2008;76:3725–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yucel OO, Berker E, Mescil L, Eratalay K, Tepe E, Tezcan I. Association of interleukin-1 beta (+3954) gene polymorphism and gingival crevicular fluid levels in patients with aggressive and chronic periodontitis. Genet Couns. 2013;24:21–35. [PubMed] [Google Scholar]
- 34.Guentsch A, Kramesberger M, Sroka A, Pfister W, Potempa J, Eick S. Comparison of gingival crevicular fluid sampling methods in patients with severe chronic periodontitis. J Periodontol. 2011;82:1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nazar Majeed Z, Philip K, Alabsi AM, Pushparajan S, Swaminathan D. Identification of gingival crevicular fluid sampling, analytical methods, and oral biomarkers for the diagnosis and monitoring of periodontal diseases: a systematic review. Dis Markers. 2016;2016:1804727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gore EA, Sanders JJ, Pandey JP, Palesch Y, Galbraith GM. Interleukin-1beta+3953 allele 2: association with disease status in adult periodontitis. J Clin Periodontol. 1998;25:781–785. [DOI] [PubMed] [Google Scholar]
- 37.Mark LL, Haffajee AD, Socransky SS, et al. Effect of the interleukin-1 genotype on monocyte IL-1beta expression in subjects with adult periodontitis. J Periodontal Res. 2000;35:172–177. [DOI] [PubMed] [Google Scholar]
- 38.Santtila S, Savinainen K, Hurme M. Presence of the IL-1RA allele 2 (IL1RN*2) is associated with enhanced IL-1beta production in vitro. Scand J Immunol. 1998;47:195–198. [DOI] [PubMed] [Google Scholar]
- 39.Nibali L, Di lorio A, Onabolu O, Lin GH. Periodontal infectogenomics: systematic review of associations between host genetic variants and subgingival microbial detection. J Clin Periodontol. 2016;43:889–900. [DOI] [PubMed] [Google Scholar]
- 40.Hajishengallis G Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15:30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agler CS, Moss K, Philips KH, et al. Biologically defined or biologically informed traits are more heritable than clinically defined ones: the case of oral and dental phenotypes. Adv Exp Med Biol. 2019;1197:179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Offenbacher S, Jiao Y, Kim SJ, et al. GWAS for Interleukin-1beta levels in gingival crevicular fluid identifies IL37 variants in periodontal inflammation. Nat Commun. 2018;9:3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giannobile WV, Braun TM, Caplis AK, Doucette-Stamm L, Duff GW, Kornman KS. Patient stratification for preventive care in dentistry. J Dent Res. 2013;92:694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]