Abstract
Multilocus genotyping was used to compare populations of Candida albicans from oral mucosa and blood. No significant differences in allele frequencies between the two samples were detected, and in a dendrogram of genotypic similarities, genotypes from both types of samples were finely interspersed. This is evidence for widespread distribution of invasive potential.
The yeast Candida albicans is a ubiquitous commensal and an important opportunistic human pathogen causing superficial infections as well as invasive fungal disease in immunocompromised patients (9, 11). This fungus is diploid and has a largely clonal population structure with no evidence for genetic differentiation along geographic lines, at least not in North America and Europe (8, 17, 18). We proposed two alternative hypotheses to explain the transition of C. albicans from superficial sites, such as oral mucosa, to the bloodstream of patients. The first is that the propensity of a strain to invade the bloodstream, when considered as a trait, is rare in C. albicans populations inhabiting superficial sites. If only a small number of strains of C. albicans have successfully moved from a superficial site to the bloodstream, then, in general, variability determined by multilocus genotyping in a sample of bloodstream isolates would be reduced compared to variability in a sample of strains from superficial sites; genetic differentiation between invasive and noninvasive samples would be expected. Alternatively, if the propensity for invasiveness is widespread in C. albicans, then many different genotypes have successfully established infections in the bloodstream and genetic variability in a sample of bloodstream isolates would not be reduced compared to a sample of strains from superficial sites; genetic differentiation between invasive and noninvasive samples would not be expected. According to the second hypothesis, the multilocus genotypes of bloodstream isolates would show close similarity to a wide range of genotypes from superficial sites.
Several investigations using different DNA typing methods have addressed the degree of strain similarity among isolates from different anatomical sites and patient populations, providing diverse results (1, 13–15, 17). On the basis of biochemical tests characterizing phenotypes, a previous study that specifically compared isolates of C. albicans from superficial and deep anatomical sites did not detect any significant differences (10). The purpose of this study was to determine if populations of C. albicans isolated from oral mucosa and blood are genetically differentiated or not.
In this study we used a multilocus genotyping system based on an assay of nucleotide polymorphisms distributed on all chromosomes. This method uses allele-specific oligonucleotide probes in Southern hybridizations with 16 PCR-amplified regions and clearly distinguishes heterozygotes from homozygotes at each polymorphic site. We and colleagues previously reported (5) on the relationship between multilocus genotypes, DNA fingerprints, and azole resistance in a sample of 81 oral isolates of C. albicans from human immunodeficiency virus-infected patients in Toronto, Canada, plus three reference strains, CAI4, WO-1, and CA (ATCC 90028). This genotyping method has proven to be reliable, with no missing data, no ambiguities, and complete reproducibility for the strains of C. albicans studied by Cowen et al. (5). Here we determined multilocus genotypes for a sample of blood isolates of C. albicans from patients with systemic infections in Toronto, Canada, and compared the allele frequencies and patterns of genetic similarity with those of the sample of oral isolates. The 39 strains of C. albicans, each isolated from the blood of a different patient, were obtained from the Ontario Ministry of Health Medical Mycology Lab and from the Hospital for Sick Children in Toronto, Canada. In addition, two reference isolates (2), CA30 and CA87, were kindly provided by G. T. Cole, Medical College of Ohio. Strain CA30 has been characterized as invasive in the infant mouse model (3).
Among the 41 isolates in the present study (39 isolates from blood cultures from patients plus 2 reference isolates), there were 39 multilocus diploid genotypes. Genotype counts at 7 out of the 16 loci were significantly different from Hardy-Weinberg expectation (Table 1). These results are similar to those obtained from the oral isolates, where 64 multilocus diploid genotypes were identified among the 84 isolates and 8 out of the 16 loci deviated significantly from Hardy-Weinberg expectation (P < 0.05 in all cases). In tests for significant deviation from Hardy-Weinberg equilibrium, results for 11 of the 16 loci in the two population samples agreed. Using a chi-square test to compare allele frequencies between blood and oral isolates for each of the 16 loci, no significant differences were detected (P > 0.9 in all cases). Seven multilocus genotypes identified in the sample of blood isolates were identical to genotypes identified in the sample of oral isolates. The DNA fingerprint of each of these isolates was determined with the species-specific probe 27A (12). Of these seven blood isolates, only one had a DNA fingerprint identical to that of an oral isolate with the same multilocus genotype. A total of 96 unique multilocus genotypes were detected among the 125 strains in the combined population samples. The reference strains CA30 and CA87 each had unique multilocus genotypes within both samples of isolates.
TABLE 1.
Genotype counts
| Locus (accession no.) | Polymorphic nucleotide positionb | Genotype (no. of strains)c |
|---|---|---|
| C15F2a (Y07668) | 174 | AA (8), AG (23), GG (10) |
| C12F10a (Y07664) | 218 | GG (35), GT (6), TT (0) |
| C2F7a (Y07669) | 95 | CC (7), CT (2), TT (32)*** |
| C2F10a (Y07666) | 151 | AA (18), AG (9), GG (14)*** |
| CHS2 (M82937) | 2055 | CC (39), CT (0), TT (2)*** |
| C2F17a (Y07665) | 206 | CC (18), CT (14), TT (9) |
| ARG4 (L25051) | 1789 | AA (12), CC (5), TT (15), AC (7), AT (2), CT (0)*** |
| CPH1 (U15152) | 1204 | AA (5), AT (22), TT (14) |
| FAS2 (L29063) | 4773 | AA (1), AG (19), GG (21) |
| GCN1 (S14D5)d | 145 | AA (2), AC (6), CC (33)** |
| PDE1 (L12045) | 1046 | AA (10), AG (25), GG (6) |
| LEU2 (AF000121) | 190 | CC (14), CT (16), TT (11) |
| CZF1 (M76586) | 728 | AA (34), AG (5), GG (2)** |
| HEX1 (L26488) | 2089 | CC (23), CT (4), TT (14)*** |
| MNS1 (265162G08.y1.seq)d | 362 | CC (2), CT (12), TT (27) |
| ERG7 (L04305) | 294 | ++ (7), +− (19), −− (15)e |
Sequences from Gräser et al. (6).
Polymorphic positions are relative to the accessioned sequences.
Diploid genotypes are followed by the number of strains in parentheses. In all cases, the total number of C. albicans strains assayed was 41. **, counts significantly different from Hardy-Weinberg expectation at P < 0.05; ***, counts significantly different from Hardy-Weinberg expectation at P < 0.001.
Accession number for the Stanford Candida albicans Sequencing Project (http://alces.med.umn.edu/candida/geneinfo.html). Other accession numbers are for GenBank.
This polymorphism is for the presence, indicated as a plus (+), in the genotypes or the absence (−) of a six-nucleotide indel (AAAGTG).
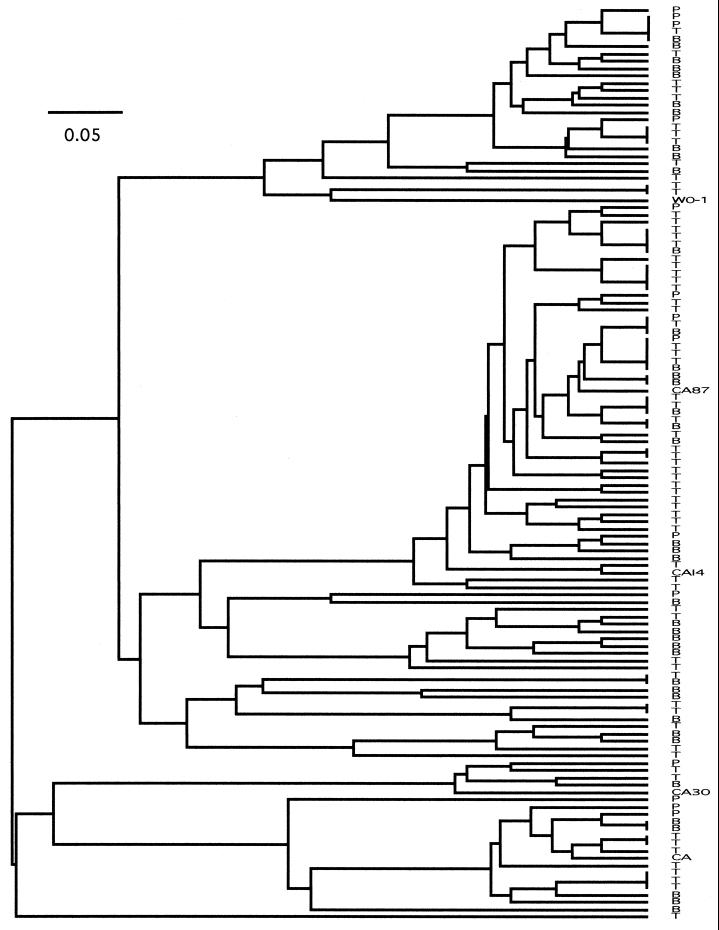
The similarity among multilocus genotypes for the 125 strains was assessed by the unweighted pair-group method with arithmetic mean (UPGMA) in PAUP, version 4. 0b2 (D. L. Swofford, Smithsonian Institution). The distance between pairs of genotypes was calculated as the sum of the numbers of allelic differences for each locus. For example, at locus 1 the distance between AA and GG is two, the distance between AG and GG is one, and the distance between AG and AA is one. UPGMA was used as the tree-building algorithm because it graphically represents similarity among genotypes and can be used without reference to any specific model of evolution. In this tree the genotypes of blood isolates were finely interspersed among genotypes from oral mucosa. Also, both reference isolates were distributed within clusters of blood and oral isolates, despite their geographically disparate origin (Fig. 1).
FIG. 1.
UPGMA tree of genotypic similarity among the 41 isolates examined in this study and the 84 isolates examined by Cowen et al. (5). Blood isolates are designated with the letter B and oral isolates are designated with either the letter P or T. The horizontal bar represents similarity as the fraction of the total range from 0 (no similarity) to 1 (identical genotypes).
In the original sample of blood isolates obtained from patients, there were two additional isolates which were highly atypical relative to the remaining isolates and were subsequently excluded from this study. For these atypical isolates most regions failed to amplify or failed to hybridize to the allele-specific probes; these two atypical isolates were later identified as Candida dubliniensis at the Toronto Public Health Laboratory. Atypical isolates were also identified at approximately the same frequency in the sample of oral isolates and were subsequently reidentified as other Candida species (5).
The results of this study are fully consistent with the hypothesis that the trait of invasiveness is widespread in C. albicans in humans. Using a multilocus genotyping system based on nucleotide polymorphisms, we detected high clonal diversity among both the oral isolates and the blood isolates but no significant differences in allele frequencies between the two samples. Also, in the dendrogram of genotypic similarity, the genotypes of the blood isolates were finely interspersed among the genotypes of the oral isolates. Furthermore, the identification of multilocus genotypes shared by both blood and oral isolates, as well as one blood isolate having both a DNA fingerprint and multilocus genotype identical to one oral isolate, provides strong evidence for one common pool of genotypes of this fungus. These observations are inconsistent with the hypothesis that the trait of invasiveness occurs rarely or is restricted to only a small fraction of the multilocus genotypes occurring in superficial sites, such as oral mucosa. The results suggest that other factors, such as the host immune system, may play a more important role than genotype in determining the invasive potential of an isolate. While our data do not address the nature of the microevolutionary changes (including gain or loss of phenotypic characteristics) that may be associated with the transition into the bloodstream, we know from experimental populations that other phenotypic characteristics are highly labile (4).
Our results are also consistent with the model of short-sighted evolution proposed by Levin and Bull (7) in which a pathogen undergoes mutation (used here in the broadest sense to include all heritable genetic changes) and clonal selection within a host and then enters and exploits microenvironments not normally available to the ancestral genotype. Evolution in these microenvironments offers no advantage to the pathogen in transmission between hosts, because the lineage is not perpetuated beyond the host. Testing whether this model is applicable to C. albicans would require (i) fine-scale sampling of the pathogen in superficial sites and in the bloodstream of the hosts, (ii) detection of the genetic variability arising with such microevolution, and (iii) fitness comparisons of isolates from superficial sites and from the bloodstream in different sites of the host. Although such detailed tests of this model have not yet been conducted for C. albicans, our results do suggest that the potential for invasiveness is widespread in this opportunistic pathogen. The potential of C. albicans to exploit a broad range of hostile host environments, including the transition between superficial sites, such as oral mucosa, and the bloodstream of patients, is consistent with other evidence of rapid adaptation by this fungus, for example, the emergence of azole resistance in clonal isolates from patients (16) and in experimental populations (4).
Acknowledgments
This work was supported by Research Grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada to L.M.K. and J.B.A. and by a grant-in-aid from Pfizer Canada Inc.
REFERENCES
- 1.Brawner D L, Cutler J E. Oral Candida albicans isolates from nonhospitalized normal carriers, immunocompetent hospitalized patients, and immunocompromised patients with or without acquired immunodeficiency syndrome. J Clin Microbiol. 1989;27:1335–1341. doi: 10.1128/jcm.27.6.1335-1341.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole G T, Lynn K T, Seshan K R. An animal model for oropharyngeal, esophageal and gastric candidosis. Mycoses. 1990;33:7–19. doi: 10.1111/myc.1990.33.1.7. [DOI] [PubMed] [Google Scholar]
- 3.Cole G T, Seshan K R, Pope L M, Yancey R J. Morphological aspects of gastrointestinal tract invasion by Candida albicans in the infant mouse. J Med Vet Mycol. 1988;26:173–185. [PubMed] [Google Scholar]
- 4.Cowen L E, Sanglard D, Calabrese D, Sirjusingh C, Anderson J B, Kohn L M. Evolution of drug resistance in experimental populations of Candida albicans. J Bacteriol. 2000;182:1515–1522. doi: 10.1128/jb.182.6.1515-1522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowen L E, Sirjusingh C, Summerbell R C, Walmsley S, Richardson S, Kohn L M, Anderson J B. Multilocus genotypes and DNA fingerprints do not predict variation in azole resistance among clinical isolates of Candida albicans. Antimicrob Agents Chemother. 1999;43:2930–2938. doi: 10.1128/aac.43.12.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gräser Y, Volovsek M, Arrington J, Schönian G, Presber W, Mitchell T G, Vilgalys R. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci USA. 1996;93:12473–12477. doi: 10.1073/pnas.93.22.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin B R, Bull J J. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol. 1994;2:76–81. doi: 10.1016/0966-842x(94)90538-x. [DOI] [PubMed] [Google Scholar]
- 8.Lott T J, Holloway B P, Logan D A, Fundyga R, Arnold J. Towards understanding the evolution of the human commensal yeast Candida albicans. Microbiology. 1999;145:1137–1143. doi: 10.1099/13500872-145-5-1137. [DOI] [PubMed] [Google Scholar]
- 9.Merz W G. Candida albicans strain delineation. Clin Microbiol Rev. 1990;3:321–334. doi: 10.1128/cmr.3.4.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odds F C, Abbott A B, Stiller R L, Scholer H J, Polak A, Stevens D A. Analysis of Candida albicans phenotypes from different geographical and anatomical sources. J Clin Microbiol. 1983;18:849–857. doi: 10.1128/jcm.18.4.849-857.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfaller M A, Lockhart S R, Pujol C, Swails W J, Messer S A, Edmond M B, Jones R N, Wenzel R P, Soll D R. Hospital specificity, region specificity, and fluconazole resistance of Candida albicans bloodstream isolates. J Clin Microbiol. 1998;36:1518–1529. doi: 10.1128/jcm.36.6.1518-1529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scherer S, Stevens D A. A Candida albicans dispersed, repeated gene family and its epidemiologic applications. Proc Natl Acad Sci USA. 1988;85:1452–1456. doi: 10.1073/pnas.85.5.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmid J, Odds F C, Wiselka M J, Nicholson K G, Soll D R. Genetic similarity and maintenance of Candida albicans strains from a group of AIDS patients, demonstrated by DNA fingerprinting. J Clin Microbiol. 1992;30:935–941. doi: 10.1128/jcm.30.4.935-941.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soll D R, Galask R, Schmid J, Hanna C, Mac K, Morrow B. Genetic dissimilarity of commensal strains of Candida spp. carried in different anatomical locations of the same healthy women. J Clin Microbiol. 1991;29:1702–1710. doi: 10.1128/jcm.29.8.1702-1710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens D A, Odds F C, Scherer S. Application of DNA typing methods to Candida albicans epidemiology and correlations with phenotype. Rev Infect Dis. 1990;12:258–266. doi: 10.1093/clinids/12.2.258. [DOI] [PubMed] [Google Scholar]
- 16.White T C, Marr K A, Bowden R A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Boyd C M, Livingston E, Meyer W, Madden J F, Mitchell T G. Species and genotypic diversities and similarities of pathogenic yeasts colonizing women. J Clin Microbiol. 1999;37:3835–3843. doi: 10.1128/jcm.37.12.3835-3843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J, Mitchell T G, Vilgalys R. PCR-restriction fragment length polymorphism (RFLP) analyses reveal both extensive clonality and local genetic differences in Candida albicans. Mol Ecol. 1999;8:59–73. doi: 10.1046/j.1365-294x.1999.00523.x. [DOI] [PubMed] [Google Scholar]