Abstract
Purpose of review
Traditional histopathology of the kidney biopsy specimen has been an essential and successful tool for the diagnosis and staging of kidney diseases. However, it is likely that the full potential of the kidney biopsy has not been tapped so far. Indeed, there is now a concerted worldwide effort to interrogate kidney biopsy samples at the cellular and molecular levels with unprecedented rigor and depth. This review examines these novel approaches to study kidney biopsy specimens and highlights their potential to refine our understanding of the pathophysiology of kidney disease and lead to precision-based diagnosis and therapy.
Recent findings
Several consortia are now active at studying kidney biopsy samples from various patient cohorts with state-of-the art cellular and molecular techniques. These include advanced imaging approaches as well as deep molecular interrogation with tools such as epigenetics, transcriptomics, proteomics and metabolomics. The emphasis throughout is on rigor, reproducibility and quality control.
Summary
While these techniques to study kidney biopsies are complementary, each on its own can yield novel ways to define and classify kidney disease. Therefore, great efforts are needed in order to generate an integrated output that can propel the diagnosis and treatment of kidney disease into the realm of precision medicine.
Keywords: Kidney biopsy, transcriptomics, large-scale imaging, tissue cytometry
Introduction
Since Nils Alwall performed the first aspiration needle kidney biopsy in 1944, traditional histopathology remains the cornerstone for biopsy-based diagnosis and classification of kidney disease (1, 2). Features of renal histopathology (e.g. fibrosis, crescents) can also guide therapy. Nevertheless, the response to therapy and clinical courses of individual patients are often unpredictable. Indeed, patients with identical diagnoses and similar pathologic features on their kidney biopsy specimens can have very dissimilar clinical courses. Therefore, a deeper examination of kidney tissue may reveal novel signatures at the cellular and molecular levels that allow a more detailed classification of renal disease. Such classification may explain the variable clinical courses of individual patients and allow for precision-based diagnostic and therapeutic approaches.
When properly handled and stored, a kidney biopsy specimen contains a wealth of information at the morphologic, cellular and molecular levels. Advances in large-scale imaging and tissue cytometry offer a novel view of kidney tissue that is both comprehensive and quantitative. In addition to classifying epithelial, stromal, immune and vascular cell types in situ, large-scale imaging techniques can also identify structural relationships in the complex architecture of the kidney. These structural relationships and their alteration in disease states can be key for a deeper understanding of renal pathophysiology. Similarly, molecular interrogation with tools such as single cell/nuclear/spatial transcriptomics, epigenetic modifications, regional proteomics and metabolomics will also refine our diagnostic spectrum and identify novel biomarkers and therapeutic targets (3**). It is hoped that the coordinated efforts of various consortia will lead to an integrated output that redefines the diagnosis and therapy of renal disease based on a spatially anchored multiplexed cellular and molecular reading of the kidney biopsy.
Text of Review
Novel technologies to interrogate kidney biopsy samples
A number of novel technologies have been utilized in kidney tissue to better understand the pathogenesis of kidney disease (Table 1). These technologies are summarized below, spanning from those which interrogate the kidney on the regional or structural level to the single cell level with and without the spatial context.
Table 1:
Renal biopsy specimen interrogation technologies
| Technology | DNA, RNA, Protein, metabolite | Tissue preservation | Spatial data (2D/3D) | Advantages | References |
|---|---|---|---|---|---|
| Sc/snRNA seq | RNA | OCT Frozen or Cryostor preservative | No | single cell gene expression at high resolution | [11–13, 15] |
| ATAC-seq | DNA | OCT Frozen | No | Regulation of gene expression at cell level | [15, 40] |
| Regional transcriptomics | RNA | OCT frozen | Yes (2D) | High sequencing depth for specific renal region | [28, 29, 35] |
| Regional proteomics | Protein | OCT frozen | Yes (2D) | Agnostic proteomics 3000–5000 proteins for specific renal regions | [35] |
| Spatial transcriptomics | RNA | OCT frozen | Yes (2D) | High sequencing depth with spatial resolution | [14–16, 27] |
| Spatial metabolomics | metabolites | Liquid nitrogen | Yes (2D) | Regional distribution of metabolites | [35] |
| 3D tissue Cytometry | Protein | OCT frozen | Yes (3D) | 8-plex, cell distributing and abundance, 3D neighborhood analysis | [15, 17, 21] |
| CODEX | Protein | OCT frozen | Yes (2D) | Multiplexing 10–60 probes | [14, 35] |
| IMC | Protein | FFPE | Yes (2D) | Multiplexing 10–60 probes | [25] |
| WGBS | DNA | OCT frozen | Yes (2D) | DNA methylation regulates gene expression across the genome | [3, 38] |
| CUT & RUN | DNA | OCT frozen or Liquid nitrogen | No | Assesses active, poised, and silent chromatin regions | [42] |
Single cell single nuclear RNA sequencing: Sc/snRNA seq; Assay for Transposase-Accessible Chromatin using sequencing: ATAC-seq; Digital Pathology: Digital Path; Co-detection by Indexing: CODEX; Spatial transcriptomics: ST; Imaging mass cytometry: IMC.
Digital pathology: Whole slide imaging and digitization is steadily transforming the field of pathology (4, 5). The availability of digital images on a cloud server for sharing and analysis have opened the way for image-based analytics and implementation of novel computational methods for automated segmentation and feature extraction. Some of the early efforts are focusing on segmenting structures such as glomeruli, tubules and the interstitium, which are also typically assessed by standard pathology (6–10). As novel machine learning tools are implemented, it is expected that sub-visual features could be automatically identified and linked with other molecular phenotypes. Hence, standard histopathology images could carry imaging-based biomarkers that will be potentially linked with molecular pathophysiology and clinical outcomes (5). Therefore, the field of pathology would further expand from diagnostics towards discovery.
Single cell/ single nuclear RNA sequencing (Sc/snRNAseq): Driven by advances in microfluidics, cell-based RNA sequencing has revolutionized medicine and science by generating gene expression data at the level of the cells. Consequently, these technologies allow cataloging of molecularly defined cell populations and subpopulations in health, and the discovery of novel cell states induced by injury (11–15**). For example, recent data from the Kidney Precision Medicine Project consortium (KPMP) suggest the presence of various cell states (e.g. altered, degenerative, repairing) of proximal and thick ascending limb cells that could be associated with kidney disease progression (15**). Such important information at the cell level is generated from dissociated tissue, but can also be spatially anchored using other technologies, thereby enhancing our understanding of the pathogenesis of kidney disease, particularly in the heterogenous microenvironments within the human kidney (14–16).
Large scale 3D imaging and tissue cytometry: This is an imaging-based technology that relies on label free imaging with second harmonic generation for detecting fibrosis and confocal multiplexed fluorescence microscopy with optical sectioning to perform 3D imaging of a thick kidney section (17–21). Such imaging is followed by tissue cytometry analysis to survey and classify all cells based on fluorescent labels or other parameters such as spatial coordinates or neighborhood membership. Tissue cytometry is typically performed using a variety of specialized software. We have recently developed a custom open-source software tool (Volumetric Tissue Exploration and Analysis, VTEA) that allows the interactive quantitative exploration of 3D imaging data (17, 20, 21). VTEA analysis can provide quantitative measures about cell abundance and distribution, and offers spatial anchoring of cell-cell or cell- structure interactions based on spatial proximity. This could be performed in a supervised and unsupervised manner (18). In addition, the 3D aspect of the imaging allows better recognition of morphological changes and classification of injury, with a number of surveyed cells ranging between 100,000 to 250,000 per kidney thick section. Consequently, the data output with all the captured features results in a big data set, yielding findings with high confidence at the tissue level (17, 18). With advancement in enhanced tissue clearing techniques and light sheet microscopy, it is possible to image thicker sections of kidney tissue, which would allow a better capture of the morphological changes in kidney disease (20, 22). However, systematic implementation of such an approach will require specialized efforts to integrate segmentations of structures and morphometric analysis in 3D, and it will likely be costly from a computational standpoint.
Co- detection by indexing (CODEX): This technology expands the ability to multiplex markers on 10 μm-thick tissue sections using DNA-conjugated antibodies (23). With CODEX imaging, these antibodies are revealed three at a time by the reversible binding of fluorescent oligonucleotide reporters. Following imaging, the fluorescent reporters are stripped from the tissue and replaced with a second set of three probes and imaged again. This process is repeated until all the antibodies in the tissue have been revealed. Images of DAPI-labeled nuclei are collected in each round to enable registration of images into a single highly multiplexed image (23). More than 40 antibodies can be used in this process, and the analysis can be quite informative to determine cell type and subtype, albeit in the 2D space.
Imaging Mass Cytometry (IMC): This approach relies on chemical imaging whereby antibodies against specific targets are conjugated to various heavy metals which can be ionized using laser ablation at a 1 μm diameter spot size. Measuring the amount of ions is performed using time of flight mass spectrometry (24). Therefore, the metal isotopes associated with each spot are simultaneously measured and indexed against the location of each spot. Line scanning is performed across the tissue, and sequential scan lines will yield an intensity map of all target proteins throughout the tissue or the region of interest. This technology is performed on formalin fixed paraffin embedded tissue and has been successfully used in kidneys by Singh et al. using 22 different markers (25). IMC can also provide a powerful multiplexing potential for > 40 markers and be very useful in detecting cell types and subtypes spatially within the kidney.
Spatial transcriptomics: Spatial transcriptomics technologies provide whole transcriptome mRNA expression with localization. Sections of frozen or formalin-fixed paraffin embedded kidney tissue are permeabilized on a specialized slide. RNA diffuses to barcoded beads underlying the tissue, after which a cDNA library is prepared and sequenced. The barcodes allow reconstruction of the spatial distribution of mRNA, aligning each unique molecular identifier to a “spot”. Multiple spatial transcriptomic platforms are available. The Visium spatial gene expression platform facilitates the mapping of over 20,000 genes detected per sample and up to 3,000 genes per spot directly over a histologic image stained with hematoxylin and eosin. This allows direct correlation of mRNA expression with renal pathology (26). Presently, the spot sizes are 55 μm in diameter. Thus, multiple cell types contribute to the expression signature of each spot. In contrast, Slide-seqV2 has greater resolution with spot sizes of 10 μm diameter or nearly single cell resolution (27*), but histology is obtained from a consecutive section. Both technologies can localize clusters and cell types derived from single cell technologies by integrating their transcriptomic signature with that of spatial transcriptomics.
Regional transcriptomics: This technology uses laser microdissection (LMD) to capture spatially defined regions in the kidney and perform transcriptomic analysis with RNA sequencing (28, 29). Using markers such as fluorescently conjugated small molecules or antibodies targeting specific cell types, LMD can faithfully dissect specific nephronal segments such as glomeruli, proximal tubules, thick ascending limbs, distal tubules/collecting duct, and the interstitium. A pipeline for RNA extraction, cDNA library generation and sequencing has been established leading to high depth sequencing data that can be used to uncover the transcriptomic signature of each of these renal regions in health and disease (28, 29). Such information could provide a spatial anchor to data obtained by dissociative technologies such as Sc/snRNA seq.
Metabolomics : Kidney molecular imaging with spatial metabolomics can be achieved using matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI) (30). Fresh-frozen kidney sections are coated with an organic matrix that facilitates desorption and ionization of endogenous molecules, and serially probed with a laser to attain mass spectral information at predefined locations. Molecular annotation coupled with data visualization and co-registration with other optical images is an important component of data processing and can be accomplished by software tools such as the METASPACE platform (developed by the EMBL) (31).
Regional proteomics: This technology delivers detailed characterization of the kidney proteome in health and disease, identifying protein markers that reflect each segment, matrix, and cell type within the tissue. LMD followed by liquid chromatography tandem mass spectrometry (LC-MS/MS) allows for direct analysis of complex protein mixtures to generate an agnostic global profile of glomerular and tubulointerstitial (TI) proteins (32–34). Both cellular and extracellular proteins are identified. The glomeruli and TI are obtained after structural identification by light microscopy on the LMD instrument. After protein extraction, peptide identification is done using mass spectrometry. Regional proteomics offers deep protein signatures at compartment level resolution that extends other spatially anchored technologies. When combined with technologies such as spatial transcriptomics, simultaneous gene and protein expression changes can be determined in the same sample, thereby identifying high confidence targets in health and disease (35).
Epigenetic interrogation methods – Gene expression is tightly regulated and adapts to physiological stresses and injury in kidney disease. Activation or silencing of gene transcription can be regulated by changes in the chromatin landscape. Three epigenomic technologies which facilitate the measurement of active, silent, or poised chromatin include Whole Genome Bisulfite Sequencing (WGBS), Cleavage Under Targets and Release Using Nuclease (CUT&RUN), and Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq). WGBS measures DNA methylation across the genome. Methylation signatures can be altered by developmental programming in utero (36, 37), metabolic or environmental insults (38), or kidney injury and disease (39). CUT&RUN lends itself to kidney biopsy sample interrogation as it can define the chromatin landscape with genome-wide mapping in a small amount of tissue as compared to Chromatin immunoprecipitation with sequencing (ChIP-seq). Chromatin profiles with histone modifications are assessed such as those of H3K27me3 (often silencing) and H3K27ac (often activating). Finally, single cell ATAC-seq identifies areas of open chromatin in specific cell clusters (15, 40). These regions represent potentially active regulatory regions; however, silenced regulatory regions often remain unidentified due to a lack of reads in these regions. Each of these three technologies can help to explain gene expression regulation within the kidney.
Kidney Precision Medicine Project (KPMP) –
One of the important challenges when applying various state of the art technologies on kidney tissue samples is to integrate the data obtained in a meaningful and interpretable manner that could provide actionable information (3**, 15**, 35, 41). For example, novel cell types and cell states uncovered by one technology should be spatially anchored and linked to a logical site within the kidney microenvironment that could explain the changes induced by disease (15). Also, these findings are optimally connected to changes in pathology and possibly others such as injury markers, signaling pathways and metabolites. Ultimately, changes at the molecular, cell and tissue levels should be linked with enhanced clinical phenotypes of the disease. The array of technologies that are used in the KPMP consortium allows Integration at various biological units: at the single cell, structure and biopsy levels (Figure 1). Integration can also occur in various ways, and this is facilitated by the fact that many technologies are performed from contiguous sections of the same biopsy core specimen (15**). Therefore, integration theoretically can occur 1) by spatial registration, which superimposes information from one technology to another based on sharing the same tissue space; 2) by alignment in the analytical space, which could allow data from various technologies to be analyzed concurrently once they are converted to a common denominator (if feasible) and 3) by a systems biology approach where dynamic pathway enrichment analyses and network mapping to biological processes critical to renal pathophysiology can be done concurrently with various technologies. A big challenge in this area is the required domain expertise in multiple technologies and in computational methods of integration across various technologies. For this reason, a large consortium such as KPMP is indeed needed, where collaborative expertise from various domains converge with clinical and pathology expertise (3**, 41).
Figure 1:
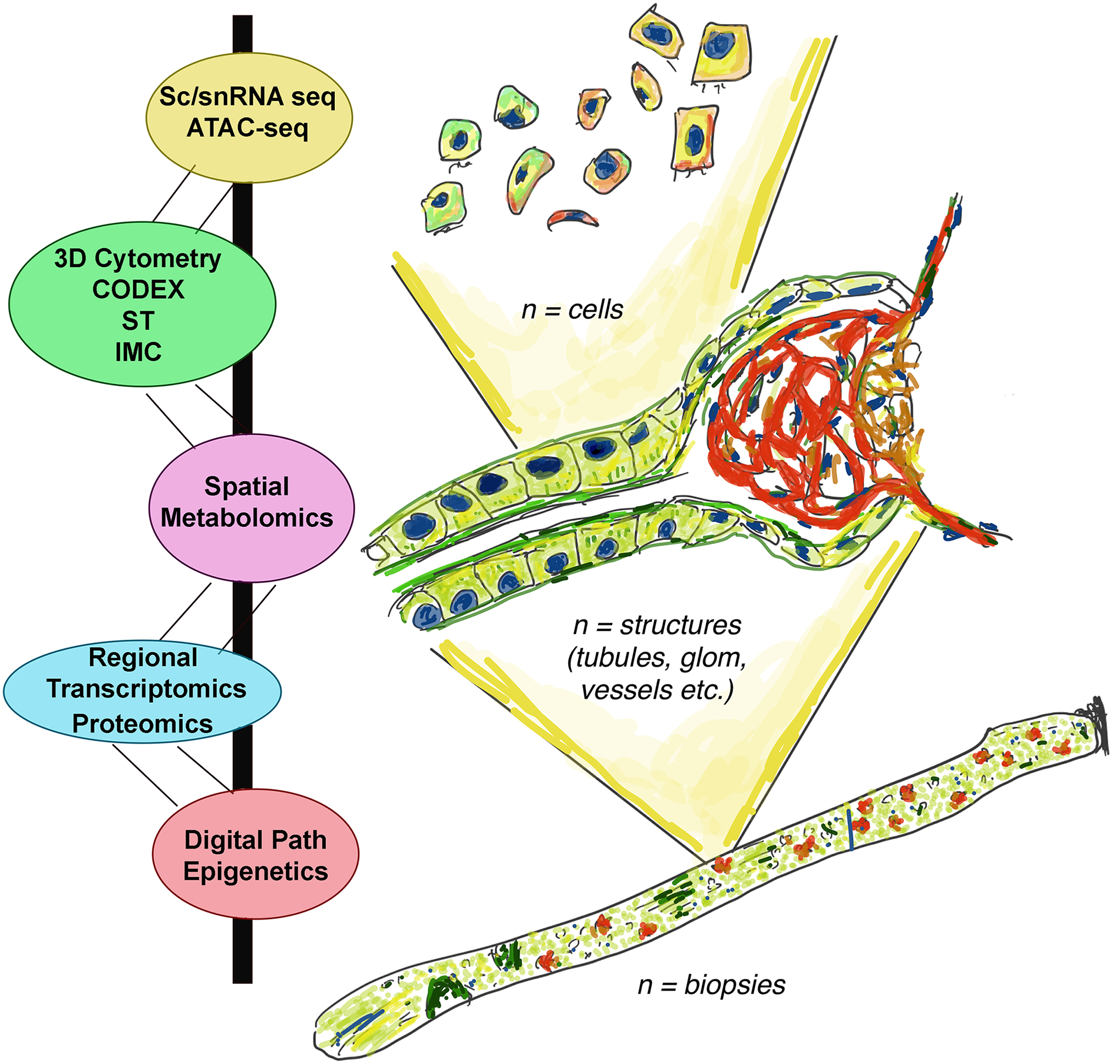
Technology integration in the KPMP.
Schematic of integration of various technologies used in the KPMP consortium at the various biological units within the kidney: cells, structures, entire biopsy specimens. Single cell single nuclear RNA sequencing: Sc/snRNA seq; Assay for Transposase-Accessible Chromatin using sequencing: ATAC-seq; Digital Pathology: Digital Path; Co-detection by Indexing: CODEX; Spatial transcriptomics: ST; Imaging mass cytometry: IMC.
Consortium contributions
Other important consortia and groups have sought to better understand the pathogenesis of kidney disease through biopsy specimen or nephrectomy interrogation. Often, each has a specific disease focus or uses different approaches to acquire and study kidney tissue. The Chan-Zuckerberg initiative’s Human Cell Atlas (HCA) and the NIH’s Human BioMolecular Atlas Program (HuBMAP) both seek to establish a reference atlas of all cells (HCA) and organs (HuBMAP) within the body. Several groups have adopted multi-omics approaches to study diabetic kidney disease, including the Transformative Research In DiabEtic NephropaThy (TRIDENT) consortia, the European cDNA bank, and new molecular studies in the Pima Indian diabetic kidney disease cohort. Biopsy interrogation of glomerulonephritidies has been undertaken by the Nephrotic Syndrome Study Network (NEPTUNE), Cure Glomerulonephropathy Network (CureGN), the European cDNA bank, and the Accelerating Medicines Partnership (AMP)-RA/SLE consortia. It is not possible to summarize all of the major contributions these groups have made to the understanding of renal disease pathogenesis here. However, brief introductions and key findings are provided below to help orient the reader to the immense body of literature available (Table 2).
Table 2:
Major consortia dedicated to renal biopsy specimen interrogation
| Kidney biopsy specimen interrogation group | Disease state focus | Unique features | References |
|---|---|---|---|
| Kidney Precision Medicine Project (KPMP) | AKI, CKD | Prospective study using protocol biopsies in subjects with AKI and CKD who may not have clinical indications for a biopsy. | [3, 41] |
| Transformative Research In DiabEtic NephropaThy (TRIDENT) | Diabetic Kidney Disease | Prospective observational cohort study of patients with diabetic kidney disease undergoing clinically indicated biopsies interrogated with multi-omics approaches. | [43–46] |
| Nephrotic Syndrome Study Network (NEPTUNE) | MCD, FSGS, MN | A prospective cohort of subjects with nephrotic syndrome with molecular kidney biopsy interrogation and long-term observation of 3–10 years. | [13, 47, 48] |
| PIMA Indian studies | Diabetic Kidney Disease | An extensively characterized cohort of diabetic kidney disease followed for over 50 years. | [49, 50] |
| Cure Glomerulonephropathy Network (CureGN) | MCD, FSGS, MN, IgA | A multi-center consortium that recruits a diverse population of glomerular disease patients. The study established a database of patients and follows clinical outcomes linked with histopathology. | [51–53] |
| Accelerating Medicines Partnership (AMP)-RA/SLE | Lupus Nephritis | An NIH and pharmaceutical partnership dedicated to define disease-specific pathways of rheumatoid arthritis and lupus. Molecular interrogation of circulating and resident kidney cells is performed. | [54, 55] |
| European cDNA bank | Diabetic Kidney Disease and other disease states | A multicenter European archive of human kidney biopsies specially processed for transcriptomic analysis. | [56, 57] |
| Human BioMolecular Atlas Program (HuBMAP) | Healthy reference | HuBMAP aims to develop a molecular map of the human body at the cellular level, with spatial context. | [58, 59] |
| Studies in transplantation1 | Acute and chronic rejection | While not a single consortium, these studies sought to understand the pathogenesis of allograft failure | [60–65] |
| Chan-Zuckerberg Initiative (Human Cell Atlas) | Healthy reference | CZI seeks to create a comprehensive, open-reference map of every cell type in the body. | [66] |
Studies in transplantation does not refer to a consortium, but recognizes seminal work in renal transplant biopsy specimen interrogation.
AKI – acute kidney injury, CKD – chronic kidney disease, MCD – minimal change disease, FSGS – focal segmental glomerulosclerosis, MN – membranous nephropathy, IgA – Immunoglobulin A nephropathy, RA – rheumatoid arthritis, LSE – systemic lupus erythematosis.
Conclusion
The novel interrogation techniques described above are ushering in a new era in the diagnosis and treatment of kidney disease. At this early stage, these interrogation techniques are best implemented and validated though organized large consortia. This is essential for rigor, validation, quality control and integration. It is hoped that the output from these consortia will redefine readouts from kidney biopsies and propel nephrology into the realm of precision medicine.
Key points:
Kidney biopsy samples have and untapped potential that can be realized through novel interrogation techniques.
These imaging, cellular and molecular techniques are best investigated through organized consortia that allow for validation and integration
Novel interrogation techniques of the kidney tissue will reshape the diagnosis and classification of kidney disease and point the way to precision renal medicine.
Acknowledgements:
The authors would like to acknowledge the National Institute of Diabetes and Digestive and the Kidney Diseases (NIDDK) Kidney Precision Medicine Project (KPMP), (www.kpmp.org).
Financial support and sponsorship:
This work is funded under NIH award number UH3DK114923.
Footnotes
Conflicts of interest:
none
References
- 1.Luciano RL, Moeckel GW. Update on the Native Kidney Biopsy: Core Curriculum 2019. Am J Kidney Dis. 2019;73(3):404–15. [DOI] [PubMed] [Google Scholar]
- 2.Kark RM, Muehrcke RC. Biopsy of kidney in prone position. Lancet. 1954;266(6821):1047–9. [DOI] [PubMed] [Google Scholar]
- 3.**.El-Achkar TM, Eadon MT, Menon R, et al. A multimodal and integrated approach to interrogate human kidney biopsies with rigor and reproducibility: guidelines from the Kidney Precision Medicine Project. Physiological genomics. 2021;53(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This article introduces the rationale, protocols, and quality control metrics of the interrogation technologies of the kidney precision medciine project
- 4.Barisoni L, Hodgin JB. Digital pathology in nephrology clinical trials, research, and pathology practice. Curr Opin Nephrol Hypertens. 2017;26(6):450–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barisoni L, Lafata KJ, Hewitt SM, et al. Digital pathology and computational image analysis in nephropathology. Nat Rev Nephrol. 2020;16(11):669–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg AZ, Palmer M, Merlino L, et al. The Application of Digital Pathology to Improve Accuracy in Glomerular Enumeration in Renal Biopsies. PLoS One. 2016;11(6):e0156441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.*.Jayapandian CP, Chen Y, Janowczyk AR, et al. Development and evaluation of deep learning-based segmentation of histologic structures in the kidney cortex with multiple histologic stains. Kidney Int. 2021;99(1):86–101. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This article tests the utility of automated segmentation in whole slide images stained with Hematoxylin & Eosin, Periodic Acid Schiff, Silver, and Trichrome.
- 8.Hermsen M, de Bel T, den Boer M, et al. Deep Learning-Based Histopathologic Assessment of Kidney Tissue. J Am Soc Nephrol. 2019;30(10):1968–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Y, Cassol CA, Jung S, et al. Deep-Learning-Driven Quantification of Interstitial Fibrosis in Digitized Kidney Biopsies. Am J Pathol. 2021;191(8):1442–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginley B, Lutnick B, Jen KY, et al. Computational Segmentation and Classification of Diabetic Glomerulosclerosis. J Am Soc Nephrol. 2019;30(10):1953–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lake BB, Chen S, Hoshi M, et al. A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat Commun. 2019;10(1):2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson PC, Wu H, Kirita Y, et al. The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc Natl Acad Sci U S A. 2019;116(39):19619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menon R, Otto EA, Hoover P, et al. Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI Insight. 2020;5(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferreira RM, Sabo AR, Winfree S, et al. Integration of spatial and single-cell transcriptomics localizes epithelial cell-immune cross-talk in kidney injury. JCI Insight. 2021;6(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.**.Lake BB, Menon R, Winfree S, et al. An atlas of healthy and injured cell states and niches in the human kidney. bioRxiv. 2021:2021.07.28.454201. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This manuscript is the atlas kidney precision medicine project atlas paper, introducing definitions of cell states in injury with their localization in kidney.
- 16.Janosevic D, Myslinski J, McCarthy TW, et al. The orchestrated cellular and molecular responses of the kidney to endotoxin define a precise sepsis timeline. Elife. 2021;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferkowicz MJ, Winfree S, Sabo AR, et al. Large-scale, three-dimensional tissue cytometry of the human kidney: a complete and accessible pipeline. Laboratory investigation; a journal of technical methods and pathology. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winfree S, Al Hasan M, El-Achkar TM. Profiling immune cells in the kidney using tissue cytometry and machine learning. Kidney 360. 2021: 10.34067/KID.0006802020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winfree S, Dagher PC, Dunn KW, et al. Quantitative Large-Scale Three-Dimensional Imaging of Human Kidney Biopsies: A Bridge to Precision Medicine in Kidney Disease. Nephron. 2018;140(2):134–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winfree S, Ferkowicz MJ, Dagher PC, et al. Large-scale 3-dimensional quantitative imaging of tissues: state-of-the-art and translational implications. Transl Res. 2017;189:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winfree S, Khan S, Micanovic R, et al. Quantitative Three-Dimensional Tissue Cytometry to Study Kidney Tissue and Resident Immune Cells. J Am Soc Nephrol. 2017;28(7):2108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jafree DJ, Long DA, Scambler PJ, et al. Tissue Clearing and Deep Imaging of the Kidney Using Confocal and Two-Photon Microscopy. Methods Mol Biol. 2020;2067:103–26. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy-Darling J, Bhate SS, Hickey JW, et al. Highly multiplexed tissue imaging using repeated oligonucleotide exchange reaction. Eur J Immunol. 2021;51(5):1262–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giesen C, Wang HA, Schapiro D, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11(4):417–22. [DOI] [PubMed] [Google Scholar]
- 25.Singh N, Avigan ZM, Kliegel JA, et al. Development of a 2-dimensional atlas of the human kidney with imaging mass cytometry. JCI Insight. 2019;4(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melo Ferreira R, Sabo AR, Winfree S, et al. Integration of spatial and single cell transcriptomics localizes epithelial-immune cross-talk in kidney injury. JCI Insight. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.*.Stickels RR, Murray E, Kumar P, et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat Biotechnol. 2021;39(3):313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This article introduces the improved Slide-seqV2 technology which has deeper transcriptomic signatures.
- 28.Barwinska D, El-Achkar TM, Melo Ferreira R, et al. Molecular characterization of the human kidney interstitium in health and disease. Sci Adv. 2021;7(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barwinska D, Ferkowicz MJ, Cheng YH, et al. Application of Laser Microdissection to Uncover Regional Transcriptomics in Human Kidney Tissue. J Vis Exp. 2020(160). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderton CR, editor Mass Spectrometry Imaging: Methodology and Applications 2017.
- 31.Palmer A, Phapale P, Chernyavsky I, et al. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat Methods. 2017;14(1):57–60. [DOI] [PubMed] [Google Scholar]
- 32.Satoskar AA, Shapiro JP, Bott CN, et al. Characterization of glomerular diseases using proteomic analysis of laser capture microdissected glomeruli. Mod Pathol. 2012;25(5):709–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satoskar AA, Shapiro JP, Jones M, et al. Differentiating Staphylococcus infection-associated glomerulonephritis and primary IgA nephropathy: a mass spectrometry-based exploratory study. Sci Rep. 2020;10(1):17179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shapiro JP, Biswas S, Merchant AS, et al. A quantitative proteomic workflow for characterization of frozen clinical biopsies: laser capture microdissection coupled with label-free mass spectrometry. J Proteomics. 2012;77:433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen J, Sealfon R, Menon R, et al. A reference tissue atlas for the human kidney. bioRxiv. 2021:2020.07.23.216507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stangenberg S, Chen H, Wong MG, et al. Fetal programming of chronic kidney disease: the role of maternal smoking, mitochondrial dysfunction, and epigenetic modfification. Am J Physiol Renal Physiol. 2015;308(11):F1189–96. [DOI] [PubMed] [Google Scholar]
- 37.Fuhrmann L, Lindner S, Hauser AT, et al. Effects of Environmental Conditions on Nephron Number: Modeling Maternal Disease and Epigenetic Regulation in Renal Development. Int J Mol Sci. 2021;22(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gluck C, Qiu C, Han SY, et al. Kidney cytosine methylation changes improve renal function decline estimation in patients with diabetic kidney disease. Nature communications. 2019;10(1):2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi K, Hishikawa A, Itoh H. DNA Damage Repair and DNA Methylation in the Kidney. Am J Nephrol. 2019;50(2):81–91. [DOI] [PubMed] [Google Scholar]
- 40.Doke T, Huang S, Qiu C, et al. Transcriptome-wide association analysis identifies DACH1 as a kidney disease risk gene that contributes to fibrosis. J Clin Invest. 2021;131(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Boer IH, Alpers CE, Azeloglu EU, et al. Rationale and design of the Kidney Precision Medicine Project. Kidney Int. 2021;99(3):498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skene PJ, Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer MB, Abedini A, Jackson C, et al. The Role of Glomerular Epithelial Injury in Kidney Function Decline in Patients With Diabetic Kidney Disease in the TRIDENT Cohort. Kidney Int Rep. 2021;6(4):1066–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abedini A, Zhu YO, Chatterjee S, et al. Urinary Single-Cell Profiling Captures the Cellular Diversity of the Kidney. J Am Soc Nephrol. 2021;32(3):614–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hogan JJ, Owen JG, Blady SJ, et al. The Feasibility and Safety of Obtaining Research Kidney Biopsy Cores in Patients with Diabetes: An Interim Analysis of the TRIDENT Study. Clin J Am Soc Nephrol. 2020;15(7):1024–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Townsend RR, Guarnieri P, Argyropoulos C, et al. Rationale and design of the Transformative Research in Diabetic Nephropathy (TRIDENT) Study. Kidney Int. 2020;97(1):10–3. [DOI] [PubMed] [Google Scholar]
- 47.Ju W, Nair V, Smith S, et al. Tissue transcriptome-driven identification of epidermal growth factor as a chronic kidney disease biomarker. Sci Transl Med. 2015;7(316):316ra193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gadegbeku CA, Gipson DS, Holzman LB, et al. Design of the Nephrotic Syndrome Study Network (NEPTUNE) to evaluate primary glomerular nephropathy by a multidisciplinary approach. Kidney Int. 2013;83(4):749–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.*.Nelson RG, Knowler WC, Kretzler M, et al. Pima Indian Contributions to Our Understanding of Diabetic Kidney Disease. Diabetes. 2021;70(8):1603–16. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This article provides a summary of over 50 years of work in a PIMA indian population with a high rate of diabetic kidney disease.
- 50.Nair V, Komorowsky CV, Weil EJ, et al. A molecular morphometric approach to diabetic kidney disease can link structure to function and outcome. Kidney Int. 2018;93(2):439–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mariani LH, Bomback AS, Canetta PA, et al. CureGN Study Rationale, Design, and Methods: Establishing a Large Prospective Observational Study of Glomerular Disease. Am J Kidney Dis. 2019;73(2):218–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Shaughnessy MM, Troost JP, Bomback AS, et al. Treatment Patterns Among Adults and Children With Membranous Nephropathy in the Cure Glomerulonephropathy Network (CureGN). Kidney Int Rep. 2019;4(12):1725–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murphy SL, Mahan JD, Troost JP, et al. Longitudinal Changes in Health-Related Quality of Life in Primary Glomerular Disease: Results From the CureGN Study. Kidney Int Rep. 2020;5(10):1679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fava A, Buyon J, Mohan C, et al. Integrated urine proteomics and renal single-cell genomics identify an IFN-gamma response gradient in lupus nephritis. JCI Insight. 2020;5(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoover P, Der E, Berthier CC, et al. Accelerating Medicines Partnership: Organizational Structure and Preliminary Data From the Phase 1 Studies of Lupus Nephritis. Arthritis Care Res (Hoboken). 2020;72(2):233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lei Y, Devarapu SK, Motrapu M, et al. Interleukin-1beta Inhibition for Chronic Kidney Disease in Obese Mice With Type 2 Diabetes. Front Immunol. 2019;10:1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cohen CD, Kretzler M. [Gene expression analyses of kidney biopsies: the European renal cDNA bank--Kroner-Fresenius biopsy bank]. Pathologe. 2009;30(2):101–4. [DOI] [PubMed] [Google Scholar]
- 58.Hu BC. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature. 2019;574(7777):187–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.*.Neumann EK, Patterson NH, Rivera ES, et al. Highly multiplexed immunofluorescence of the human kidney using co-detection by indexing. Kidney Int. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This article is the first human kidney CODEX publication.
- 60.Wu H, Malone AF, Donnelly EL, et al. Single-Cell Transcriptomics of a Human Kidney Allograft Biopsy Specimen Defines a Diverse Inflammatory Response. J Am Soc Nephrol. 2018;29(8):2069–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moreso F, Sellares J, Soler MJ, et al. Transcriptome Analysis in Renal Transplant Biopsies Not Fulfilling Rejection Criteria. Int J Mol Sci. 2020;21(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang B, Sylvius N, Luo J, et al. Identifying Biomarkers from Transcriptomic Signatures in Renal Allograft Biopsies Using Deceased and Living Donors. Front Immunol. 2021;12:657860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Connell PJ, Zhang W, Menon MC, et al. Biopsy transcriptome expression profiling to identify kidney transplants at risk of chronic injury: a multicentre, prospective study. Lancet. 2016;388(10048):983–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hauser P, Schwarz C, Mitterbauer C, et al. Genome-wide gene-expression patterns of donor kidney biopsies distinguish primary allograft function. Laboratory investigation; a journal of technical methods and pathology. 2004;84(3):353–61. [DOI] [PubMed] [Google Scholar]
- 65.Mas VR, Archer KJ, Yanek K, et al. Gene expression patterns in deceased donor kidneys developing delayed graft function after kidney transplantation. Transplantation. 2008;85(4):626–35. [DOI] [PubMed] [Google Scholar]
- 66.Regev A, Teichmann SA, Lander ES, et al. The Human Cell Atlas. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]


