Abstract
Background:
Chloride intracellular channel 4 (CLIC4) is a multifunctional metamorphic protein with a growing body of evidence supporting a major role in the brain’s molecular and behavioral response to ethanol. Although key to understanding the functional biology underlying this role, little is known about the cellular and subcellular expression patterns of CLIC4 in brain and how they are affected by ethanol.
Methods:
qRT-PCR was used to assess Clic4 mRNA expression in medial prefrontal cortex (mPFC) of C57BL/6J mice in the absence and presence of acute ethanol exposure. Two complementary immunohistochemical techniques were employed to assess the subcellular localization of CLIC4 protein, as well as its pattern of expression across brain cell types in mPFC in the absence and presence of acute ethanol.
Results:
Through immunohistochemical and stereological techniques, we show that CLIC4 protein is robustly expressed by oligodendrocytes (most abundant), microglia, and astrocytes, with minimal expression in neurons. Following acute ethanol, we observed a rapid increase in Clic4 mRNA expression in female but not male mice and an overall increase in the number of oligodendrocytes and astrocytes expressing CLIC4 protein.
Conclusions:
These findings suggest a function for Clic4 as an early response gene for acute ethanol in brain, which likely underlies its ability to modulate ethanol behavior. Furthermore, our results suggest the role of CLIC4 in the brain’s response to ethanol is likely mediated through oligodendrocytes.
Keywords: CLIC4, ethanol, mice, qRT-PCR, immunohistochemistry
Introduction
Chloride intracellular channels are a small family of evolutionarily conserved metamorphic proteins capable of altering their three-dimensional structure under specific redox conditions (Littler et al., 2004; Littler et al., 2005). Although the functions of these proteins are still being elucidated, repeated studies have documented that one of the best characterized members of this family, CLIC4, has diverse roles in development (Chalothorn et al., 2009), apoptosis (Fernandez-Salas et al., 2002; Suh et al., 2004), and membrane trafficking (Chou et al., 2016; Maeda et al., 2008). CLIC4 has been shown to translocate to the nucleus as an early responder of cell stress (Suh et al., 2004) and to modulate transcription in the transforming growth factor beta (TGF-β) pathway (Shukla et al., 2009). The protein has also been implicated in ion channel activity and glutaredoxin-like enzymatic activity (Littler et al., 2005; Al Khamici et al., 2015) through its ability to interconvert between membrane-integrated and soluble globular states.
Clic4 expression is fairly ubiquitous in vertebrate tissues with highest expression in heart, lung, liver, kidney, and skin (Padmakumar et al., 2014). Expression in vascular tissues and kidney are thought to be related to the protein’s role in membrane trafficking and tubulogenesis (Chalothorn et al., 2009; Chou et al., 2016) whereas its expression in skin has been implicated in TGF-β signaling during wound healing (Padmakumar et al., 2012). In contrast to nearly ubiquitous expression in peripheral tissues, Clic4 expression in mouse brain is low, except in regions of neurogenesis and axonal tracts of the forebrain and cerebellum (Padmakumar et al., 2014). Its role in brain is currently unknown, but it has been identified in white matter proteomic screening (Ishii et al., 2009) and organized into myelin gene networks in genomic studies of brain tissue (Kerns et al., 2005; Bogenpohl et al., 2019), suggesting a function in oligodendrocytes. While these studies suggest CLIC4 protein is functionally expressed in brain and likely oligodendrocytes, a systematic evaluation of expression at the cellular level has not been performed. Furthermore, little is known about CLIC4 expression in other central nervous system cell types such as neurons, astrocytes, and microglia.
CLIC4 contains a nuclear localization sequence in its C-terminus and has been shown to translocate to the nucleus following TNF-α signaling and various cellular stresses including DNA damage, metabolic inhibitors, and inhibitors of transcription and translation (Suh et al., 2004; Fernandez-Salas et al., 2002). CLIC4 also has a p53 response element in its promoter and is upregulated by p53 pro-apoptotic signaling where its translocation to the nucleus accelerates apoptosis (Suh et al., 2004). Additionally, the protein has been shown to translocate to the nucleus in order to promote TGF-β signaling by stabilizing phospho-SMAD transcription factors (Malik et al., 2010; Shukla et al., 2009). Subcellular expression patterns are key to elucidating protein function and have shed much light on the role of CLIC4 in peripheral tissues. Despite these studies, CLIC4 expression characteristics in central nervous system cells have not been described, obscuring current understanding of its role in brain.
While the biological function of Clic4 in brain remains unknown, increasing evidence points to an important role in alcohol use disorder (AUD). Clic4 has been identified as an ethanol-responsive gene in several genomic studies conducted in brain tissue of mice, monkeys, and humans (Kerns et al., 2005; Mulligan et al., 2006; Liu et al., 2006; Wolen et al., 2012; Farris and Miles, 2013; Wolstenholme et al., 2011; Ponomarev et al., 2012; Bogenpohl et al., 2019). Most notably, Clic4 mRNA has been shown to be inducible by acute ethanol exposure in medial prefrontal cortex (mPFC) of DBA/2J (D2) mice (Kerns et al., 2005; Bhandari et al., 2012). The mPFC is a brain region associated with many executive functions including impulsivity, compulsivity, and decision making and is key to development of addictions, including alcohol use disorder (Klenowski, 2018). Overexpression of Clic4 in the D2 mouse mPFC and RNAi knockdown and transposon interruption of invertebrate Clic4 orthologs have been shown to alter ethanol sensitivity (Bhandari et al., 2012; Chan et al., 2014; Weston et al., 2021). These findings of altered sensitivity are remarkable in light of the correlation between low initial ethanol sensitivity and development of AUD in humans (Schuckit, 1994; Schuckit and Smith, 1996). Together this suggests a potentially important role for Clic4 in ethanol behaviors, although the cell types mediating these effects are not yet known. Additionally, while induction of Clic4 expression by ethanol has been demonstrated in D2 mice, which have a high sensitivity to ethanol, it has not been observed in other strains such as C57BL/6J (B6) mice. B6 mice show a lower sensitivity but high preference for voluntary ethanol consumption, thus representing a potentially more generalizable model organism for humans with AUD.
In this study, we extend current knowledge of CLIC4’s expression pattern in brain and its response to ethanol. We use qRT-PCR to show ethanol-responsive Clic4 expression in the mPFC of B6 mice. Two complementary methods of immunohistochemistry are employed to describe CLIC4 protein cell type-specific expression and subcellular localization. We quantify CLIC4 protein expression patterns in individual CNS cell types through double-labeling immunohistochemistry and stereology and assess the response to acute ethanol. These experiments characterize the cellular patterns of CLIC4 localization in brain (mPFC) and identify oligodendrocytes as the most likely cell type underlying its modulation of the brain’s response to ethanol.
Materials and Methods
Animals
All procedures using live animals were approved by the Virginia Commonwealth University Institutional Animal Care and Use Committee and were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
For CLIC4 qRT-PCR, nuclear translocation, and subcellular localization studies, C57BL/6J mice were obtained from Jackson Laboratories and maintained on ad libitum food and water access. For qRT-PCR studies of acute ethanol, male and female B6 mice (8–12 weeks old; n=5–6 per group) were given i.p. injections of either normal saline, 0.5, 2.0, or 4.0 g/kg ethanol in normal saline (20% v/v). 4 hours after injection, animals were euthanized by cervical dislocation and multiple brain regions including mPFC were rapidly microdissected and flash frozen in liquid nitrogen.
For nuclear translocation studies, male B6 mice (8–12 weeks old; n=3 per group) were given i.p. injections of either normal saline or 2 g/kg ethanol in normal saline (20% v/v) and were perfused 30 minutes later under isoflurane as described in the tissue preparation section below. In the subcellular localization studies, male B6 mice (8–12 weeks old; n=3) were anesthetized with isoflurane and perfused as described in the tissue preparation section below.
For stereology studies, CLIC4-GFP mice were obtained from Dr. Stuart Yuspa (Padmakumar et al., 2014). After backcrossing onto a C57BL6/J background, these mice were used to create a breeding colony at Virginia Commonwealth University. At 3–5 months of age, heterozygous CLIC4-GFP mice (n=6) or wild type littermates (n=2) of both sexes were given an i.p. injection of 4 g/kg of ethanol (20% v/v in saline) or 0.9% sterile saline and were sacrificed 4 hours later by perfusion.
Antibodies and cell-type markers
Information on the cell-type markers used in this study and the antibodies used for immunohistochemistry is found in Table 1.
Table 1 –
Cell type marker definitions and antibody information for IHC experiments
| Stereology Studies | |||
| Marker Symbol | Marker Name | Cell Type | Antibody Information |
| APC | Adenomatous polyposis coli | Oligodendrocytes (mature) | Millipore #OP80; 1:1000 |
| GFP | Green Fluorescent Protein | N/A | Abcam #ab6662, FITC conjugated; 1:300 |
| GS (GLUL) | Glutamine synthetase (Glutamate-ammonia ligase) | Astrocytes | Abcam #ab49873; 1:5000 |
| IBA1 | Ionized calcium binding adaptor molecule 1 | Microglia | FUJIFILM Wako #019–19741; 1:1000 |
| NEUN (RBFOX3) | Neuronal Nuclei (RNA binding protein fox-1 homolog 3) | Neurons | Abcam #ab177487; 1:2000 |
| OLIG2 | Oligodendrocyte transcription factor 2 | Oligodendrocytes | Millipore #AB9610; 1:1500 |
| Secondary | AlexaFluor 594 anti-rabbit | N/A | Abcam #ab150084; 1:500 |
| Subcellular Localization Studies | |||
| Marker Symbol | Marker Name | Cell Type | Antibody Information |
| APC | Adenomatous polyposis coli | Oligodendrocytes (mature) | Millipore #OP80; 1:200 |
| CLIC4 | Chloride Intracellular Channel 4 | N/A | Cell Signaling Technology #D2A7D; 1:100 |
| CNP | 2’,3’-cyclic nucleotide 3’ phosphodiesterase | Oligodendrocytes (myelin) | Abcam #ab6319; 1:400 |
| GS (GLUL) | Glutamine synthetase (Glutamate-ammonia ligase) | Astrocytes | Abcam #ab64613; 1:200 |
| GFAP | Glial fibrillary acidic protein | Astrocytes | Abcam #ab4674; 1:1000 |
| IBA1 | Ionized calcium binding adaptor molecule 1 | Microglia | GeneTex #GT10312; 1:200 |
| NFH | Neurofilament heavy chain | Neuronal axons | Abcam #ab4680; 1:1000 |
| Secondary | AlexaFluor 488 anti-mouse | N/A | ThermoFisher Scientific #A21121; 1:1000 |
| Secondary | AlexaFluor 488 anti-chicken | N/A | ThermoFisher Scientific #A11039; 1:300 |
| Secondary | AlexaFluor 594 anti-rabbit | N/A | ThermoFisher Scientific #A11012; 1:300 |
qRT-PCR analysis
Total RNA was isolated from mPFC of acute ethanol-treated B6 mice by Qiagen RNeasy Mini Kit (#74104). RNA quality and concentration were assessed by NanoDrop spectrophotometry and only samples with 260/280 ratios > 1.8 were analyzed further. cDNA was synthesized using the Bio-Rad iScript cDNA Synthesis Kit (#1708891). PCR was performed with iQ SYBR Green Supermix (#1708880) and a Bio-Rad CFX Connect thermocycler. Primer sequences for Clic4 were as follows: Forward – TTGTCAAGGTGGTGGCCAAA and Reverse – TGTTGGTGAACTCGTCCCTG. Expression values were calculated by the delta-delta Ct method using Ublcp1 and B2M as reference genes. Statistical analysis was performed in R v3.6.2 (R Development Core Team & Team, 2016) using R Studio v 1.2.5033 (RStudio Team, 2016) and ggplot2 package for plotting (Wickham, 2016). Two-way ANOVAs across sex and treatment with Tukey’s HSD post-hoc testing were used for statistical analysis.
Tissue preparation and immunohistochemistry
We used two complementary methods for immunolabeling of CLIC4. For qualitative observations on subcellular localization, we used straightforward immunohistochemistry (IHC) with an antibody raised against native CLIC4 to directly detect the protein. Since CLIC4 belongs to a family of six homologous CLIC proteins, we used an antibody directed against an antigenic peptide targeted around CLIC4 His111 (D2A7D; Cell Signaling Technologies, personal communication), a portion of the protein that is divergent among the CLIC family. For stereology studies revealing the cell type expression pattern, however, we used IHC against Green Fluorescent Protein (GFP) in a knock-in mouse line expressing a CLIC4-GFP fusion protein (Padmakumar et al., 2014). This approach involved simpler tissue processing and was even more effective at avoiding any risk of CLIC protein family cross-reactivity. The CLIC4-GFP transgenic line contains the first 24 amino acids of CLIC4 fused in frame to GFP, which replaces the remainder of the CLIC4 coding region at the native Clic4 gene locus, leaving all Clic4 regulatory elements intact. Heterozygous mice express both the fusion protein and unmodified CLIC4, and thus develop normally (Padmakumar et al., 2014). GFP was detected by immunofluorescence (#ab6662; Abcam; Cambridge, UK) to increase signal strength.
For subcellular localization and nuclear translocation studies, mice were perfused with cold 1x PBS followed by 4% paraformaldehyde before brain extraction and 24-hour post-fixation in 4% paraformaldehyde. Cryoprotection was performed in 30% sucrose, and brains were flash-frozen before cutting on a Leica 3050S cryostat at 20 μm per section. Antigen retrieval was performed for 15 minutes at 80°C in 10 mM citrate buffer (pH6) before blocking with a 10% normal goat serum and 0.2% Triton X-100 solution for 30 minutes at room temperature. Free floating sections were incubated overnight with primary antibodies at 4°C and secondary antibodies applied for 2 hours at room temperature. Mounting was performed using VECTASHIELD Hardset with DAPI (Vector Laboratories; Burlingame, CA). Sample sizes include 3 animals with a minimum of 3 sections per animal per label. Antibodies and dilutions are as described above.
For stereology studies, CLIC4-GFP mice and wild type littermates were perfused with cold 1x PBS followed by 2% paraformaldehyde in PBS before brain extraction and 24-hour post-fixation in 2% paraformaldehyde. Brains were cut coronally on a vibrating microtome into 60 μm sections and stored in antifreeze solution (30% ethylene glycol, 30% glycerol in PBS) at −20°C until use. For immunohistochemistry, brain sections underwent antigen retrieval with 1% sodium borohydride for 20 min before preincubation with 10% normal goat serum, 1% bovine serum albumen, and 0.3% Triton X for 30 min at room temperature. Free floating sections were incubated with gentle agitation overnight with primary antibodies and then for 2 hours with secondary antibodies at room temperature. In separate tissue from that being quantified, TrueBlack lipofuscin autofluorescence quencher reagent (Biotium; San Francisco, CA) was applied after secondary antibody incubations, per manufacturer instructions. Quantification was done in tissue free of TrueBlack because post-treatment with this reagent can result in lower fluorescence signal from antibodies (Figure S2D), as noted in the product insert, and because lipofuscin puncta were easily differentiable from the cell-filling antibody labeling. Sections were mounted with VECTASHIELD Antifade Mounting Medium with DAPI (Vector). Antibodies and dilutions are described in Table 1.
Microscopy
For subcellular localization and nuclear translocation studies, images were captured on a Zeiss LSM700 with a 63x oil objective. Z-stacks were sampled at Nyquist density (43×43×130nm voxels) and a scan zoom set at either 1x or 4.6x (290x total). Deconvolution of 290x z-stacks was performed with Scientific Volume Imaging Huygens Suite software (v19.04) using default settings and experimentally obtained point spread function files for each color channel generated using fluorescent beads. Maximum intensity projection and montage preparations were prepared with FIJI distribution (Schindelin et al., 2012) of ImageJ (Rasband, 1997–2018). Nuclear abundance was calculated as the ratio of the mean nuclear fluorescence to cytosolic fluorescence. Images analyzed in this study were taken from mPFC brain regions including prelimbic, infralimbic, and anterior cingulate cortex.
For stereology studies, images were acquired on a Nikon Eclipse Ti C2 confocal microscope and analyzed using Nikon NIS-Elements Imaging Software (Nikon; Melville, NY). Z-stacks measuring 200 × 200 x ~60 μm were acquired with a 60x oil objective in a checkerboard pattern across the mPFC at two A-P levels per animal, corresponding approximately to bregma +1.5 and +0.5. At least twelve Z-stacks per brain section were counted, resulting in a very ample 57.6 million cubic microns of mPFC tissue counted per animal per label. Fluorescent cells were counted by a blinded technician using standard stereological methods with an optical fractionator. As colocalization of each cell type marker with CLIC4-GFP was assessed in separate IHC reactions, no statistical comparisons were made across markers, and the Student’s t-test was used to compare ethanol vs. saline within each reaction.
Results
Ethanol-responsive expression of Clic4
Clic4 is known to be regulated by acute ethanol in D2 mouse mPFC (Kerns et al., 2005; Bhandari et al., 2012), but this has not been thoroughly evaluated in B6 mice. To investigate this possibility, qRT-PCR was performed on mPFC tissue isolated from wild type male and female B6 mice 4 hours after an i.p. injection of saline, 0.5, 2.0, or 4.0 g/kg ethanol. A significant interaction was identified between sex and treatment such that females receiving larger doses of ethanol showed the highest expression of Clic4 (Figure 1; F(3, 39) = 6.61, p = 0.001). Females displayed higher expression of Clic4 compared to males overall (F(1, 39) = 111.26, p < 0.001), which was largely due to significant induction of Clic4 at each of the three ethanol doses (Tukey’s HSD, p < 0.001). In contrast, there was only a trend towards a difference in Clic4 expression between sexes in the saline-treated animals (Tukey’s HSD, p = 0.11). In females, Tukey’s HSD post-hoc analysis revealed an upregulation of Clic4 in animals receiving a 4.0 g/kg injection when compared to the 2.0 g/kg (p < 0.001), 0.5 g/kg (p < 0.001), and saline-treated (p < 0.001) groups. Males showed no significant differences in Clic4 expression between treatments (Tukey’s HSD, p > 0.05). Ethanol-induced upregulation of mPFC Clic4 expression was diminished at 8 hours post injection and back to the level of control animals at 24 hours, and no upregulation of Clic4 was seen 24 hours after last access to ethanol in a five-week two-bottle choice drinking study (data not shown). In summary, these data show that an acute dose of 4 g/kg of ethanol induced Clic4 expression in female mice.
Figure 1 –
Clic4 mRNA expression with ethanol exposure. qRT-PCR analysis of Clic4 mRNA in wild type B6 mouse mPFC 4 hours after acute i.p. injection of saline or ethanol. Error bars represent SEM. Significance levels indicated by **p<0.001.
Brain cell type expression pattern of CLIC4-GFP
CLIC4-GFP expression was assessed in neurons, oligodendrocytes, astrocytes, and microglia in the mPFC using cell type specific antigen markers. In agreement with experiments described below where CLIC4 was directly immunolabeled, CLIC4-GFP expression was identified in all glial cell types examined, as well as in vascular cells, but not neurons (Figure S1A, S2). The CLIC4-GFP fusion protein had a consistent diffuse cytoplasmic and nuclear pattern of expression within each cell type observed (Figure S2). While this pattern was generally absent in neurons, sparse granule-like puncta fluorescing in multiple channels were occasionally observed within neuronal cell bodies (Figure S2, S3A–C). This granular fluorescence was consistent with lipofuscin deposition and was correspondingly eliminated by TrueBlack Lipofuscin Autofluorescence Quencher (Figure S3D). Wild type CLIC4-GFP littermates showed no immunoreactivity for GFP in any cell type (Figure S3B). Similarly, omission of primary antibodies resulted in no fluorescence in the red (cell type specific markers) or green (CLIC4-GFP) channels, other than lipofuscin deposits, highlighting the specificity of the secondary labeling (Figure S3C). These results and control experiments demonstrate reliable expression of CLIC4-GFP protein in each major brain cell type with the exception of neurons.
While CLIC4-GFP labeling was minimal in neurons, CLIC4-GFP immunoreactivity in the mPFC was abundant in glia (Figure S1A). Two different markers were used to identify oligodendrocytes: OLIG2, which is expressed along the entire differentiation pathway from neural progenitor cell to mature oligodendrocyte (Meijer et al., 2014), and APC, which is primarily expressed in mature myelinating oligodendrocytes (Bin, Harris and Kennedy, 2016). As expected, acute ethanol treatment did not alter the density of any of the cell type markers (Figure S1B).
Focusing on the stereologically quantified percentages of identified cell types that contained CLIC4-GFP, 33.6% of OLIG2+ oligodendrocytes in the mPFC contained CLIC4-GFP under control conditions, and ethanol treatment significantly increased this figure to 57.2% (p=0.032, Student’s t-test; Figure 2A). Similarly, 15.0% of GS+ astrocytes contained CLIC4-GFP under control conditions, which was increased to 32.0% by ethanol treatment (p=0.0052; Figure 2D). The percentages of APC+ oligodendrocytes (~77%) and IBA1+ microglia (~50%) containing CLIC4-GFP were not affected by ethanol (p=0.69 and 0.49, respectively; Figures 2B, 2E). CLIC4-GFP labeling in NEUN+ neurons was minimal, with an average of 0.78% of neurons containing green immunofluorescence (Figure 2C). The inverse comparisons of this data, showing the percentages of CLIC4-GFP+ cells containing each glial marker are shown in Figure S4. The most abundant CLIC4-GFP expressing cell type was oligodendrocytes. In summary, CLIC4-GFP was found in all types of glia (but not in neurons), and alcohol treatment increased the proportions of oligodendrocytes and astrocytes expressing CLIC4-GFP.
Figure 2 –
Quantification of CLIC4-GFP expression in identified cell types. Stereologically quantified percentages of identified cell types expressing CLIC4-GFP are shown for oligodendrocytes (A, B), neurons (C), astrocytes (D), and microglia (E) in the absence and presence of acute ethanol. Acute ethanol treatment was an i.p. injection of 4 g/kg 4 hours before sacrifice. N=3 mice per group; error bars represent SEM; *p<0.05, Student’s t-test.
CLIC4 subcellular localization
Native CLIC4 protein expression in mPFC of wild type mice was also characterized in oligodendrocytes, neurons, astrocytes, and microglia by combining immunofluorescent co-labeling of CLIC4 and cell type specific markers with high resolution deconvolution confocal microscopy. To maximize specificity for CLIC4, an antibody was selected that targets a peptide with minimal sequence identity among the other five CLIC paralogs and has been shown by the manufacturer to not produce signal in extracts prepared from 293 cells transfected with CLIC1 or CLIC5 (Cell Signaling Technologies, personal communication). CLIC1, CLIC4 and CLIC5 are the only CLIC family members detected at any abundance in mouse brain by in situ hybridization (http://mouse.brain-map.org).
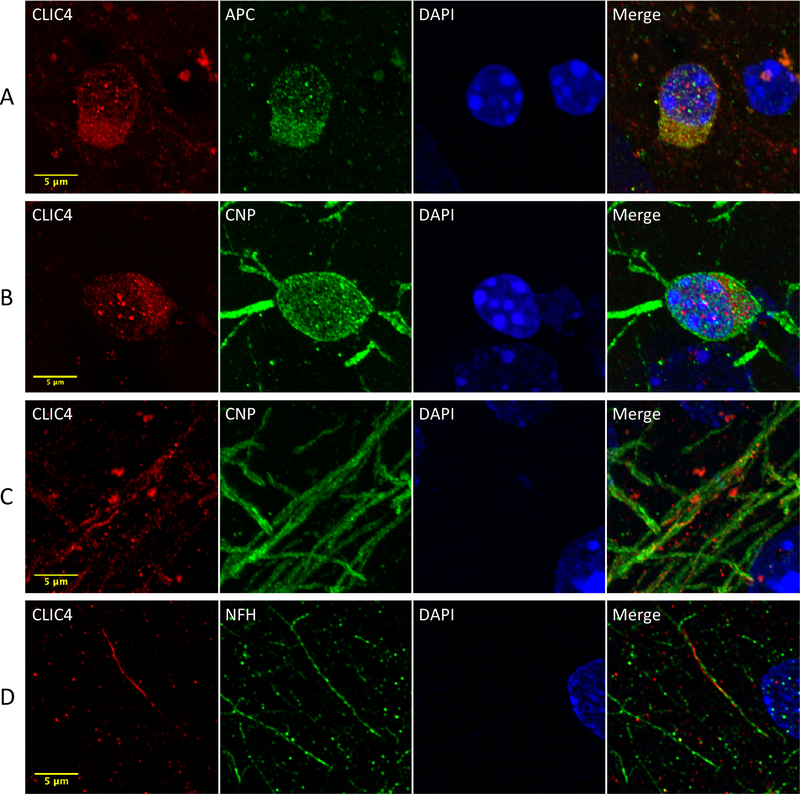
Similar to CLIC4-GFP, native CLIC4 protein was found to be robustly expressed in mPFC oligodendrocytes (Figure 3A). In brain, APC and CNP are both expressed primarily in differentiated and myelinating oligodendrocytes. However, they localize to different subcellular compartments, with APC being largely cytosolic and the lipid-anchored CNP localizing to membranes and especially myelin. Dense punctate expression of CLIC4 was detected within both cytosolic and nuclear compartments of both CNP+ and APC+ oligodendrocytes (Figure 3A, 3B), with cytosolic expression being 38% higher than nuclear (Figure S5). Expression of CLIC4 did not appear to co-localize with CNP at the plasma membrane. CLIC4 was also found in a pattern of small puncta and short linear segments within CNP+ myelin (Figure 3C). Correspondingly, a small proportion of axons labeled with NFH could be found apposed to CLIC4 in an adjacent but non-overlapping configuration, likely representing localization within ensheathing myelin (Figure 3D).
Figure 3 –
CLIC4 protein expression in oligodendrocytes. Confocal microscopy images depicting CLIC4 expression alongside oligodendrocyte markers APC and CNP in cell bodies (A, B) and myelin (C). (D) CLIC4 immunolabeling apposed to axonal marker NFH. Images represent 290x deconvolved z-stack maximum intensity projections. Scale bars represent 5 μm.
Many IBA1-labeled microglia displayed expression of CLIC4 protein in the mPFC (Figure 4A). Expression of CLIC4 in microglia tended to display a pattern of a single dense, well-circumscribed region of cytoplasm (Figure 4B). In GFAP+ and GS+ astrocytes, CLIC4 was found to display diffuse localization in the soma and processes (Figure 5). While GFAP is a common marker for astrocytes, it labels only a small volume of the cell due its association with intermediate filaments of the cytoskeleton. The enzyme GS labels astrocytes more uniformly, having a diffuse cytosolic localization. CLIC4 expression was not detected in all GFAP+ and GS+ cells, but was generally localized to the nucleus when observed (Figure 5B). Vascular cells robustly expressed CLIC4 and were commonly detected and distinguished by morphology (Figure 5A). However, due to adequate documentation in published literature, vascular expression was not formally evaluated in this study. In summary, CLIC4 protein was detected in the cytoplasm and nuclei of glial cells, and it sometimes also extended into fine processes.
Figure 4 –
CLIC4 protein expression in microglia. Confocal microscopy images depicting CLIC4 expression alongside microglial marker IBA1 in two 63x slices (A) and a 290x deconvolved z-stack maximum intensity projection (B). Yellow arrow indicates a CLIC4+ blood vessel surrounded my microglial processes.
Figure 5 –
CLIC4 protein expression in astrocytes. Confocal microscopy images depicting CLIC4 expression alongside astrocyte markers GFAP and GS in 63x slices (A-B) and a 290x deconvolved z-stack maximum intensity projection (C). Yellow arrows indicate CLIC4+ blood vessels encircled by astrocyte processes.
Discussion
Using two complementary immunohistochemistry methods, this work has described both the subcellular localization and cell-type expression pattern of CLIC4 protein in the mouse mPFC, showing a nuclear/cytoplasmic localization primarily in glial cells with very little expression in neurons. Furthermore, we have demonstrated that acute ethanol treatment induces Clic4 expression at both the mRNA and protein levels, in oligodendrocytes and astrocytes. These data, in concert with other recent findings discussed below, suggest that Clic4 may be an important modulator of cellular and behavioral responses to ethanol.
Cell type expression pattern and ethanol response of Clic4
At the mRNA level, mPFC Clic4 expression was transiently and dose-dependently upregulated by acute ethanol treatment in B6 mice. This agrees with previous findings from a similar study in D2 mice (Bhandari et al., 2012). Thus, we have shown that the ethanol response of Clic4 expression is conserved across two strains of mice that differ greatly in sensitivity and preference for ethanol. The responsiveness of this gene to ethanol is influenced by animal sex and strain in a complex fashion, an intriguing characteristic that is explored further in the following section.
Indeed, Clic4 was significantly induced at an acute dose of 4 g/kg, which is akin to an average-sized adult male human consuming 500 mL of 80-proof liquor (Pruett et al. 2020). So, our expression and localization data must be interpreted in the context of a large dose of ethanol that may be expected during heavy binge drinking, which is common among individuals with AUD. Trends of increased expression of Clic4 at doses of 0.5 and 2 g/kg in mice (Figure 1) suggest that similar changes in CLIC4 protein may be present under doses more typical of human alcohol consumption, perhaps to a lesser degree. Published genomic studies from our laboratory have shown induction of Clic4 in DBA2/J male mice following 2 or 4 g/kg i.p. ethanol (Kerns et al., 2005; Bhandari et al., 2012), as well as regulation by 3 g/kg i.p. in C57BL/6J:129S F2 mice (Farris and Miles, 2013). Furthermore, prior microarray studies from our laboratory on prefrontal cortex tissue from male rhesus macaques showed that voluntary access to ethanol for 12 months caused decreased CLIC4 expression in a manner highly correlated with a number of myelin-related genes (Bogenpohl et al., 2019). Finally, genomic studies on postmortem human alcoholic brain tissue have also found changes in CLIC4 mRNA expression (Liu et al., 2006). Thus, there is abundant evidence for Clic4 mRNA regulation by acute and chronic ethanol across multiple species.
At the protein level, we have quantified the expression of CLIC4-GFP across a variety of histologically identified brain cell types. Under control conditions, substantial proportions of oligodendrocytes, astrocytes, and microglia contained the CLIC4-GFP, and none of the cell type markers themselves changed in density with ethanol treatment. Ethanol induced a significant increase in the percentages of OLIG2+ oligodendrocytes and GS+ astrocytes that contained CLIC4-GFP, pointing to induction of CLIC4 in those cell types, since there was no change in the overall densities of OLIG2+ or GS+ cells. This suggests that the qRT-PCR documented ethanol-induced increases in Clic4 mRNA (Figure 1) were manifested, at least in part, by novel expression in oligodendrocytes and astrocytes that previously had little to no CLIC4. These data do not rule out a contribution from increased expression of Clic4 in cells already expressing the gene, in response to ethanol. This latter idea is supported by the finding of no change in the overall density of CLIC4-GFP labeling in response to ethanol (Figure S1A). The increased percentages of oligodendrocytes and astrocytes expressing CLIC4-GFP (Figures 2A, 2D) may appear at odds with the finding of no change in the overall density of glial CLIC4-GFP. The increased percentages in oligodendrocytes and astrocytes may have been offset by non-significant decreases in microglia expressing CLIC4-GFP, non-significant decreases in the overall densities of oligodendrocytes and astrocytes, the existence of ethanol-responsive CLIC4-GFP+ cells that are negative for all cell type markers used, and/or the presence of glial cells expressing more than one of the cell type markers.
Immunolabeling for both OLIG2 and APC allowed for further characterization of the oligodendrocytic response to ethanol in Clic4 expression. APC is a marker of mature and myelinating oligodendrocytes, whereas OLIG2 shows sustained expression throughout oligodendrocyte differentiation (Meijer et al., 2014). Despite a higher proportion of APC+ cells expressing CLIC4 basally, only OLIG2+ oligodendrocytes showed a response to acute ethanol treatment. This implies that CLIC4 is expressed during oligodendrocyte development as well as in the mature state and that ethanol can induce novel expression in differentiating or young oligodendrocytes. Taking a step further, the ethanol response of CLIC4 may be an important event triggering maturation or novel myelinogenesis in oligodendrocyte progenitor cells (OPCs), possibly contributing to functional myelination and reward pathway remodeling. Alternatively, the induction of Clic4 might be protective in oligodendrocytes and especially OPCs, either by regulating or responding to oxidative stress (Al Khamici et al., 2015; Littler et al., 2004), which increases in the brain after ethanol exposure (Zhong et al., 2012; Haorah et al., 2008). Supporting this latter hypothesis, a recent transcriptomic study performed in Drosophila found that knockdown of the sole Clic4 ortholog, Clic, altered expression of a large group of redox genes (Weston et al., 2021). The same study also showed that modulation of ethanol sensitivity by Clic knockdown could be reversed by hyperoxia, suggesting Clic4-like proteins serve a common role within these two biological processes. A protective role of CLIC4 could be important in vertebrate oligodendrocytes, in light of their particular susceptibility to oxidative stress, which results from a high burden of reactive oxygen species produced as a byproduct of copious lipid metabolism (McTigue and Tripathi, 2008). Additionally, CLIC4 might function to stimulate the differentiation/maturation of OPCs in order to induce myelinogenesis aimed at repairing disruptions in white matter, which chronic ethanol has been shown to cause (Chanraud et al., 2007; Pfefferbaum et al., 2000).
These findings support the growing body of evidence for a role of CLIC4 in myelin, particularly under conditions of ethanol exposure, and they warrant further study of its function in astrocytes. Furthermore, given that experimental modulation of CLIC4 or its orthologs results in changes in ethanol behaviors (Bhandari et al., 2012), these data highlight the importance of glial cells in the generation of complex behaviors, a process canonically ascribed to neurons.
Sex and strain differences in Clic4 expression
Employing gene expression microarrays, Kerns et al. (2005) found that Clic4 expression was induced by acute ethanol in male D2 mouse mPFC but not in male B6 mice, despite B6 mice showing higher overall expression. If Clic4 expression correlates with acute ethanol sensitivity, as has previously been shown (Bhandari et al., 2012), this finding may not be surprising when considering B6 mice have lower sensitivity to ethanol than D2 (Linsenbardt et al., 2009; Lister, 1987; Phillips, Dickinson and Burkhart-Kasch, 1994). Indeed, our purpose for utilizing B6 mice in this study was to evaluate a mouse model with a low ethanol sensitivity and high consumptive drive in order to better model human AUD (Schuckit, 1994; Schuckit and Smith, 1996). Our qRT-PCR results reproduce the findings of Kerns and male B6 mice again do not show induction of Clic4 following acute ethanol. However, novel to this study, we found female B6 mice show a robust dose response of Clic4 expression with ethanol exposure. Intriguingly, in B6 mice, females are known to have a higher sensitivity to ethanol than males (Becker and Koob, 2016), representing a similar dichotomy to D2 versus B6 males. This complex interaction between strain and sex in ethanol sensitivity correlates with Clic4 expression, suggesting either a causal relationship or an underlying mechanistic co-regulation.
CLIC4 subcellular localization
There is little published data on the localization of CLIC4 in the mammalian brain, but Ishii et al. (2009) identified enrichment of CLIC4 in white matter over grey matter through large-scale proteomics and Padmakumar et al. (2014) described expression along white matter tracks in Allen Brain Atlas in-situ hybridization data. Further evidence for white matter expression comes from genomic data, where studies have shown correlation of Clic4 mRNA expression with known myelin genes in both macaques (Bogenpohl et al., 2019) and mice (Kerns et al., 2005). We extend these past findings with high power confocal microscopy and show enrichment of CLIC4 expression in mouse APC+ and CNP+ mature oligodendrocytes. CLIC4 was identified both in oligodendrocyte cytoplasm and nucleus, as well as CNP-labeled myelin sheaths, adjacent to axonal marker NFH. These expression patterns may reflect the known metamorphic properties of CLIC4, which permit transitioning between soluble and membrane-bound states (Littler et al., 2005). This variety of localization suggests dynamic roles for CLIC4, possibly related to myelin structure and function or signaling oxidative stress from ethanol. A possible mechanism for this might be through the TGF-beta signaling pathway, which is known to drive oligodendrocyte differentiation and promote myelination (Santos et al., 2019). Outside of brain, CLIC4 has been shown to enhance TGF-beta signaling by translocating into nuclei and stabilizing key transcription factors (Shukla et al., 2009). CLIC4 induction in oligodendrocytes may therefore influence myelin gene expression acutely after ethanol, although this would be less likely to affect acute ethanol sensitivity than it would chronic behavior, which has yet to be evaluated. Alternatively, presence of CLIC4 in oligodendrocyte nuclei may be related to its known functions in cell stress signaling (Fernandez-Salas et al., 2002; Suh et al., 2004). However, we did not observe a shift in the nuclear to cytoplasmic ratio of CLIC4 localization 30 minutes after an acute behaviorally activating dose of ethanol, but it is worth noting that this experiment was performed in male mice, which did not show the same degree of ethanol-induced Clic4 expression as females (Figure 1). Also worth noting is that the ethanol dose used in our nuclear translocation experiment was 2 g/kg. A larger dose, such as the 4 g/kg that induced Clic4 expression, may be more likely to induce cellular stress and the concomitant nuclear translocation of CLIC4. Thus, this is an interesting direction for future experiments. It would also be useful to correlate CLIC4 localization with degree of cellular stress, which could be done through an ethanol exposure time course and quantitative immunofluorescence. Alternatively, an in vitro approach could provide finer temporal resolution and could be paired with readily available assays for assessing oxidative stress.
Although not a direct aim of this study, CLIC4 was commonly detected in vascular endothelial cells, which has been previously reported, and CLIC4 has a known role in angiogenesis (Chalothorn et al., 2009; Ulmasov et al., 2009). Novel to this study, CLIC4 expression was also identified in astrocytes and microglia. Astrocytes were found to express CLIC4 in sparse cytoplasmic and nuclear puncta while microglia showed distinct highly enriched well-circumscribed regions of plasma membrane-adjacent cytoplasm. This microglial expression pattern may represent phagocytic CLIC4-enriched oligodendrocyte debris, such as myelin, or potentially functional expression of CLIC4 in microglial phagosomes. Supporting this potential role in the endosomal degradation pathway, CLIC4 has recently been shown to associate with late endosomes in retinal pigment epithelial cells, where it regulates sorting of metalloproteinases for extracellular matrix breakdown (Hsu et al., 2019). Clic4 knockout mice show dysregulation of endolysosomal biogenesis in renal proximal tubule epithelial cells (Chou et al., 2016). Additionally, CLIC4 paralog CLIC1 has been observed in peripheral macrophages to associate with phagosomes where it promotes acidification (Jiang et al., 2012).
These findings provide novel characterization of CLIC4 cellular and subcellular expression in mPFC and evidence for acute ethanol-induced upregulation of Clic4 in female B6 mice. Oligodendrocytes have been identified as the most abundant CLIC4 expressing cell type in mPFC, and significant expression can be found in nuclei, cytoplasm, and myelin. While these results imply a fundamental role for CLIC4 in oligodendrocytes, they also suggest oligodendrocytes may be key to CLIC4’s role in modulating ethanol sensitivity. Future work will be necessary to identify how CLIC4 expression in oligodendrocytes and other cell types influences ethanol drinking behavior in mice and how its modulation might impact myelin long term.
Supplementary Material
Acknowledgements
We thank Dr. Stuart Yuspa for providing CLIC4-GFP mice. We thank Dr. Andrew van der Vaart, Dr. Guy Harris, Morgan Driver, Eric English, Ayanna Limaye, Amy Doody, Charlie Nelson, Alyssa Wright, Felicity Saylor, and Laura Roy for technical support.
Support: This work was supported by NIAAA grants R01AA020634 and P50AA027581 to MFM and F30AA026497 to RMW.
References
- Al Khamici H, Brown LJ, Hossain KR, Hudson AL, Sinclair-Burton AA, Ng JP, Daniel EL, Hare JE, Cornell BA, Curmi PM, Davey MW and Valenzuela SM (2015) ‘Members of the chloride intracellular ion channel protein family demonstrate glutaredoxin-like enzymatic activity’, PLoS One, 10(1), pp. e115699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JB and Koob GF (2016) ‘Sex Differences in Animal Models: Focus on Addiction’, Pharmacol Rev, 68(2), pp. 242–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari P, Hill JS, Farris SP, Costin B, Martin I, Chan CL, Alaimo JT, Bettinger JC, Davies a. G., Miles MF and Grotewiel M (2012) ‘Chloride intracellular channels modulate acute ethanol behaviors in Drosophila, Caenorhabditis elegans and mice’, Genes, brain, and behavior, 11(4), pp. 387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin JM, Harris SN and Kennedy TE (2016) ‘The oligodendrocyte-specific antibody ‘CC1’ binds Quaking 7’, J Neurochem, 139(2), pp. 181–186. [DOI] [PubMed] [Google Scholar]
- Bogenpohl JW, Smith ML, Farris SP, Dumur CI, Lopez MF, Becker HC, Grant KA and Miles MF (2019) ‘Cross-Species Co-analysis of Prefrontal Cortex Chronic Ethanol Transcriptome Responses in Mice and Monkeys’, Front Mol Neurosci, 12, pp. 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalothorn D, Zhang H, Smith JE, Edwards JC and Faber JE (2009) ‘Chloride intracellular channel-4 is a determinant of native collateral formation in skeletal muscle and brain’, Circ Res, 105(1), pp. 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RF, Lewellyn L, DeLoyht JM, Sennett K, Coffman S, Hewitt M, Bettinger JC, Warrick JM and Grotewiel M (2014) ‘Contrasting influences of Drosophila white/mini-white on ethanol sensitivity in two different behavioral assays’, Alcohol Clin Exp Res, 38(6), pp. 1582–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanraud S, Martelli C, Delain F, Kostogianni N, Douaud G, Aubin HJ, Reynaud M and Martinot JL (2007) ‘Brain morphometry and cognitive performance in detoxified alcohol-dependents with preserved psychosocial functioning’, Neuropsychopharmacology, 32(2), pp. 429–38. [DOI] [PubMed] [Google Scholar]
- Chou SY, Hsu KS, Otsu W, Hsu YC, Luo YC, Yeh C, Shehab SS, Chen J, Shieh V, He GA, Marean MB, Felsen D, Ding A, Poppas DP, Chuang JZ and Sung CH (2016) ‘CLIC4 regulates apical exocytosis and renal tube luminogenesis through retromer- and actin-mediated endocytic trafficking’, Nat Commun, 7, pp. 10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris SP and Miles MF (2013) ‘Fyn-dependent gene networks in acute ethanol sensitivity’, PloS one, 8(11), pp. e82435–e82435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Salas E, Suh KS, Speransky VV, Bowers WL, Levy JM, Adams T, Pathak KR, Edwards LE, Hayes DD, Cheng C, Steven AC, Weinberg WC and Yuspa SH (2002) ‘mtCLIC/CLIC4, an organellular chloride channel protein, is increased by DNA damage and participates in the apoptotic response to p53’, Mol Cell Biol, 22(11), pp. 3610–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haorah J, Ramirez SH, Floreani N, Gorantla S, Morsey B and Persidsky Y (2008) ‘Mechanism of alcohol-induced oxidative stress and neuronal injury’, Free Radic Biol Med, 45(11), pp. 1542–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu KS, Otsu W, Li Y, Wang HC, Chen S, Tsang SH, Chuang JZ and Sung CH (2019) ‘CLIC4 regulates late endosomal trafficking and matrix degradation activity of MMP14 at focal adhesions in RPE cells’, Sci Rep, 9(1), pp. 12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii A, Dutta R, Wark GM, Hwang SI, Han DK, Trapp BD, Pfeiffer SE and Bansal R (2009) ‘Human myelin proteome and comparative analysis with mouse myelin’, Proc Natl Acad Sci U S A, 106(34), pp. 14605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Salao K, Li H, Rybicka JM, Yates RM, Luo XW, Shi XX, Kuffner T, Tsai VW, Husaini Y, Wu L, Brown DA, Grewal T, Brown LJ, Curmi PM and Breit SN (2012) ‘Intracellular chloride channel protein CLIC1 regulates macrophage function through modulation of phagosomal acidification’, J Cell Sci, 125(Pt 22), pp. 5479–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW and Miles MF (2005) ‘Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice’, The Journal of neuroscience : the official journal of the Society for Neuroscience, 25(9), pp. 2255–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenowski PM (2018) ‘Emerging role for the medial prefrontal cortex in alcohol-seeking behaviors’, Addict Behav, 77, pp. 102–106. [DOI] [PubMed] [Google Scholar]
- Linsenbardt DN, Moore EM, Gross CD, Goldfarb KJ, Blackman LC and Boehm SL (2009) ‘Sensitivity and tolerance to the hypnotic and ataxic effects of ethanol in adolescent and adult C57BL/6J and DBA/2J mice’, Alcohol Clin Exp Res, 33(3), pp. 464–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister RG (1987) ‘The effects of ethanol on exploration in DBA/2 and C57Bl/6 mice’, Alcohol, 4(1), pp. 17–9. [DOI] [PubMed] [Google Scholar]
- Littler DR, Assaad NN, Harrop SJ, Brown LJ, Pankhurst GJ, Luciani P, Aguilar M-I, Mazzanti M, Berryman M. a., Breit SN and Curmi PMG (2005) ‘Crystal structure of the soluble form of the redox-regulated chloride ion channel protein CLIC4’, The FEBS journal, 272(19), pp. 4996–5007. [DOI] [PubMed] [Google Scholar]
- Littler DR, Harrop SJ, Fairlie WD, Brown LJ, Pankhurst GJ, Pankhurst S, DeMaere MZ, Campbell TJ, Bauskin AR, Tonini R, Mazzanti M, Breit SN and Curmi PM (2004) ‘The intracellular chloride ion channel protein CLIC1 undergoes a redox-controlled structural transition’, J Biol Chem, 279(10), pp. 9298–305. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK and Mayfield RD (2006) ‘Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals’, Neuropsychopharmacology, 31(7), pp. 1574–82. [DOI] [PubMed] [Google Scholar]
- Maeda K, Haraguchi M, Kuramasu A, Sato T, Ariake K, Sakagami H, Kondo H, Yanai K, Fukunaga K, Yanagisawa T and Sukegawa J (2008) ‘CLIC4 interacts with histamine H3 receptor and enhances the receptor cell surface expression’, Biochemical and biophysical research communications, 369(2), pp. 603–8. [DOI] [PubMed] [Google Scholar]
- Malik M, Shukla A, Amin P, Niedelman W, Lee J, Jividen K, Phang JM, Ding J, Suh KS, Curmi PMG and Yuspa SH (2010) ‘S-nitrosylation regulates nuclear translocation of chloride intracellular channel protein CLIC4’, The Journal of biological chemistry, 285(31), pp. 23818–23828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTigue DM and Tripathi RB (2008) ‘The life, death, and replacement of oligodendrocytes in the adult CNS’, J Neurochem, 107(1), pp. 1–19. [DOI] [PubMed] [Google Scholar]
- Meijer DH, Sun Y, Liu T, Kane MF, Alberta JA, Adelmant G, Kupp R, Marto JA, Rowitch DH, Nakatani Y, Stiles CD and Mehta S (2014) ‘An amino terminal phosphorylation motif regulates intranuclear compartmentalization of Olig2 in neural progenitor cells’, J Neurosci, 34(25), pp. 8507–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF and Bergeson SE (2006) ‘Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis’, Proceedings of the National Academy of Sciences of the United States of America, 103(16), pp. 6368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumar V, Masiuk KE, Luger D, Lee C, Coppola V, Tessarollo L, Hoover SB, Karavanova I, Buonanno A, Simpson RM and Yuspa SH (2014) ‘Detection of differential fetal and adult expression of chloride intracellular channel 4 (CLIC4) protein by analysis of a green fluorescent protein knock-in mouse line’, BMC Dev Biol, 14, pp. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumar VC, Speer K, Pal-Ghosh S, Masiuk KE, Ryscavage A, Dengler SL, Hwang S, Edwards JC, Coppola V, Tessarollo L, Stepp MA and Yuspa SH (2012) ‘Spontaneous skin erosions and reduced skin and corneal wound healing characterize CLIC4(NULL) mice’, The American journal of pathology, 181(1), pp. 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan EV, Hedehus M, Adalsteinsson E, Lim KO and Moseley M (2000) ‘In vivo detection and functional correlates of white matter microstructural disruption in chronic alcoholism’, Alcohol Clin Exp Res, 24(8), pp. 1214–21. [PubMed] [Google Scholar]
- Phillips TJ, Dickinson S and Burkhart-Kasch S (1994) ‘Behavioral sensitization to drug stimulant effects in C57BL/6J and DBA/2J inbred mice’, Behav Neurosci, 108(4), pp. 789–803. [DOI] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA and Mayfield RD (2012) ‘Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence’, J Neurosci, 32(5), pp. 1884–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruett S, Tan W, Howell GE, and Nanduri B (2020) ‘Dosage Scaling of alcohol in binge exposure models in mice: An empirical assessment of the relationship between dose, alcohol exposure, and peak blood concentrations in humans and mice’, Alcohol, 89, pp. 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband W 1997–2018. ImageJ. Bethesda, Maryland, USA: U. S. National Institutes of Health. [Google Scholar]
- Santos AK, Vieira MS, Vasconcellos R, Goulart VAM, Kihara AH and Resende RR (2019) ‘Decoding cell signalling and regulation of oligodendrocyte differentiation’, Semin Cell Dev Biol, 95, pp. 54–73. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P and Cardona A (2012) ‘Fiji: an open-source platform for biological-image analysis’, Nat Methods, 9(7), pp. 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA (1994) ‘Low level of response to alcohol as a predictor of future alcoholism’, The American journal of psychiatry, 151(2), pp. 184–9. [DOI] [PubMed] [Google Scholar]
- Schuckit MA and Smith TL (1996) ‘An 8-year follow-up of 450 sons of alcoholic and control subjects’, Archives of general psychiatry, 53(3), pp. 202–10. [DOI] [PubMed] [Google Scholar]
- Shukla A, Malik M, Cataisson C, Ho Y, Friesen T, Suh KS and Yuspa SH (2009) ‘TGF-beta signalling is regulated by Schnurri-2-dependent nuclear translocation of CLIC4 and consequent stabilization of phospho-Smad2 and 3’, Nature cell biology, 11(6), pp. 777–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh KS, Mutoh M, Nagashima K, Fernandez-Salas E, Edwards LE, Hayes DD, Crutchley JM, Marin KG, Dumont R. a., Levy JM, Cheng C, Garfield S and Yuspa SH (2004) ‘The organellular chloride channel protein CLIC4/mtCLIC translocates to the nucleus in response to cellular stress and accelerates apoptosis’, The Journal of biological chemistry, 279(6), pp. 4632–4641. [DOI] [PubMed] [Google Scholar]
- Ulmasov B, Bruno J, Gordon N, Hartnett ME and Edwards JC (2009) ‘Chloride intracellular channel protein-4 functions in angiogenesis by supporting acidification of vacuoles along the intracellular tubulogenic pathway’, The American journal of pathology, 174(3), pp. 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart A, Meng X, Bowers MS, Batman AM, Aliev F, Farris SP, Hill JS, Green TA, Dick D, Wolstenholme JT, Miles MF and Consortium, a. t. C. (2018) ‘Glycogen synthase kinase 3 beta regulates ethanol consumption and is a risk factor for alcohol dependence’, Neuropsychopharmacology, 43(13), pp. 2521–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston RM, Schmitt RE, Grotewiel M and Miles MF (2021) ‘Transcriptome analysis of chloride intracellular channel knockdown in Drosophila identifies oxidation-reduction function as possible mechanism of altered sensitivity to ethanol sedation’, PLoS One, 16(7), pp. e0246224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H 2016. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York. [Google Scholar]
- Wolen AR, Phillips C. a., Langston M. a., Putman AH, Vorster PJ, Bruce N. a., York TP, Williams RW and Miles MF (2012) ‘Genetic Dissection of Acute Ethanol Responsive Gene Networks in Prefrontal Cortex: Functional and Mechanistic Implications’, PLoS ONE, 7(4), pp. e33575–e33575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstenholme JT, Warner J. a., Capparuccini MI, Archer KJ, Shelton KL and Miles MF (2011) ‘Genomic analysis of individual differences in ethanol drinking: Evidence for non-genetic factors in C57Bl/6 mice’, PLoS ONE, 6(6), pp. e21100–e21100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Dong G, Luo H, Cao J, Wang C, Wu J, Feng YQ and Yue J (2012) ‘Induction of brain CYP2E1 by chronic ethanol treatment and related oxidative stress in hippocampus, cerebellum, and brainstem’, Toxicology, 302(2–3), pp. 275–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.