Abstract
Caenorhabditis elegans is a ubiquitous free-living nematode that feeds on bacteria. The organism was introduced into a laboratory setting in the 1970s and has since gained popularity as a model to study host-bacteria interactions. One advantage of using C. elegans is that its intestine can be colonized by the bacteria on which it feeds. Quantifying the bacterial load within C. elegans is an important and easily obtainable metric when investigating host-bacteria interactions. Although quantification of bacteria harbored in C. elegans via whole-worm lysis is not a novel assay, there is great variation between existing methods. To lyse C. elegans, many protocols rely on the use of a hand-held homogenizer, which could introduce systematic error and subsequent variation between researchers performing the same experiment. Here, we describe a method of lysing the intestines of C. elegans to quantify the bacterial load within the intestine. Our method has been optimized for removing exogenous bacteria while maintaining worm paralysis, to ensure no bactericidal agents are swallowed, which could kill bacteria within the intestine and affect results. We utilize and compare the efficiency of two different homogenization tools: a battery-powered hand-held homogenizer, and a benchtop electric homogenizer, where the latter minimizes variability. Thus, our protocol has been optimized to reduce systematic error and decrease the potential for variability among experimenters.
Graphic abstract:
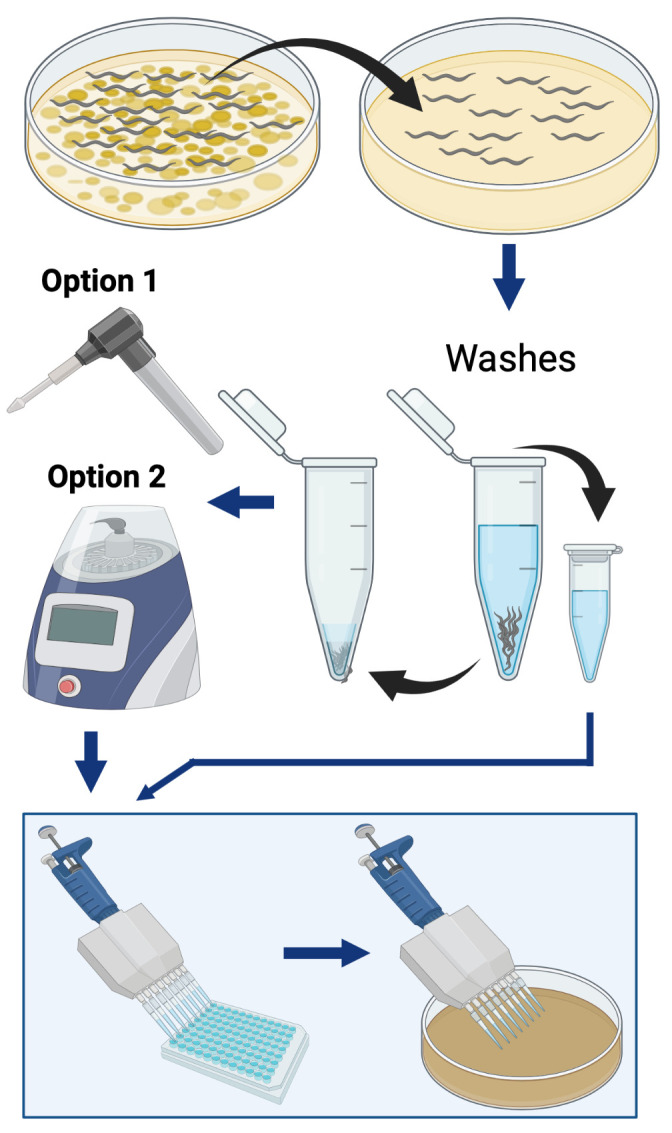
Simplified overview of the procedure used to quantify the bacterial load within C. elegans. The two different methods are herein described for worm lysis: “Option 1” is a hand-held homogenizer, and “Option 2” is a benchtop homogenizer.
Keywords: Caenorhabditis elegans, Bacteria, Colonization, Intestine, CFU, Quantification, Enumeration
Background
The nematode Caenorhabditis elegans is a bacterivore that is widely used as a model to study host-bacteria interactions, due to its simple morphology, high homology of its genome to the human genome, and the amenability of its microbiome ( Zhang et al., 2017 ). These nematodes use their pharynx to intake bacteria from their surrounding environment (Avery and You, 2018). In a laboratory setting, germ-free larvae are obtained by isolating embryos from gravid adults. Once hatched, larvae (L1s) will begin to feed on bacteria (Brenner, 1973). Hence, it is easy to colonize the C. elegans gut with bacterial strain(s) of interest. Studies involving host-bacteria interactions often call for the enumeration of the bacterial load within the intestine. One of the most common and reliable bacterial quantification techniques is plating for colony forming units (CFUs). Quantification of bacteria within C. elegans is achieved via the physical disruption of worm bodies to release intestinal content, and plating the resulting lysate on solid medium to support microbial growth and subsequent enumeration of CFUs. However, there is variation between existing protocols for the aforementioned assay. For example, prior to the homogenization process, worms are washed several times in a microcentrifuge tube to eliminate external bacteria from the sample. During these washes, worms must be allowed to settle to the bottom of the tube before the supernatant is aspirated, and the researcher must be aware of the location of the worms within the tube, to ensure worms are not aspirated out with the supernatant—because worms tend to stick to pipette tips, this is a critical precautionary measure, which is not emphasized in many existing protocols. Many protocols also call for gentamicin or bleach washes, to eliminate bacteria attached to the cuticle of the worm prior to homogenization. However, if swallowed, bactericidal agents could affect the viability of intestinal bacteria, skewing results. To circumvent these risks, we add levamisole, an alkaline phosphatase inhibitor that reversibly paralyzes C. elegans, to our gentamicin and non-gentamicin washing solutions. This step stops pharyngeal pumping and decreases the risk of the germicidal agent making its way to the intestine. Other techniques used to prevent ingestion of bactericidal agents include lowering the temperature of the samples, and adding reagents that retard motility and minimize pharyngeal pumping (Vega and Gore, 2017; Ortiz et al., 2021 ). Temperature-mediated inhibition of motility is an attractive approach that can be incorporated into our protocol, for applications that may want to avoid using levamisole. Manual homogenization of worms introduces the most variability. For example, moving the pestle rapidly in the sample tube could cause bubbling and subsequent spillage of the sample. The time and force exerted on the pestle could also affect reproducibility between experimenters. We describe detailed steps in our protocol, such as the position of the pestle and the movement of the hand-held homogenizer, to mitigate the aforementioned risks. Other more hands-off methods of disrupting worm bodies include the automated higher-throughput worm lysis that likely limits the risk of human-induced variation (Vega and Gore, 2017). As such, we describe a benchtop homogenizer-based automated method that diminishes the risk of human error and variation. A flow-chart of the described method is illustrated in Figure 1. Three researchers in our lab performed the protocol described herein on the same day, using worms taken from the same sample. Quantification of the bacterial load within C. elegans varied between researchers using a hand-held homogenizer (Figure 2A), whereas results were nearly identical when a benchtop homogenizer was used (Figure 2B). Hence, option 2 should be the method of choice, as it provides the most consistent results. It is worth noting that this protocol can be modified to permit quantification of other microbes harbored within C. elegans, or any experiment that requires worm lysis to be consistent between samples.
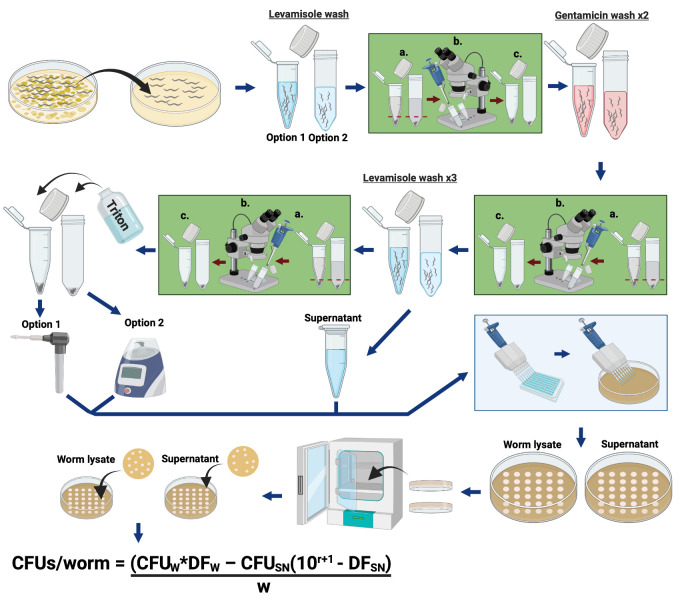
Figure 1. Schematic illustrating the protocol “Quantification of Bacterial Loads in C. elegans”.
Detailed methods can be found in “Procedure”. Explanation of CFU calculations can be found in “Data Analysis”.
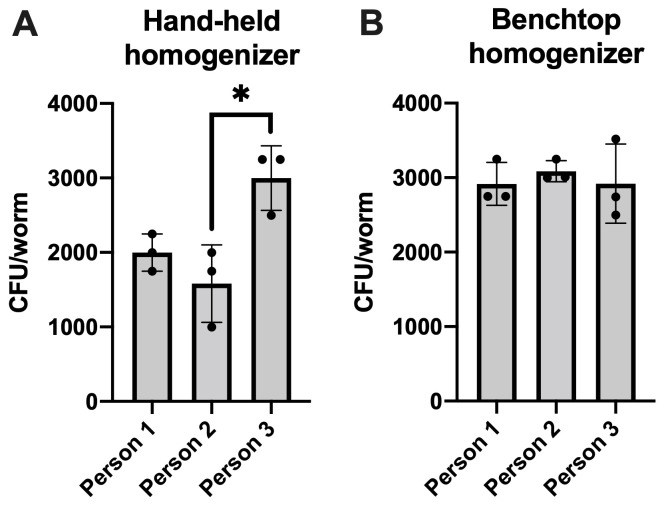
Figure 2. Quantification of bacteria within C. elegans.
Reproducibility between three experimenters conducting the protocol described herein using (A) a hand-held homogenizer, and (B) a benchtop homogenizer to lyse C. elegans. Data are represented as the average bacterial load per C. elegans intestine. Each bar represents one independent experiment with three internal replicates. Error bars represent standard error of the mean (SEM). Significance was calculated using one-way analysis of variance (ANOVA) followed by multiple comparison Tukey’s post-hoc test (*P < 0.05).
Materials and Reagents
Note: Unless otherwise specified, all reagents are stored at room temperature.
Erlenmeyer flasks (2,000 mL and 1,000 mL)
6 cm Petri dishes (Genesee Scientific, catalog number: 32-105G)
10 cm Petri dishes (Genesee Scientific, catalog number: 32-107G)
Pestle and tube (Bel-Art F650000006, catalog number: 03-421-221), if following “Option 1”; or empty 2 mL non-skirted screw cap micro tubes with conical bottom (Thermo Scientific 3468, catalog number: 21-403-200), if following “Option 2” in Step C of “Procedure”
Bead Bug 1.5 mm zirconium beads (Benchmark Scientific, catalog number: D1032-15)
Pipette tips (P1000, P200, P20, or P10)
Magnetic stir bar
Autoclave tape
500 mL Olympus vacuum filter flasks (Genesee Scientific, catalog number: 25-227)
Lennox Luria broth powder (Genesee Scientific, catalog number: 11-125)
Agar (Fisher Scientific, catalog number: BP1423)
Double distilled water (ddH2O)
NaCl (Fisher Scientific, catalog number: S671-500)
Trypticase-peptone (Gibco, catalog number: 211921)
Cholesterol (MP Biomedicals, catalog number: ICN10138201)
Ethanol, 200-proof (Decon Laboratories, catalog number: 04-355-223)
CaCl2·2H2O (Fisher Scientific, catalog number: C79-500)
MgSO4·7H2O (Fisher Scientific, catalog number: A14491)
KH2PO4 (Fisher Chemical, catalog number: P285-3)
NA2HPO4·7H2O (Fisher Scientific, catalog number: S373-500)
Levamisole hydrochloride (MP biomedicals, catalog number: 155228)
Triton X-100 (Promega, catalog number: H5142)
Phosphate-buffered saline (PBS), 10× (Fisher Scientific, catalog number: J62036)
Gentamicin sulfate solution (Cassion Labs, catalog number: ABL03)
LB agar plates (see Recipes)
Phosphate Buffered Saline (PBS), 1× (see Recipes)
M9 Minimal Medium (see Recipes)
Levamisole Stock Solution (see Recipes)
M9/Levamisole Solution (see Recipes)
Gentamicin Stock Solution (see Recipes)
Gentamicin Wash Solution (see Recipes)
Cholesterol Stock Solution (see Recipes)
1 M CaCl2 Stock Solution (see Recipes)
1 M MgSO4 Stock Solution (see Recipes)
1 M KH2PO4, pH 6.0 Stock Solution (see Recipes)
NGM Plates (see Recipes)
1% Triton X-100 solution (see Recipes)
Equipment
Microtube Homogenizer System (Bel-Art, catalog number: F65000-0000)
BeadBug3 Microtube Homogenizer (Benchmark Scientific, catalog number: D1030)
Zeiss Stemi 305 stereo microscope (Zeiss, catalog number: 435063-9010-000)
Incubators for bacteria (37°C) and C. elegans (15-25°C)
Pipette (Eppendorf, models: P1000, P200 or P20, multichannel models P200, P10 or P20)
Platinum wire worm pick (made in-house as described in Wollenberg et al., 2013 )
Ethanol candle (DWK Life Sciences, catalog number: 04-245-1)
Autoclave
Biological safety cabinet
Denver Instrument Summit SI-4002 Summit Series Toploading Balance (Denver instruments, catalog number: EW-11422-13)
pH meter (Thermo Electric Corporation)
Fisher Scientific Isotemp Stirrer (Fisher Scientific, catalog number: 11-100-49S)
Software
Microsoft Excel (http://office.microsoft.com/excel)
Graphpad Prism v8.4.3. (Graphpad Software, Inc., https://www.graphpad.com)
BioRender (www.biorender.com)
Procedure
Notes:
Unless otherwise specified, all steps are conducted outside of a biological safety cabinet (BSC) using aseptic techniques.
This protocol assumes nematodes have already been cultured and subsequently colonized with bacteria of interest.
This protocol describes obtaining worms from solid nematode growth media (NGM). Worms can also be obtained from a liquid culture.
-
Worm preparation (pre-wash)
To remove excess bacteria, pick 12 or more worms from a plate of interest onto a plain, unseeded NGM plate. Let sit for approximately 30 min. It may be necessary to periodically move worms around the plate away from the transferred bacteria, to prevent them from staying in transferred bacteria.
-
Washing worms to remove external bacteria
-
Transfer 10 worms from the plain NGM plate from Step A1 into 1 mL of M9/levamisole solution.
Note: If “Option 2” in Step C will be followed, worms must be placed in a tube that is compatible with the homogenizer machine, such as a 2 mL screw-cap non-skirted tube with a conical bottom.
-
Allow worms to settle to the bottom of the tube and gently aspirate 950 μL of the supernatant.
Note: To ensure worms are not aspirated, it is recommended that a dissecting microscope be used to look at the worms during this process. Because large pipette tips can increase the chance of aspirating worms along with the supernatant, it is recommended to remove no more than approximately 800 μL using a 1 mL pipette, followed by removal of 150 μL using a smaller volume pipette and pipette tip, approximately 50 μL at a time. There should be 50 μL remaining in the tube at the end of this step.
Perform a gentamicin wash, using 950 μL of gentamicin wash solution. Allow worms to bathe in gentamicin solution for 15-20 min.
Aspirate 950 μL of supernatant following the precautions mentioned in Step B2 “Note”.
Repeat Steps B3 and B4.
Wash worms with 950 μL of M9/levamisole solution and aspirate 950 μL of the supernatant, following the precautions mentioned in Step B2 “Note”.
Repeat Step B6.
-
Perform a third and final wash as described in Step B6, but keep the 950 μL of removed supernatant in a sterile microcentrifuge tube. This will be used to quantify remaining external bacteria, to obtain the actual number of intestinal CFUs in the final calculations.
Note: To ensure that no worms were lost during washes, worms should be counted before homogenization.
-
-
Lysing C. elegans
Note: We describe two ways to lyse C. elegans: Option 1 uses a hand-held battery-powered homogenizer, and Option 2 employs an electric benchtop homogenizer.
Option 1: Lysing C. elegans using a hand-held homogenizer
Note: A video demonstration of “Option 1” Steps 2-3 can be found in Video 1.
Add 200 μL of 1% Triton X-100 to the worms and let sit for 5 min. Allow worms to sink to the bottom of the tube. The total volume in each tube should be 250 μL.
Push the pestle towards the base of the microcentrifuge tube very gently to avoid resuspending the worms. Mash the worms using as little upward motion as possible. A twisting motion is best to grind worms.
Attach a hand-held homogenizer to the pestle without lifting the pestle away from the base of the tube. Grind worms until they appear shredded when viewed under a microscope, such as the Zeiss Stemi. Homogenization time should be kept as consistent as possible between samples. This step often introduces variability due to duration, force, motion, tube, and pestle type.
Option 2: Lysing C. elegans using an electric benchtop homogenizer
Add 200 μL of 1% Triton X-100 to the worms and let sit for 5 min. The total volume in each tube should be 250 μL.
Add 30 1.5 mm sterile zirconium beads to each tube.
-
Sequentially run at 4,000 rotations per minute (rpm) for 90 s, twice.
Note: A video demonstration of the benchtop homogenizer at work can be found in Video 2.
-
Plating for CFUs
-
In a BSC, perform serial dilutions of saved supernatant from Step B8 and homogenized worm mixture, using sterile PBS. Begin by transferring 200 μL of each undiluted sample into the top row of a 96-well plate and perform at least five tenfold dilutions. Dilutions are performed by taking 20 μL of solution from the previous well and transferring into subsequent wells filled with 180 μL of sterile PBS.
Note: If bubbles are present after homogenization, briefly centrifuge tubes to get rid of bubbles.
-
Plate 10 μL of each dilution, as well as non-diluted samples, onto agar plates containing appropriate medium (i.e., LB), let dry, and place upside down at 37°C. The incubation time will depend on the bacteria used, but usually 16-24 h incubation is sufficient.
Note: plate a minimum of three internal replicates per sample.
-
-
Enumerating CFUs
Count the number of CFUs from each 10 μL spot and record the corresponding row. Calculations and analysis can be found in “Data analysis”.
Video 1. Demonstration of whole-worm lysis using a hand-held homogenizer.
Video 2. Demonstration of whole-worm lysis using a benchtop homogenizer.

Data analysis
The total number of CFUs/worm in each internal replicate can be calculated with the following equation: CFUs/worm = [CFUW*DFW – CFUSN(10r+1 – DFSN)]/w , where CFUW = CFUs on plate from whole worm lysate, CFUSN = CFUs on plate from supernatant, DFW = dilution factor of whole worm lysate, DFSN = dilution factor of supernatant, r = row number, and w = number of worms in the sample. Dilution factors (DF) can be found in Table 1 and correspond to the row in which the CFUs were counted. As stated in “Procedure” Step D2 “Note”, it is recommended that samples are plated in triplicates, in which case the average CFU/worm would be calculated by taking the mean of CFUs/worm of all internal replicates within each sample.
Statistical analysis depends on the sample set. When calculating statistical significance between pairs, the Student’s t-test should be used. When comparing colonization of multiple test bacteria to control bacteria, statistical significance should be calculated using one-way analysis of variance (ANOVA) followed by multiple comparison with Dunnett’s post-hoc test. If there is no single control set, significance should be calculated using one-way ANOVA followed by multiple comparison with Tukey’s post-hoc test. Significance should be demonstrated within graphs using asterisks, where * P < 0.05, ** P < 0.01, *** P < 0.001, ****P < 0.0001.
Table 1. Dilution Factors.
| Row | Dilution factor | Row | Dilution factor |
| 1 (undiluted sample) | 25 | 1 (undiluted sample) | 95 |
| 2 | 250 | 2 | 950 |
| 3 | 2,500 | 3 | 9,500 |
| 4 | 25,000 | 4 | 95,000 |
| 5 | 250,000 | 5 | 950,000 |
| 6 | 2,500,000 | 6 | 9,500,000 |
Notes
All reagents, pipette tips, microcentrifuge tubes, beads, and pestles should be sterile to avoid contamination of samples. All other precautionary notes are contained within corresponding steps of the protocol in “Procedure”.
Recipes
-
LB agar plates
In a 2 L flask, combine 20 g of Luria broth powder and 15 g of agar.
Fill with ddH2O up to 1 L.
Add a magnetic stir bar.
Place on stir plate and let reagents mix for several minutes.
Place cap (loosely screwed on) or cover the bottle opening with aluminum foil.
Adhere autoclave tape.
Autoclave using a fluid cycle for 30 min at 121°C.
Cool to 50°C and pour into 10 cm Petri dishes.
Dry plates for one day, put back in sleeves, and store upside down at 4°C.
-
Phosphate Buffered Saline (PBS), 1×
In a sterile container, dilute 10× sterile PBS in sterile ddH2O for a final concentration of 1× PBS. Store at room temperature (RT).
-
M9 Minimal Medium
Combine 5.8 g Na2HPO4·7H2O, 3.0 g KH2PO4, 5.0 g NaCl, and 0.25 g MgSO4·7H2O in a 1 L flask.
Add a magnetic stir bar and fill to 1 L with ddH2O.
Place on stir plate and let mix until all reagents are dissolved.
Filter sterilize into two 500 mL bottles.
Aliquot into smaller sterile bottles if desired, and store at RT.
-
Levamisole Stock Solution
Dissolve levamisole powder in ddH2O for a final stock concentration of 240.75 mg/mL (1 M).
Filter sterilize, aliquot, and store at -20°C until use.
-
M9/Levamisole Solution
Dilute levamisole stock solution in M9 to a final concentration of 80 μM levamisole.
Note: Make fresh the day of use.
-
Gentamicin Stock Solution
Dilute gentamicin (50 mg/mL) in sterile ddH2O to a final concentration of 5 mg/mL.
Aliquot and store at -20°C until use.
-
Gentamicin Wash Solution (M9/Levamisole/Gentamicin Solution)
Add gentamicin stock solution to M9/Levamisole solution for a final concentration of 100 μg/mL gentamicin.
Note: Make fresh the day of use.
-
Cholesterol Stock Solution
Dissolve cholesterol in 200-proof ethanol to a final concentration of 5 mg/mL cholesterol. Store at -20°C until use.
-
1 M CaCl2 Stock Solution
Add 147 g CaCl2·2H2O to a 1 L flask.
Fill to 1 L with ddH2O.
Add a magnetic stir bar.
Place on stir plate until reagent dissolves.
Aliquot and screw caps loosely on bottles.
Adhere autoclave tape.
Autoclave fluid cycle for 30 min.
Let cool, fully screw caps on, and store at RT until use.
-
1 M MgSO4 Stock Solution
Add 246.5 g MgSO4·7H2O to a 1 L flask.
Fill to 1 L with ddH2O.
Add a magnetic stir bar.
Place on stir plate until reagent dissolves.
Aliquot and screw caps loosely on bottles.
Adhere autoclave tape.
Autoclave using a fluid cycle for 30 min at 121°C.
Let cool, fully screw caps on, and store at RT until use.
-
1 M KH2PO4, pH 6.0 Stock Solution
Add 136.1 g KH2PO4 to a 1 L flask.
Dissolve in 800 mL of ddH2O.
Add a magnetic stir bar.
Place on stir plate until reagent dissolves.
Adjust pH to 6.0 with 10 g KOH. Bring up to 1 L.
After reagents are completely dissolved, and pH is confirmed to be 6.0, aliquot, and screw caps loosely on bottles.
Adhere autoclave tape.
Autoclave using a fluid cycle for 30 min at 121°C.
Let cool, fully screw caps on, and store at RT until use.
-
NGM Plates
In a 2 L flask, combine 3.0 g NaCl, 2.5 g trypticase-peptone, and 17.0 g agar.
Fill ddH2O up to 1 L.
Add a magnetic stir bar.
Place on stir plate and let reagents mix for several minutes.
Place cap (loosely screwed on) or foil on the opening of the bottle.
Adhere autoclave tape.
Autoclave using a fluid cycle for 30 min at 121°C.
Cool to 50°C.
-
Add 1 mL of 5 mg/ mL cholesterol stock solution, 1 mL of 1 M CaCl2 stock solution, 1 mL of 1 M MgSO4 stock solution, and 25 mL of 1M KH2PO4, pH 6.0 stock solution.
Note: Let cholesterol stock solution (5 mg/mL) reach room temperature and ensure cholesterol re-dissolves in solution before adding to NGM.
Cover and put back on stir plate for one minute.
Pour into 60 mm Petri dishes.
Let cool and and store upside down in a plastic box, with lid at 4°C.
-
1% Triton X-100 solution:
Dilute Triton X-100 in M9 for a total concentration of 1% Triton X-100.
Note: If both reagents are not kept sterile, filter sterilize and store at RT until use.
Acknowledgments
This work was supported in part by the Infectious Diseases Society of America and Start-up funding provided by the Microbiology and Cell Science Department at the University of Florida Institute of Food and Agricultural Sciences to DMC. Walker et al. (2021) is the original research paper from where this protocol was derived.
Competing interests
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors have declared that no competing interests exist.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1. Avery L. and You Y. J.(2018). C. elegans feeding. WormBook: The Online Review of C. elegans Biology[Internet]. WormBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brenner S.(1973). The genetics of behaviour. Br Med Bull 29(3): 269-271. [DOI] [PubMed] [Google Scholar]
- 3. Ortiz A., Vega N. M., Ratzke C. and Gore J.(2021). Interspecies bacterial competition regulates community assembly in the C. elegans intestine . ISME J 15(7): 2131-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vega N. M. and Gore J.(2017). Stochastic assembly produces heterogeneous communities in the Caenorhabditis elegans intestine . PLoS Biol 15(3): e2000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walker A. C., Bhargava R., Vaziriyan-Sani A. S., Pourciau C., Donahue E. T., Dove A. S., Gebhardt M. J., Ellward G. L., Romeo T. and Czyz D. M.(2021). Colonization of the Caenorhabditis elegans gut with human enteric bacterial pathogens leads to proteostasis disruption that is rescued by butyrate. PLoS Pathog 17(5): e1009510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wollenberg A. C., Visvikis O., Alves A. M. F. and Irazoqui J. E.(2013). Staphylococcus aureus Killing Assay of Caenorhabditis elegans . Bio-protocol 3(19): e916. [Google Scholar]
- 7. Zhang J., Holdorf A. D. and Walhout A. J.(2017). C. elegans and its bacterial diet as a model for systems-level understanding of host-microbiota interactions. Curr Opin Biotechnol 46: 74-80. [DOI] [PMC free article] [PubMed] [Google Scholar]