Abstract
Emerging studies suggest that monocytes can be trained by bacterial endotoxin to adopt distinct memory states ranging from low-grade inflammation to immune exhaustion. While low-grade inflammation may contribute to the pathogenesis of chronic diseases, exhausted monocytes with pathogenic and immune-suppressive characteristics may underlie the pathogenesis of polymicrobial sepsis including COVID-19. However, detailed processes by which the dynamic adaption of monocytes occur remain poorly understood. Here we exposed murine bone-marrow derived monocytes to chronic lipopolysaccharide (LPS) stimulation at low-dose or high-dose, as well as a PBS control. The cells were profiled for genome-wide H3K27ac modification and gene expression. The gene expression of TRAM-deficient and IRAK-M-deficient monocytes with LPS exposure was also analyzed. We discover that low-grade inflammation preferentially utilizes the TRAM-dependent pathway of TLR4 signaling, and induces the expression of interferon response genes. In contrast, high dose LPS uniquely upregulates exhaustion signatures with metabolic and proliferative pathways. The extensive differences in the epigenomic landscape between low-dose and high-dose conditions suggest the importance of epigenetic regulations in driving differential responses. Our data provide potential targets for future mechanistic or therapeutic studies.
Subject terms: Chronic inflammation, Epigenomics
Lynette Naler and Yuan-Pang Hsieh et al. evaluate epigenomic and transcriptomic differences in mouse bone marrow-derived macrophages following exposure to high or low doses of LPS. Their results suggest that both low- and high-grade inflammation involve TRAM-dependent pathways.
Introduction
Forming a close co-habitat with bacteria, mammalian hosts are intimately impacted by the presence of varying dosages of bacterial endotoxin lipopolysaccharide (LPS) within their circulation. While high-dose LPS triggers multi-organ injury and sepsis, through inducing pathogenic inflammation and immuno-suppression, lower dose LPS sustains a low-grade, or chronic inflammation, playing an important role in the onset or exacerbation of many chronic diseases, including Alzheimer’s disease1, depression2, diabetes3, and cancers4. Despite this, there still are no canonical markers for low-grade inflammation, partly due to a correlation of inflammatory activity with age5,6. Some recent studies in humans have attempted more sophisticated integrated approaches that show promise, but more research is required to identify key biomarkers and how they should be interpreted6–8.
Low-grade inflammation is often associated with certain lifestyle choices, like Western-style eating practices9, sedentary behavior10,11, sleep deprivation12, and social stress13. In fact, individuals who underwent major childhood stressors have elevated mortality and morbidity of immune or chronic diseases later in life, and epigenetic programming has been proposed14. This is further supported by twin studies that showed most variation in the immune system is due to non-heritable (i.e. epigenetic) influences15.
In a bacterial infection, components of the pathogen16 are recognized by pattern-recognition receptors (PRRs) that are found on immune and non-immune cells17–20. This, in turn, leads to a signaling cascade that is used to recruit immune cells, such as neutrophils and monocytes/macrophages, which ingest the microbes or release anti-microbials21,22. Once the threat is eliminated, pro-inflammatory macrophages uptake spent neutrophils, which reprogram the macrophages to an anti-inflammatory state23,24. Anti-inflammatory macrophages help promote healing by protecting against tissue damage, clearing out debris, and producing growth factors24. However, when macrophage and monocyte inflammatory activity is not resolved then it can lead to low-grade inflammation and chronic diseases25–30.
Lipopolysaccharide (LPS) is an endotoxin and component of the cell wall of Gram-negative bacteria, such as Escherichia coli, that also has been found at low levels in individuals who have chronic diseases or detrimental lifestyle choices as mentioned previously31,32. It has been used to study inflammation both in vivo and in vitro, although more often with high-doses of LPS. While persistent low-dose LPS has been shown to lead to a low-grade inflammatory state, the mechanism by which it does so is still not entirely known, though could be due to sustained activity of the inflammatory Nuclear Factor Kappa B (NF-κB)32–35.
LPS stimulation activates several different pathways, which both have pro- and anti-inflammatory aspects. First, LPS binds to Toll-like Receptor 4 (TLR4) and activates the Myeloid Differentiation Primary Response (MyD88)-dependent pathway32,36,37. The MyD88-dependent signaling cascade activates Mitogen-Activated Protein Kinases (MAPKs), NF-κB translocation to the nucleus, and transcription factors cAMP Response Element-Binding Protein (CREB) and Activator Protein 1 (AP-1), as well as induces expression of pro-inflammatory genes such as Tumor Necrosis Factor alpha (Tnfa), Interleukin 6 (Il6), and Prostaglandin-Endoperoxide Synthase 2 (Ptgs2)37. However, it is also involved in the production of anti-inflammatory cytokines like Interleukin 10 (IL10). In addition, MyD88-dependent signaling leads to an increase in glycolysis, synthesis of acetyl-CoA, and synthesis of fatty acids. Next, TLR4 can also trigger the Translocation Associated Membrane Protein (TRAM)/TIR-domain-containing adapter-inducing interferon-β (TRIF)-dependent pathway37. The TRIF-dependent pathway activates Interferon Regulatory Factor 3 (IRF3) and Interferon Regulatory Factor 7 (IRF7) to induce expression of type I interferons (IFNs), C-C Motif Chemokine Ligand 5 (Ccl5), and C-X-C Motif Chemokine Ligand 10 (Cxcl10). It is also involved in anti-inflammatory cytokine IL10 production and is important for Tnfa expression37,38. Furthermore, in macrophages, perhaps independent of TRAM, the TRIF-mediated pathway also upregulates cell-surface Cluster of Differentiation 40 (Cd40), Cluster of Differentiation 80 (Cd80), and Cluster of Differentiation 86 (Cd86) which are necessary for antigen presentation for T lymphocytes, bridging the gap between the innate and adaptive immune system37,39. The exact role of TRAM, however, is not well understood in the cellular responses to either low or high doses of LPS. Finally, both pathways are involved in the canonical activation of the NLR Family Pyrin Domain Containing 3 (NRLP3) inflammasome, which is responsible for the activation of the inflammatory cytokine, Interleukin 1 beta (IL-1β), and can cause pyroptosis, a form of programmed cell-death37.
The epigenome plays a large role in the behavior and identity of macrophages. For example, the epigenome of tissue-resident macrophages is affected by their local microenvironment and they can even be reprogrammed by transplanting them to a different location40. They also have varied transcriptional signatures during efferocytosis, a process where apoptotic cells are cleared41. LPS-stimulation has also been shown to have a significant, and sometimes lasting, effect on the histone modifications of macrophages42. LPS-induced tolerance affects the epigenome of macrophages by inhibiting induction of inflammatory genes while leaving antimicrobial genes unaffected43. Research into means of targeting and altering the epigenome of inflammatory macrophages into an anti-inflammatory state has also increased in recent years, with studies showing possible therapeutic potential44. Despite this, there is little research on how low-doses of LPS affect the epigenome45–48 or transcriptome48–52 of monocytes or macrophages differently compared to high-doses of LPS.
In this study, we profiled the histone mark H3K27ac and performed RNA-seq analysis of murine bone marrow-derived monocytes that are exposed to low and high-dose levels of LPS, as would be seen in low-grade inflammation and immune exhaustion respectively. Low-input technologies including Microfluidic Oscillatory Washing ChIP-seq (MOWChIP-seq53,54) and Smart-seq255,56 were used for the epigenomic and transcriptomic profiling, respectively. We compared the conditions to extract the effects of LPS-dosage on the epigenome and, in turn, the transcriptome of immune cells. Furthermore, we also analyzed the effects of Tram deletion, in order to clarify the role of this less-studied TLR4 adapter, as well as Interleukin 1 Receptor Associated Kinase M (Irak-m) deletion. Although IRAK-M is a known inhibitor of TLR4 pathway, the exact role of IRAK-M in modulation of low-grade inflammation and monocyte exhaustion is not well defined57. We revealed that TRAM is at least partially involved in the generation of both low-grade inflammatory monocytes, as well as the development of exhaustion, through facilitating Signal Transducer and Activator of Transcription 1 (STAT1) / IRF7 activation and induction of selective exhaustion markers such as S100 Calcium Binding Protein A8 (S100A8), often seen in patients with COVID-19. The impacts of Irak-m deletion are more nuanced, by exacerbating low-grade inflammation and refraining the development of exhaustion. Altogether, our analyses provide population-level adaptation of monocytes to the persistent challenges of LPS with varying signal strengths.
Results
TRAM facilitates inflammatory monocyte generation under both low-dose and high-dose LPS conditions
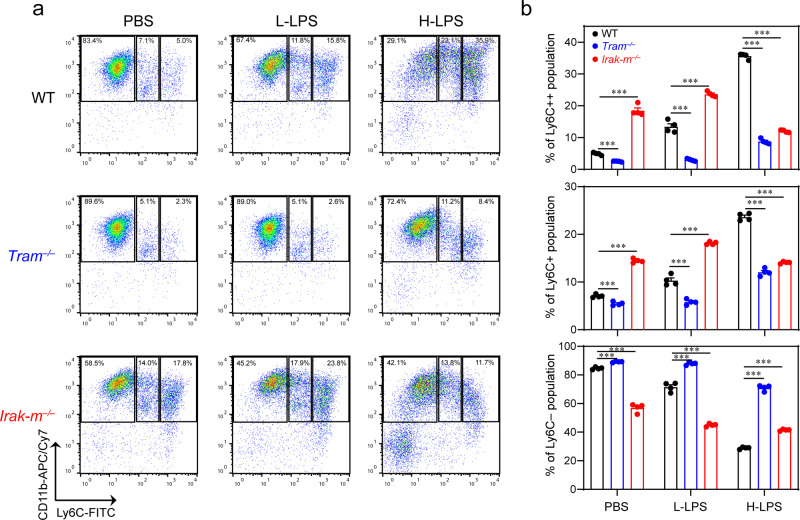
Bone marrow-derived monocytes (BMDMs) were isolated from mice (Suppl. Fig. 1) and dosed with PBS, low-dose LPS (100 pg/mL), or high-dose LPS (1 µg/mL) for a 5 day period as we performed previously58,59. Persistent challenges with low-dose LPS enabled the generation of inflammatory monocytes (both the intermediate Ly6C + and the classical Ly6C++ monocytes) (Fig. 1), consistent with our previous reports58,60. In contrast, exhaustive stimulations with high-dose LPS almost completely depleted the Ly6C- non-classical monocytes, and led to a drastic expansion of the Ly6C++ monocytes (Fig. 1a). The sharp reduction of non-classical monocytes was also observed in septic patients including COVID-19 patients, suggesting the development of innate immune exhaustion61–64. However, the detailed profiles and underlying mechanisms of both low-grade inflammatory monocytes and exhausted monocytes are not well defined. Given our previous findings that TRAM is involved in the generation of low-grade inflammatory monocytes33,60,65, we then evaluated the development of both low-grade inflammation (with low-dose LPS) and exhaustive inflammation (with persistent challenges of high-dose LPS) comparing WT and TRAM-deficient monocytes. Indeed, we observed that the expansion of Ly6C+ and Ly6C++ inflammatory monocytes were completely ablated in TRAM-deficient monocytes when challenged with low-dose LPS (Fig. 1b, Suppl. Data 1). Interestingly, the drastic depletion of non-classical Ly6C- monocytes and severe expansion of exhausted monocytes seen in WT cells challenged with high dose LPS were also attenuated in TRAM-deficient monocytes, suggesting a role of TRAM in both events. We also examined the involvement of IRAK-M during the generation of low-grade vs exhausted monocytes. Consistent with previous findings, we observed that Irak-m deletion enabled the significant expansion of Ly6C+ and Ly6C++ inflammatory monocytes under low-grade inflammatory condition (Fig. 1b). In contrast, Irak-m deletion attenuated the drastic expansion of Ly6C+ and Ly6C++ inflammatory monocytes under the exhausted high-dose LPS challenge. Our data suggest that TRAM may function as a gate-keeper for sensing LPS challenge, while IRAK-M may serve as a downstream fine tuner for the adaption of monocytes.
Fig. 1. The generation of inflammatory monocytes under persistent low-grade or exhaustive challenges, facilitated by TRAM and finely modulated by IRAK-M.
Primary murine monocytes from WT, Tram−/− and Irak-m−/− mice were treated with either PBS, low-dose LPS (100 pg/mL) or high-dose LPS (1 µg/mL) for 5 days, and monocyte subsets were analyzed with flow cytometry. a Representative staining profile of the monocyte subsets. b Quantification of the populations of non-classical (Ly6C−); intermediate (Ly6C+); and classical monocytes (Ly6 + +). Data are representative of three independent experiments, and error bars represent means ± SEM. ***P < 0.001; one-way ANOVA (n = 4 for each group).
Higher LPS dosage causes larger overall change in the epigenome
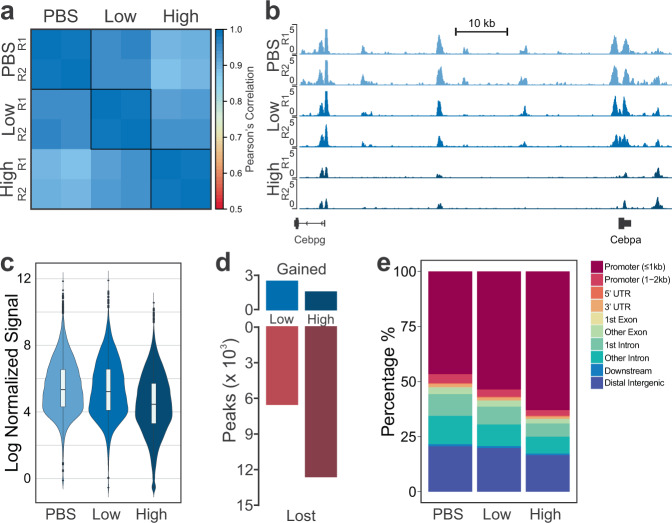
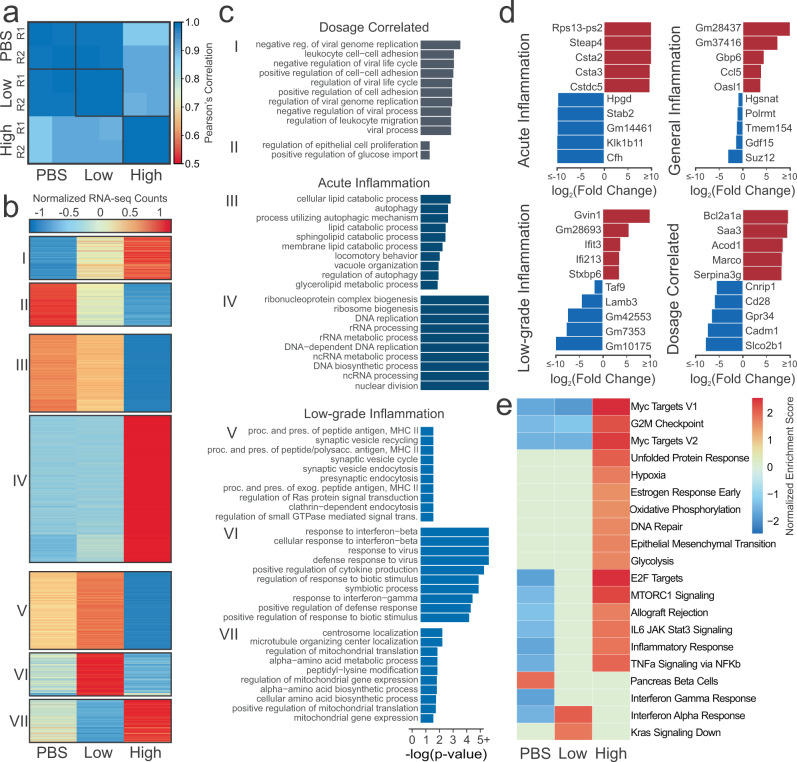
To characterize the global profiles of epigenetic alterations, we then profiled H3K27ac using MOWChIP-seq54 and performed RNA-seq with two replicates per condition (Suppl. Tables 1 and 2). We found very high average correlation between ChIP-seq replicates of 0.986 (Fig. 2a). Between conditions, the highest correlation is between PBS and low-dose (0.957) and the lowest correlation between PBS and high-dose (0.896), with low-dose and high-dose falling in-between (0.939). This suggests that the increasing LPS dosage causes a concomitant change to H3K27ac signal. When looking at genome-wide H3K27ac signal, we see that increasing the dose of LPS tends to reduce the H3K27ac signal (Fig. 2b). In fact, the overall H3K27ac signal at peaks also tends to go down with increasing LPS-dosage (Fig. 2c, Suppl. Data 2). Furthermore, the number of peaks present in each sample was decreased with increasing dosage (PBS = 25,679, Low = 21,597, High = 14,659). As such, many of the peaks that were present in PBS samples were not present in low-dose samples (6,566) and even more (12,602) were not present in high-dose (Fig. 2d, Suppl. Data 2). However, low-dose samples gained a small number of peaks (2,484) while high-dose samples gained even fewer (1,582). In addition, the fraction of peaks near promoters increases with increasing LPS dosage, yet this primarily was due to a reduction of distal peaks, rather than an increase of proximal peaks (Fig. 2e, Suppl. Data 2). However, some of the peaks gained by low-dose and high-dose conditions were proximal to gene promoters. Since H3K27ac also localizes to active promoters, this suggests that LPS dosage is activating genes that are not active under normal conditions66.
Fig. 2. Overview of epigenomic data for murine BMDMs dosed with PBS, low-dose LPS, or high-dose LPS.
a Pearson’s correlation of normalized H3K27ac signal around promoter regions (TSS ± 2 kb). b Tracks of normalized H3K27ac signal for replicates dosed with PBS, low-dose LPS, or high-dose LPS (from top to bottom). Tracks are aligned in mm10 and the region displayed is chr7:35,042,108-35,138,618. c Distribution of normalized H3K27ac signal at peaks (n = 29,071). The middle bar of the boxplot denotes the median. The upper and lower bounds of the box correspond to the 75th and 25th percentile, respectively. Each whisker spans up to the range of 1.5 * interquartile range from the edge of the box. d Number of peaks gained or lost in low-dose or high-dose conditions compared to PBS. e Percentage of peaks at genomic locations.
Low-dose and high-dose LPS stimulations have different effects on enhancers
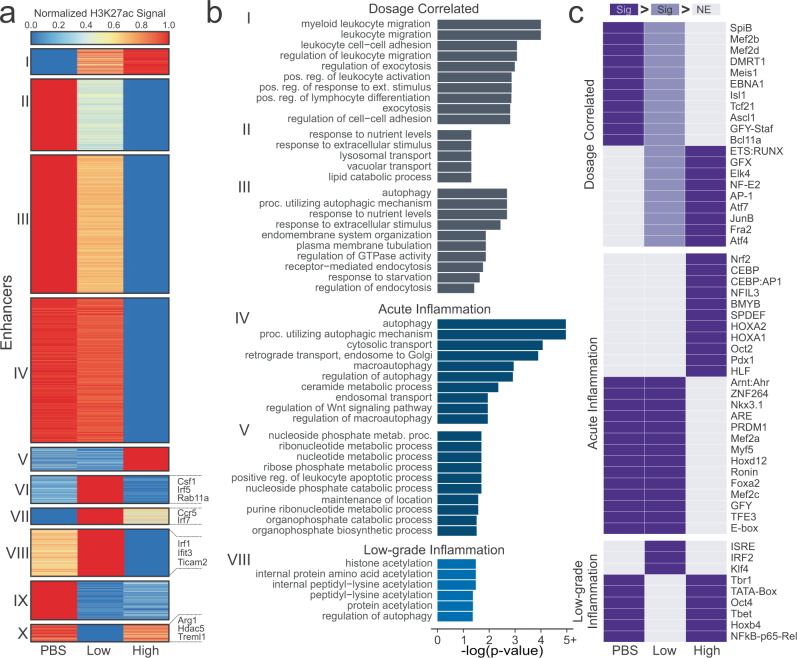
Although H3K27ac does mark active transcription, it plays a more pivotal role in long-range gene regulation at enhancer regions. Therefore, we determined the locations of active enhancers, which were defined as H3K27achigh regions that do not overlap with regions near transcription start sites (TSS) and can be linked to genes in conjunction with RNA-seq data (see Methods). Thus, each of the active enhancers we describe have a statistically significant link to their gene. As these active enhancers very likely regulate their linked genes, we use these genes to determine significantly enriched pathways. This allows us to leverage and integrate our epigenomic and transcriptomic data. For simplicity, we will refer to these active enhancers simply as ‘enhancers’. The number of enhancers also decreased with increasing LPS-dosage (PBS = 5,738, Low = 4,400, High = 2,711). Normalized H3K27ac signal for the conditions at each enhancer was clustered with k-means clustering (Fig. 3a, Suppl. Data 3). The genes linked to the enhancers in each of the clusters were analyzed for overrepresentation of Gene Ontology biological process gene sets (Fig. 3b, Suppl. Data 3). We then separated these clusters into three groups: dosage correlated (I, II, III), acute inflammation (IV and V) and low-grade inflammation (VIII).
Fig. 3. Effect of LPS dosage on enhancers.
a K-means clustering of enhancers present in PBS, Low-dose, or High-dose samples. b Significant (FDR < 0.05) Gene Ontology biological process gene sets for clusters from (a). c Motifs that are significantly enriched (p < 1 × 10−6) in at least one condition compared to another. Dark purple cells denote that motif is significantly enriched (p < 1 × 10−6) in that experimental condition in relation to any light purple or gray cells. Light purple cells are used as a middle ground such that dark purple is more significant than light purple, and light purple is more significant than gray. Non-enriched gray cells (NE) do not have a p-value cutoff, as they refer to the conditions with the lowest enrichment of that motif.
The dosage correlated group, including clusters I, II, and III, had either increased or decreased H3K27ac signal with increased LPS-dosage. Most obviously, we see that Cluster I, which increases in expression from PBS to high-dose, is filled with leukocyte-related (which include monocytes and macrophages) gene ontologies, which play a key role in inflammation67,68. As such, it is reasonable that with increased LPS-dosage we would see a stronger inflammatory response. In Clusters II and III, which decreased from PBS to high-dose, we see several pathways related to autophagy and endocytosis. It has been shown in literature that LPS-stimulation induces autophagy as a means of modulating the inflammatory response69–72. Upon further investigation into the enhancer-linked genes in these autophagy-related processes, we find genes such as Trem273, Sesn174, and Nrbf275, all of which inhibit the inflammatory effect of macrophages. Differential H3K27ac expression at gene promoters also shows that increased LPS-dosage reduces expression at genes that negatively regulate components of the TLR4 signaling pathway, such as the MAPK cascade (Suppl. Fig. 2, Cluster III)37,76,77.
The exhausted group, where the signal was either substantially lower or higher in high-dose LPS compared to low-dose LPS and PBS, consists of clusters IV and V. These enhancers are recognized as highly characteristic of the high-dose condition and associated innate exhaustion. In Cluster IV, we once again see autophagy-related pathways. Gene promoters with substantially lower H3K27ac signal in high-dose cells were enriched in pathways for IL6 and IL8 production, which can have pathogenic inflammatory and immuno-suppressive effects characteristic of exhausted monocytes (Suppl. Fig. 2, Cluster IV)76,77. In Cluster V, we see many nucleotide-associated metabolic and catabolic processes, which have been shown to be increased with LPS stimulation in literature78,79. It is unclear why these processes are not increased in low-dose cells.
The low-grade inflammation group, in which signal was either substantially increased or decreased in low-dose LPS when compared to high-dose LPS or PBS, consists of clusters VI, VII, VIII, IX, and X. These enhancers are recognized as highly characteristic of the low-dose condition and associated low-grade inflammation. Only Cluster VIII had any significantly enriched terms, most of which were related to histone and protein acetylation, primarily acetylation at lysines. LPS stimulation has been shown in literature to affect histone acetylation80, but we do not have enough information to determine which modifications (other than H3K27ac) are affected uniquely by low-dose LPS, though aberrant histone acetylation has been found in multiple chronic inflammatory diseases81,82. However, the enhancers within this grouping were linked to some interesting genes within the immune system. For example, enhancers who had much higher signal in low-dose were linked to genes like Csf1, Irf5, Rab11a, Ccr5, Irf7, Ticam2, Ifit3, and Irf1, while enhancers with lower signal in low-dose were linked to genes like Arg1, Hdac5, and Treml1. RAB11A is responsible for transporting TLR4 from the endocytic recycling compartment to forming phagosomes37. This transport triggers the TRIF-dependent signaling pathway, which requires TRAM (Ticam2). TRAM/TRIF-dependent signaling leads to increases in interferon-alpha and interferon-beta production which, in turn, activate interferon-induced genes such as Ifit3. Hdac583 and Treml184 both regulate the inflammatory response. Furthermore, reduction of Hdac5 expression was also associated with an increase of Irf1 and transcription of interferon-beta83. In addition, Arg1, which is a typical marker of anti-inflammatory macrophages49, is reduced in low-dose while Irf5, shown to promote pro-inflammatory macrophage polarization85, is increased in low-dose. Together, these suggest that, while the low-dose cells are markedly different from high-dose cells, they do have a pro-inflammatory phenotype, which is consistent with previous studies33. It is interesting to note that Csf1, which is associated with anti-inflammatory macrophages, is highly increased in enhancer signal in low-dose cells, however, previous studies have shown an association between Csf1 and chronic inflammation86.
Since enhancers are hotbeds of transcription factor binding activity, the enhancer regions were then scanned for transcription factor binding motifs that were separated into motifs uniquely and significantly enriched or diminished in low-grade inflammation, acute inflammation, or if the enrichment was correlated to LPS-dosage (Fig. 3c). Among the dosage-dependent affected motifs, we see multiple immune-related transcription factors that are increased with increasing LPS-stimulated inflammation such as JUNB87, AP-188,89, ATF490, and NFEE251.
In exhaustive inflammation, we see several transcription factors that are important in the TLR-signaling pathway such as NFIL391,92, HOXA293, NRF294, CEBP:AP195,96, and OCT297,98, all of which are activated with LPS stimulation in literature. In fact, the differential enrichment of NRF2 between high-dose and low-dose is also supported by previous research. NRF2 is activated by LPS stimulation via the reduction of the protein KEAP1, however, KEAP1 protein has been shown to accumulate in low-dose conditions33,94. The MEF2 family (MEF2A, MEF2B, MEF2C, MEF2D) of transcription factors motifs are significantly deficient in high-dose cells, but are present in low-dose cells. It has been show that the MEF2 family is initially upregulated by LPS-stimulation but are soon downregulated51. It is possible that, in the low-dose condition, the LPS-stimulation is not enough to lead to the downregulation of the MEF2 family.
Low-grade inflammation leads to fewer enriched motifs than deficient motifs. Motifs that are enriched only in low-dose are ISRE and IRF2. IRF299,100 is an inflammation regulator while ISRE is an IFN-I stimulating response element which, when bound, activates genes in the inflammation pathway101. This is consistent with previously discussed upregulated enhancer-linked genes. Additionally, IRF2 has been shown to positively regulate the non-canonical inflammasome pathway, which, in turn, leads to increased Gsdmd expression102. In fact, we do see increased Gsdmd expression in our enhancer data, where it is located in Cluster VI. Low-dose also has reduced P65 motifs, a NF-κB subunit that is part of the canonical pathway involved in inflammation103. However, in monocytic cells that have already been stimulated with a Gram-negative bacteria, a second stimulation shows reduced P65 activity104,105. There is a possibility that sustained low-dosage of LPS may lead to such a reaction. Furthermore, it appears that reduced P65 expression can be somewhat compensated for106.
Tram−/− has more profound effects on enhancer activity than Irak-m−/−
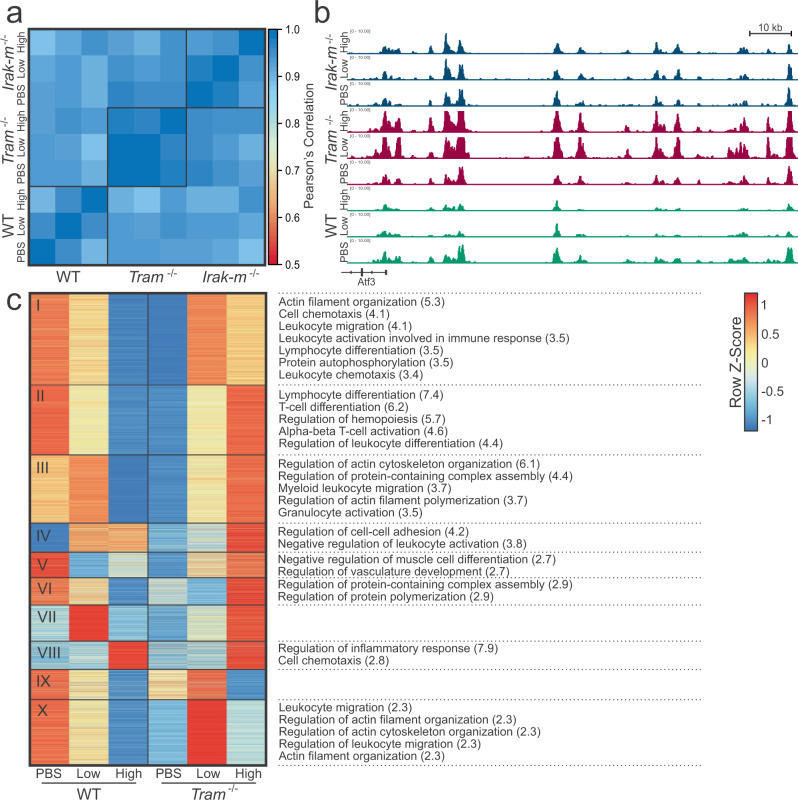
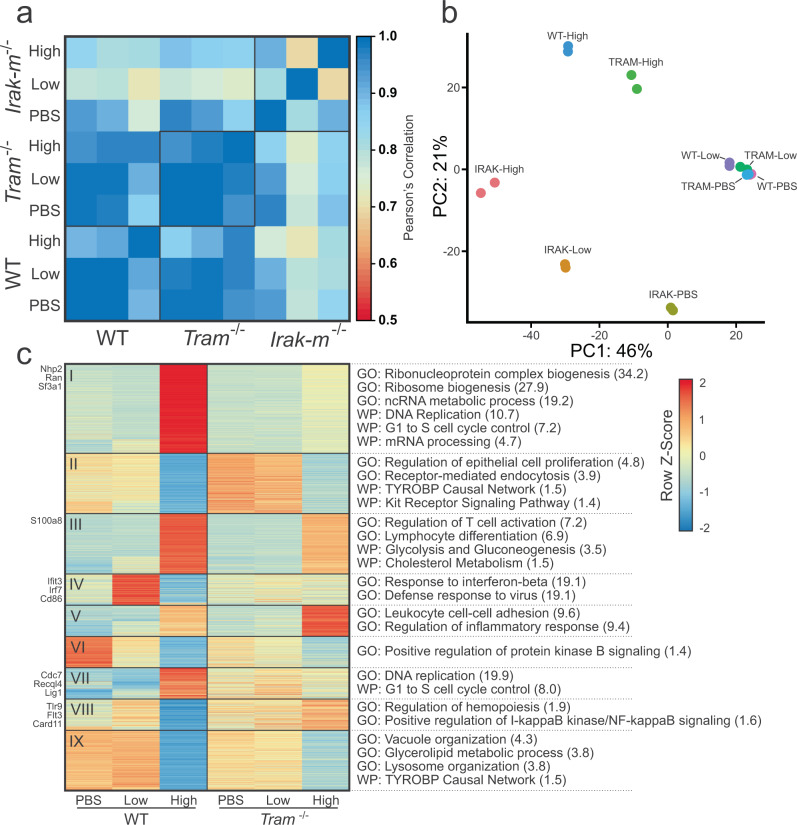
Next, we analyzed how TRAM-deficiency and IRAK-M deficiency alters the effects of increasing LPS dosage on H3K27ac signal and enhancer activity (Fig. 4). Similar to WT, the PBS and low-dose conditions for both TRAM-deficient and IRAK-deficient cells were more correlated with one another (0.990 and 0.980, respectively) than with the high-dose condition (average 0.967 and 0.943, respectively), though the differences were small (Fig. 4a). Correlation among the experimental conditions (PBS = 0.933, low-dose = 0.931, high-dose = 0.938) was worse than within genomic conditions, but was comparable to inter-genomic comparisons (WT vs. TRAM = 0.926, WT vs. IRAK = 0.918, TRAM vs. IRAK = 0.942). This can also be seen in the representative H3K27ac tracks, where the pattern of the H3K27ac signal is similar across the WT and mutant cells, though also with some clear differences (Fig. 4b). To understand what effect that TRAM-deficiency has on WT enhancers, we analyzed the normalized H3K27ac signal of TRAM-deficient cells at the location of the WT enhancers, and compared the WT signal with the TRAM-deficient signal (Fig. 4c). We see that almost all of the enhancers have an opposite or differing pattern in TRAM-deficient cells than in WT cells, suggesting that TRAM-deficiency has an effect on enhancers in both chronically inflamed and exhausted monocytes. In Clusters I, II, and III, we see that TRAM plays a role in actin organization, leukocyte migration, and lymphocyte differentiation in both low-dose and high-dose, though often to different degrees. However, in Cluster X, we found that some of the enhancers associated with leukocyte migration and actin organization are uniquely increased in low-dose conditions in TRAM-deficient cells. In Cluster IV, we also see that TRAM is necessary for regulating leukocyte activation, which appears to be needed for low-dose conditions but not high-dose. This suggests that perhaps TRAM plays a role in preventing the overactivation of the immune system in low-grade inflammation.
Fig. 4. Effect of LPS on H3K27ac in TRAM-deficient and IRAK-M-deficient cells.
a Pearson’s correlation of normalized H3K27ac signal at promoter regions (TSS ± 2 kb) of WT, Tram−/−, and Irak-m−/−. b Representative tracks of normalized H3K27ac signal from WT, Tram−/−, and Irak-m−/−. Tracks are aligned in mm10 and the region displayed is chr1:191,173,497-191,334,210. c Normalized H3K27ac signal at WT enhancers in WT and TRAM-deficient cells. Significant biological process gene ontologies (FDR < 0.05) listed on the right, with log(FDR) in parentheses.
We next performed a similar analysis of the effect of IRAK-M-deficiency on enhancers (Suppl. Fig. 3a). We did find noticeable differences with significant pathway enrichment in most of the clusters. In Cluster V, we see that the Myd88 pathway normally inhibits the transcription of primary miRNAs—this might be worth future investigation, as miRNAs play a role in regulating gene expression. In Cluster VIII, we see that, while the overarching pattern is the same, IRAK-deficiency shifts the low-dose profile closer to the high-dose profile, suggesting that IRAK-M has a small role in regulation here with regards to migration, reactive oxygen species metabolic processes, and endocytosis. In Cluster XI, we see that peptidyl-tyrosine phosphorylation is substantially reduced in IRAK-M-deficient low-dose and increased in high-dose. Tyrosine phosphorylation is a notable part of the LPS signaling cascade. It is possible that IRAK-M is necessary in low-grade inflammation for prolonged activation and in high-dose for inhibition. Cluster XII shows that IRAK-M deletion also appears to have an affect on enhancers associated with CD4+ T cell activation. One of the key genes in this cluster is Tnfrsf1b, which encodes Tnfr2 and has been shown to promote survival in monocytes at low concentrations in a pathogenic environment107. Since the expression increases in low-dose IRAK-M-deficient cells, this suggests perhaps that the Myd88 pathway has a mild protective effect on monocytes in low-grade inflammation conditions. We also see that this cluster is related to Cluster VI. In both clusters, IRAK-deficiency leads to increased lymphocyte activation (or decrease negative regulation) in low-dose conditions, and to decreased activation in high-dose conditions. We also noticed that there seemed to be more consensus between H3K27ac signal of WT and IRAK-M-deficient cells at WT enhancer locations, and confirmed that TRAM-deficient cells primarily negatively correlated with WT cells, while IRAK-M-deficient cells positively correlated (Suppl. Fig. 3b). To determine if this was an artifact of only examining the enhancer signal at WT enhancer locations, we analyzed the distribution of signal at each of the respective, individual enhancer locations (i.e., WT, Tram−/−, Irak-m−/− etc.) and found that this trend holds in their respective enhancers as well (Suppl. Fig. 3c). While enhancer signal decreases with increasing LPS dose for WT and IRAK-M-deficient cells, enhancer signal increases with increasing LPS dose for TRAM-deficient cells. This suggests that the decrease in H3K27ac signal with increasing LPS dosage is possibly TRAM dependent.
Transcriptomic changes due to low-dose and high-dose LPS exposure mirror epigenomic changes
We analyzed RNA-sequencing data to further understand the effect of LPS on murine immune cells. We see a similar pattern of RNA-seq data correlation as was in the ChIP-seq data (Fig. 5a). Average replicate correlation was 0.996 and the correlation between PBS and low-dose was similarly high (0.987). The high-dose replicates had approximately similar correlations with either PBS (0.892) or low-dose (0.914). Unlike the H3K27ac signal, there was not a decrease in RNA signal with increasing LPS-dosage (Suppl. Fig. 4a). Much like the differential peaks, the number of differentially expressed genes (DEGs) was lowest between PBS and low-dose (155) with much greater differences between PBS and high-dose (3,249) closely followed by low-dose and high-dose (3,061) (Suppl. Fig. 4b, Suppl. Data 4). However, differentially expressed genes were largely equally split into up- and down-regulated genes for each comparison, except between PBS and low-dose, where there were more genes upregulated in low-dose than PBS samples. Many genes (~63%) that are differentially expressed in one comparison, are differentially expressed in multiple comparisons (Suppl. Fig. 4c).
Fig. 5. Effect of LPS dosage on gene expression.
a Pearson’s correlation of normalized RNA-seq counts at genes b Heatmap of normalized gene expression of DEGs present in more than one comparison. c Significant biological process gene ontologies for genes that are differentially expressed in more than one comparison. d Top upregulated and downregulated genes for each condition. e Gene-set enrichment analysis. Color denotes normalized expression score if that pathway was significant (FDR < 0.05) in sample vs. rest.
The normalized RNA-seq expression data of DEGs were visualized in a heatmap (Fig. 5b, Suppl. Data 5). Each of these clusters were analyzed for enrichment of Gene Ontology biological process gene sets (Fig. 5c, Suppl. Data 5). Dosage correlated Clusters I and II, which increased or decreased with added LPS-dosage, are consistent with enhancer analysis. Both analyses identified leukocyte-related pathways were increased with increasing dosage. In addition, negative viral process was also increased with additional LPS. While LPS is from a bacteria, TLR4 can also be activated by viral ligands, thus there is large overlap in the genes in each of these ontologies108. We also see an reduction of glucose import with increased LPS-dosage, which is also consistent with literature109. Clusters specific to acute inflammation, Clusters III and IV, had similar autophagy and nucleic acid-related processes as the corresponding enhancer clusters. However, Cluster III also had more lipid metabolic processes that were down-regulated in high-dose conditions, which is consistent with previous research showing LPS-stimulation to decrease lipid catabolism110.
Finally, Clusters V, VI, and VII were specific to low-dose. In Clusters V and VI, we see ontologies that are significantly upregulated in low-dose compared to high-dose or PBS. These include MHC II antigen processing and response to interferon-beta. MHC II antigen processing is the means by which an exogenous antigen is prepared and presented on the cell surface for the activation of CD4+ T cells. These cell surface molecules that present the antigen are only upregulated in the TRIF-dependent pathway. Furthermore, what the macrophage secretes can also affect CD4+ T cell function and aberrant CD4+ T cells have been implicated in inflammatory diseases111,112. Interferon-beta related genes and motifs were also found to be enriched or have increased enhancer expression in low-dose cells compared to high-dose or PBS. This suggests that epigenetics play a notable role in the etiology of low-grade inflammation. This is further supported by the peptidyl-lysine modification process in Cluster VII, which is down in low-dose and high in high-dose. With closer inspection, Hdac2 and Hdac4 are included within this clusters. Hdac2 is considered to be crucial to the LPS inflammatory response while also mediating it, and decreased Hdac2 levels have been found in COPD patients113–115. Hdac4 is necessary for LPS-stimulated production of pro-inflammatory cytokines, and degradation leads to secretion of HMGB1, which is believed to play a substantial role in sepsis116,117.
The fold-changes of the top genes significantly expressed or repressed in exhaustive Acute Inflammation (high-dose vs. rest), Low-grade inflammation (low-dose vs. rest), General Inflammation (PBS vs. rest), and Dosage-specific (increasing or decreasing from PBS to high-dose) were inspected (Fig. 5d, Suppl. Data 5). In each, we see some genes that have been pointed out in literature before as affected by inflammation or LPS in some way, such as Steap4118, Bcl2a1a119, Saa3120, Marco121, Cfh122, Ifit3123, Lrrc14b124, Ccl5125, and Cd28126. We also see multiple predicted genes that might be worth further in vivo or in vitro functional investigation.
Gene-set enrichment analysis was then performed using each of the conditions compared to the rest (Fig. 5e). The first aspect of note is that high-dose is significantly enriching many pathways that have been found to be enriched by LPS-stimulation in literature such as Myc127,128, hypoxia129, glycolysis130, and unfolded protein response131. We also see oxidative phosphorylation, which is generally believed to be reduced in LPS-stimulation as the cell transfers to glycolysis, though there is some conflicting evidence132. Yet, there are two reasons that it might be enriched in high-dose. First, oxidative phosphorylation is necessary for inflammatory resolution133. Or, it could be as a result of glucose starvation, which can cause cells to shift back towards oxidative phosphorylation134. In addition, there are also pathways that seem to be somewhat dosage-dependent, such as E2F targets135, MTORC1136, IL6 JAK Stat3 signaling137,138, and TNF-α139,140 that are hallmarks of inflammation. However, it is interesting to note that it appears low-dose cells are not undergoing as much replication, as seen in the reduced G2M checkpoint and glycolysis. Furthermore, it appears that, while slight interferon-gamma enrichment141,142 occurs in both low-dose and high-dose (seen by deficiency in PBS), that low-dose has stronger enrichment of interferon-alpha and Kras signaling down. Interestingly, interferon-alpha overexpression, which studies suggest can lead to chronic inflammation143, is a characteristic of systemic lupus eruthematosus, an autoimmune disease144,145. The reduction in Kras signaling, which is part of the Ras/MAPK pathway, would also point to reductions in the cell-cycle of low-dose cells146.
As differential transcript usage (DTU), such as alternative splicing, has been shown to be affected in the macrophage inflammatory response147, we also chose to examine DTU among the three experimental conditions as differential transcript usage can effect the function of the genes involved. There were a total of 309 genes that had significant DTU with predicted functional consequences across the three comparisons (PBS vs. low-dose = 82, PBS vs. high-dose = 171, low-dose vs. high-dose = 172) (Suppl. Fig. 5a). Genes with DTUs in PBS versus High-dose were entirely associated with RNA-splicing ontologies (Suppl. Fig. 5b), as well as many in low-dose vs high-dose (Suppl. Fig. 5c). However, low-dose versus high-dose did have several immune related ontologies, suggesting that DTU may play a role in the differing responses. In particular, we see response to interferon-beta and one of the DTU genes that was involved, IFI204, plays a key role in interferon-beta release148. We also found that, of the 172 DTUs in low-dose vs. high-dose, 111 had (FDR < 0.05) differential H3K27ac signal at their promoters (expected = 40) and 38 were linked to enhancers (expected = 22) for a total of 121 DTUs associated epigenetic regulation. This further suggests that epigenetic regulators may be playing a role in the differences between acute and low-grade inflammation.
Tram−/− and Irak-m−/− each have roles under low-dose and high-dose LPS conditions
The increase that we see of type I interferons-alpha and beta-related processes in low-dose cells, both epigenetically and transcriptomically, as well as increased epigenetic enhancement of TRAM (RNA-seq FDR < 0.05, FC ≈ 1.8) led us to postulate that low-dose LPS might lead to more use of the TRIF-dependent pathway (which requires TRAM), while high-dose might lead to more use of the MyD88-dependent pathway (Fig. 3a and Fig. 5c, e)37. In order to better understand the mechanistic pathways that might be differentially affected by low-dosage of LPS, we performed additional experiments using TRAM-deficient and IRAK-M-deficient BMDMs (Fig. 6a).We see that WT-high-dose cells correlate the strongest with TRAM-deficient-high (r = 0.961) followed by the low-dose and PBS conditions (r = 0.866 – 0.915), while the IRAK-M samples had the lowest correlation (High = 0.815, PBS = 0.762). Average correlation between the WT and TRAM-deficient low-dose and PBS samples was very high at r = 0.986. IRAK-M-deficient cells had the lowest average intra-group correlation (IRAK-M = 0.803, TRAM = 0.969, WT = 0.934) and the lowest average inter-group correlation (IRAK-MvWT = 0.819, IRAK-MvTRAM = 0.847, WTvTRAM = 0.952). We can also see this pattern in the representative RNA-seq tracks of all three groups (Suppl. Fig. 6a). When we perform principle component analysis of the most variable genes, we see fairly similar results (Fig. 6b). These data were consistent with our initial independent flow-cytometry based studies, demonstrating that the expansion of low-grade inflammatory monocytes by low-dose LPS was completely TRAM dependent (Fig. 1), and that the exhaustion of monocytes induced by higher dose LPS was also partially TRAM dependent. In contrast, IRAK-M deficiency led to an expansion of the low-grade inflammatory monocyte when challenged with low-dose LPS, while IRAK-M deletion attenuated the magnitude of monocyte exhaustion in cells treated with high dose LPS. Due to the substantial differences in IRAK-M-deficient cells, we chose to focus more on the changes present in TRAM-deficient cells.
Fig. 6. Effect of TLR4 pathway on LPS-dosage response.
a Pearson’s correlation of RNA-seq signal between conditions. b Principle Component Analysis of replicates using top 500 variable genes. c Heatmap of WT and TRAM-deficient gene expression at varied LPS-dosage using DEGs from the WT comparisons. Significant pathways on right are from Gene Ontology or WikiPathways. Log(FDR) for each in parentheses.
We then analyzed gene expression across the three dosing conditions for both WT and TRAM-deficient cells, though we restricted the analysis to genes that were differential expressed between the WT conditions (Fig. 6c, Suppl. Data 6). A decent number of genes were moderately affected by Tram deletion, but most maintained a similar expression pattern to WT. Despite this, there are 4 clusters of interest (Clusters I, IV, VII, and VIII) that stand out. Clusters I and VII both show a failure to upregulate DNA replication pathways in TRAM-deficient high-dose cells. While there is evidence of macrophage renewal due to inflammatory insult, it is not well understood, but it is possible that it is TRAM-dependent149,150. We also noticed that S100a8 was preferentially induced by high dose LPS in WT cells, and less in TRAM deficient monocytes.
On the other hand, Clusters IV and VIII both have a pattern of low-dose having the highest expression and high-dose the lowest in WT. In Cluster VIII, Tlr9 is an intracellular toll-like receptor that recognizes unmethylated CpG motifs in bacterial or viral DNA151. However, TLR9 activation has been shown to occur after LPS stimulation152 and TLR9 inhibition helps suppress excessive inflammation in bacterial sepsis153–155. Furthermore, it helps regulate antigen presentation in macrophages156 and participates in interferon-alpha production through IκB kinase signaling157. Interestingly, TLR9 is found to be activated through the NF-κB, ERK, JNK, and p38 MAPK pathways which are not TRIF-dependent, so it is unclear why there is not an increase in TRAM-deficient cells as well152,158.
Another critical gene is Irf7, which is upregulated in WT-low but not in TRAM-low, that is important for the production of type I interferons159. Furthermore, IRF7 expression can lead to a feed-forward loop of type I interferon production, much like we see in WT-low160,161. However, research has shown that IRF7 is necessary for the IL1B and the type I interferon response elicited through TLR4, and that IRF7 is induced through the TRIF-dependent pathway, consistent with the lack of IRF7 increase in TRAM-deficient cells. Trim28 is a negative regulator of IRF7159 and is significantly reduced in WT-low compared to WT-high cells (FDR < 0.05, FC < 2), while remaining unchanged across the TRAM-deficient cells. If TRIM28 is phosphorylated at serine 473 through a PKR/p38 MAPK/MSK1 signaling cascade, it is no longer capable of inhibiting IRF7162. Although knockdown of activated Msk1 does increase production of some inflammatory cytokines in neuroinflammation, TRIM28 is responsible for regulating a staggering number of genes, so it is unclear if it is involved162,163. In addition, phosphorylation of TRIM28 is not necessarily equivalent to reduced gene expression. FOXO3164 and CFLAR165 have also been shown to inhibit IRF7 but neither had notable differences in gene expression.
We performed a similar analysis of comparing the expression of WT DEG genes in WT and IRAK-M-deficient cells (Suppl. Fig. 6b). We see that Clusters I, II, IV, V, and VI had the most clear differences between the two cell-types, though Cluster III did not have any significantly enriched pathways. In Cluster IV, we see that there is much higher signal in low-dose IRAK-M-deficient cells, and that these genes are associated with a defense response to virus and response to interferon-beta. We previously saw that this was increased in low-dose WT cells (Fig. 5c), but it seems to be even further increased in IRAK-M-deficient low-dose, suggesting that the MyD88 pathway may have a role in regulating the low-dose response. Clusters V and VI generally show a loss of catabolic activity in PBS and low-dose conditions. Clusters II and VII show increases in metabolic pathways (shown as a decrease of negative regulation in Cluster VII) in high-dose conditions. In DNA replication, mTORC1 is responsible for inhibiting catabolic processes and possibly involved in purine and pyrimidine synthesis166. We note that WT high-dose already had increased mTORC1 signaling (Fig. 5e) and lower catabolic activity (Suppl. Fig. 6b). Therefore, this suggests that Irak-m deletion causes an increase in DNA replication activity across all conditions.
TRAM is involved in the generation of exhausted monocytes
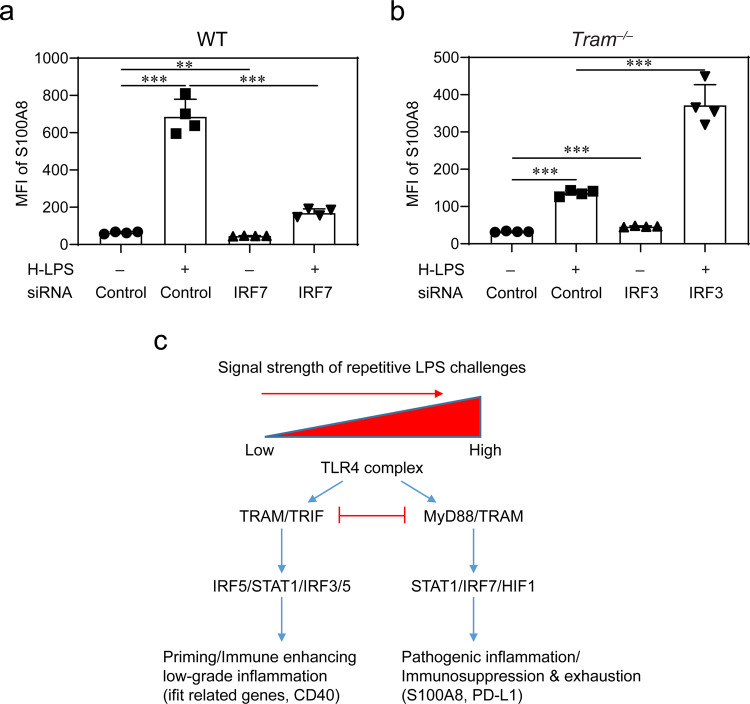
Although the immune-enhancing effects of monocytes may be generated through TRIF167, it is not clear how monocyte exhaustion may develop. Our data suggest that with higher dose LPS challenge, TRAM, in contrast to TRIF, may distinctly mediate the generation of exhausted monocytes. Exhausted monocytes treated with high-dose LPS express pathogenic mediators such as S100A8, independently identified in monocytes from patients suffering from sepsis or COVID-19168–170. We observed that TRAM-deficient monocytes expressed less S100A8 following persistent challenges with high-dose LPS. We independently confirmed such findings with flow cytometry analyses (Fig. 7a, b, Suppl. Data 7). Based on the clue that IRF7 elevation is dependent upon TRAM, we further performed siRNA studies to confirm the causal connection between elevated IRF7 with the expression of S100A8171. As shown in Fig. 7, siRNA knock-down significantly suppressed the induction of S100A8 in WT monocytes challenged with high-dose LPS. IRF3 and IRF7 form a competitive circuit171, and reduction of IRF3 was shown to cause enhanced activation of IRF7 (Fig. 7). We then performed siRNA studies to knockdown IRF3 in TRAM-deficient monocytes. Figure 7 also shows that TRAM-deficient monocytes with Irf3 knockdown robustly expressed S100A8, as compared to control TRAM-deficient monocytes.
Fig. 7. Causal contribution of TRAM-mediated IRF7 during the generation of exhausted monocytes.
a WT monocytes were treated with high-dose LPS (1 µg/mL) in the presence of IRF7 siRNA or control siRNA for 5 days. The expression of S100A8 was determined with flow cytometry. b Tram−/− monocytes were treated with high-dose LPS (1 µg/mL) in the presence of IRF3 siRNA or control siRNA for 5 days. The expression of S100A8 was determined with flow cytometry. Data are representative of three independent experiments, and error bars represent means ± SEM. **P < 0.01, and ***P < 0.001; one-way ANOVA (n = 4 for each group). c A schematic summary of the potential dynamics involved in the programming of inflammatory monocytes and exhausted monocytes.
Together, our data revealed that TRAM is uniquely involved in the generation of not only the inflammatory monocytes, but also the exhausted monocytes, likely through sustaining IRF7 activation (Fig. 7c).
Discussion
Our genome-wide study of gene expression and histone modification changes suggest that low-grade inflammation condition (with persistent low-dose LPS challenges) leads to the generation of low-grade inflammatory monocytes with interferon signatures, while exhaustive inflammation (with persistent high-dose LPS stimulations) leads to a near complete depletion of non-classical monocytes and polarization of Ly6C++ monocytes, differentially modulated by TRAM. First, we found that these changes are seen in the variation of enhancers, the regulators of gene expression. Enhancers with increased signal in low-dose samples were linked to genes related to the TRAM/TRIF-dependent pathway. In addition, promoters and motifs enriched in low-dose samples were also associated with the interferon-beta response. Second, we showed that genes with increased expression preferentially in low-dose LPS, such as Irgm1, Ifit3, and Bst2, were significantly involved in interferon-beta and interferon-alpha responses. Furthermore, genes with differential transcript usage between low-dose and high-dose were also associated with interferon-beta response. Finally, we compared the gene expression of WT samples to TRAM-deficient mice to determine that TRAM-deficiency abrogates the interferon-beta associated genes, indicating that the low-dose increases in interferon-beta specifically rely on TLR4-associated TRAM/TRIF-dependent pathway, as opposed to MyD88 pathway. In the context of exhaustive inflammation, we observed that TRAM-mediated STAT1/IRF7 circuitry may be involved in the expansion of Ly6C++ monocytes with elevated induction of S100a8, a key signature gene elevated in septic patients including COVID-19 patients.
There is increasing appreciation for signal-strength dependent programming of both innate and adaptive immune systems, enabling complex and dynamic host responses to changing landscapes of infectious and inflammatory conditions172,173. Although much progress has been made with the dynamic programming of T helper cells exhibiting multi-staged activation and exhaustion under signal-strength and history dependent challenges174, the similar scenario of innate immune cell adaptation is still less understood. Due to limited systems approaches, even the most well-known concept of endotoxin tolerance regarding innate cell adaptation to repeated LPS challenges fails to clarify the complex innate immune adaptation dynamics. Past studies regarding endotoxin tolerance overly focused on dampened gene expression of limited inflammatory mediators32, and failed to address augmented induction of diverse immune-; metabolic-; and proliferative related genes involved in complex adaptation to higher dosages of endotoxin as increasingly recognized as the exhausted phenotype with dual features of pathogenic inflammation and immune suppression. Much less is known about innate responses to pathologically relevant subclinical low-dose LPS, highly prevalent in humans with chronic conditions due to mild leakage of mucosal barriers175. The lack of systems and clear understanding of signal strength and history-dependent adaptation to LPS underlies our limited translational success in treating related diseases ranging from acute sepsis to chronic cardiovascular diseases. Our current study provides a comprehensive assessment of gene expression dynamics as well as corresponding epigenetic variations in monocytes challenged with rising dosages of LPS.
Confirming limited previous studies, our collected data reveal that higher doses of LPS not only cause suppression of certain subsets of inflammatory genes, but also potently induce wide arrays of genes involved in altered immune metabolism and proliferative potential. Further characterization of these altered gene expression landscape may help better explain the compound phenotypes of pathogenic inflammation and immune-suppression observed in septic leukocytes collected from human sepsis patients and model murine septic animals176,177. In contrast, low-dose LPS preferentially induces inflammatory interferon responsive genes, recently shown to be expressed in inflammatory monocytes collected in vivo from various chronic inflammatory disease models including lupus and atherosclerosis145,178,179.
Our integrated analysis of epigenomic changes at enhancer and promoter regions complements our gene expression data, in further revealing the preferential usage of TRAM/TRIF pathway by low-dose LPS. Our finding is consistent with limited previous studies showing that the TRAM/TRIF-dependent pathway is favored in low-dose LPS conditions and critical for lesion development in atherosclerosis33. In addition, we identified signature transcription factors involved in monocyte activation by low-dose LPS such as IRF1, 5, and 7, with IRF5 previously reported to be involved in monocyte priming by low-dose LPS59. The preferential enrichment of additional transcription factors such as IRF2 and KLF4 is also interesting, and may provide additional insight regarding the regulation of low-grade inflammatory monocytes. Low-dose LPS also enhanced the expression of Rab11a, a molecule involved in endocytic recycling of TLR4, providing a further mechanistic explanation for our previous observation that internalization of TLR4-LPS complex and the activation of TRAM/TRIF pathway are required for sensing low-dose LPS35. It is worth to note that TRIF may play a beneficial role under septic conditions by preventing immune exhaustion and enhancing immune functions180–182. In contrast, our data suggest that TRAM may diverge with TRIF under the exhaustive condition with high dose LPS challenge, and facilitate monocyte exhaustion through elevated STAT1/IRF7 circuitry. Further studies are warranted to tease out the complex coordination and competition between MYD88, TRAM, TRIF and other related TLR4 adaptors in directing the adaption of monocytes in signal strength and history dependent fashion.
Collectively, our integrative systems study further clarifies the highly complex and dynamic adaptation of macrophages to rising dosages of LPS, and reveals more of the inner workings of underlying mechanisms, yet much more research will be required to fully understand how immune pathways and components interact in the dynamic ontogeny of macrophage activation states related to the etiology of low-grade inflammation. Studies that utilize time courses, additional LPS concentrations, and other transgenic mice all would be beneficial for truly unveiling complex dynamics of monocyte programming and memory including the relative stability and plasticity of reprogrammed cells both in vitro and in vivo. Although ample studies reveal the temporal and spatial dependent memory of adaptive immune cells such as T helper cells183, the extent of innate immune memory and its potential reversal still remains largely unknown. Information revealed in this report complements emerging studies that monocytes may adopt diverse states and/or intermediate states with overlapping features far exceeding the original simple paradigm of M1/M2 states184. Future extensive studies will be needed to address these important questions. Additional epigenomic studies may also reveal the causal relationships at play and possible therapeutic targets. Together, this could lead to identification of relevant molecular targets in human immune cells for future clinical applications.
Methods
Mice
C57/BL6 mice purchased from the Jackson Laboratory were bred and maintained in pathogenic-free conditions. Male 8–12-week-old mice were used in this study. The Institutional Animal Care and Use Committee (IACUC) approved all procedures performed on the mice. The TRAM-deficient and IRAK-M-deficient mice used were also on a C57/BL6 background.
Cell culture
Crude BM cells were isolated from the mice and cultured as previously published33. Briefly, cells were pooled from 6 to 7 mice and split into three separate plates. The cells were cultured in RPMI complete media (RPMI 1640 with 10% FBS, 2 mM L-glutamine, and 1% penicillin/streptomycin) along with M-CSF (10 ng/mL) and either PBS, low-dose LPS (100 pg/mL), or high-dose LPS (1 µg/mL). Fresh LPS was added every two days and the cells were harvested after 5 days.
Flow cytometry analyses
For analyzing subset proportions, the monocytes treated with LPS for 5 days were harvested and incubated with anti-CD16/-CD32 antibodies (1:100 dilution, BD Biosciences, no. 553141) to block Fc-receptors. The cells were stained with fluorochrome-conjugated anti-CD11b (1:200 dilution, BioLegend, no. 101226) and anti-Ly6C (1:200 dilution, BioLegend, no. 128006) antibodies, and PI (Sigma-Aldrich) was added before flow cytometry. For examining S100A8 expression, the monocytes were treated with high-dose LPS (1 µg/mL), and IRF7 siRNA (Thermo Fisher Scientific), IRF3 siRNA (Thermo Fisher Scientific), or control siRNA (Thermo Fisher Scientific) was mixed with Lipofectamine reagent to transfect monocyte cultures (25 pmol siRNA/well). Fresh LPS and siRNA were added every two days. After 5 days, the cells were fixed, permeabilized, stained with fluorochrome-conjugated anti-S100Ab antibody (1:100 dilution, Novus, no. NBP2-27067AF647), and then analyzed by flow cytometry.
Chromatin shearing
Samples containing 106 cells were centrifuged at 1600 g for 5 min at 4 °C. Each sample was washed twice with cold 1 mL PBS and resuspended in 9.375 mL of PBS. Then, 0.625 mL of 16% formaldehyde was added and the samples were incubated on a shaker for 5 min at room temperature. The samples were then quenched with 0.667 mL of 2 M glycine and incubated for 5 min at room temperature on a rotator. The cells were then centrifuged at 1600 g for 5 min and washed twice with 1 mL cold PBS. The pellet was resuspended in 130 µL of Covaris sonication buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.1% SDS and 1× protease inhibitor cocktail (PIC)) and sonicated with a Covaris S220 sonicator using 75 W peak incident power, 5% duty factor, and 200 cycles per burst for 16 min at 4 °C. Sonicated samples were centrifuged at 16,100 × g for 10 min at 4 °C before the supernatant containing sheared chromatin was removed to a fresh tube. 2.4 µL of sheared chromatin was then mixed with 46.6 µL of IP buffer (20 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% (w/v) sodium deoxycholate, 0.1% SDS, 1% (v/v) Triton X-100, 1% freshly added PMSF and PIC) to generate a 50 µL sample containing chromatin from 20,000 cells for MOWChIP-seq.
Bead preparation
IP buffer (20 mM Tris-HCl [pH 8], 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% (w/v) sodium doxycholate, 0.1% SDS, 1% (v/v) Triton-100X in Milli-Q water) was used to wash protein A-coated Dynabeads (Life Technologies). The beads were resuspended in 150 μL of IP buffer with 0.5 μg of H3K27ac (abcam, cat: ab4729, lot: GR312651-2) antibody, then rotated for 2 h at 4 °C. After rotation, the beads were washed with IP buffer three times before being resuspended in 5 μL of IP buffer and placed on ice.
MOWChIP-seq
Sonicated chromatin samples of 20,000 cells per assay were profiled for H3K27ac (abcam, 0.5 µg per assay, cat: ab4729, lot: GR312651-2) with MOWChIP-seq as described in our previous publications53,185. Briefly, IP buffer is flowed into the device at a rate of 200 μL/min (30 psi) until all air is expelled. The sieve valve was closed and the antibody-coated beads were loaded into the device to form a packed bed. Then, the chromatin was flowed through at a rate of 1.5 μL/min. The beads were then washed in an oscillatory manner using first 50 μL of Low Salt washing buffer (20 mM Tris-HCl [pH 8], 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% (v/v) Triton-100X in Milli-Q water) for five minutes at ~0.65 psi with the valve open and then another five-minute with High Salt washing buffer (20 mM Tris-HCl [pH 8], 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% (v/v) Triton-100× in Milli-Q water). IP buffer was used to flow out the beads. Finally, the ChIP DNA was isolated from the beads by phenol-chloroform extraction. Libraries for sequencing were prepared using the Accel-NGS 2S Plus DNA Library Kit (Swift-Bio) and samples were sequenced using an Illumina HiSeq 4000 with single-end 50 nt reads.
RNA-seq
In total 10,000 cells were used to produce each RNA-seq library, with two replicates for each genotype and experimental condition. RNA was extracted into a 30-µL volume using the RNeasy Mini Kit (74104, Qiagen) and RNase-Free DNase Set (79254, Qiagen), following the manufacturer’s instruction. The extracted mRNA was then concentrated by ethanol precipitation and resuspended in 4.6 μL of RNase-free water. Next, we used the SMART-seq2 protocol55, with minor modifications, to prepare cDNA. 2 μL of oligo-dT primer (100 μM) and 2 μL of dNTP mix (10 mM) were added to 2 ng of mRNA in 4.6 µL of water. The mRNA solution was denatured at 72 °C for 3 min, then immediately placed on ice. Next, 11.4 μL of reverse transcription mix [1 μL of SuperScript II reverse transcriptase (200 U/μL), 0.5 μL of RNAse inhibitor (40 U/μL), 4 μL of Superscript II first-strand buffer, 1 μL of DTT (100 mM), 4 μL of 5 M Betaine, 0.12 μL of 1 M MgCl2, 0.2 μL of TSO (100 μM), 0.58 μL of nuclease-free water] was added to the mRNA solution. For the reverse transcription reaction, the solution was incubated at 42 °C for 90 min, followed by 10 cycles of 50 °C for 2 min, 42 °C for 2 min, then inactivation at 70 °C for 15 min. 20 μL of the resulting solution (first-strand mixture) was then mixed with 30 μL of PCR mix [25 μL KAPA HiFi HotStart ReadyMix, 0.5 μL IS PCR primers (10 μM), 0.5 μL Evagreen dye, and 4 μL nuclease-free water] and amplified using the program 98 °C for 1 min, followed by 9–11 cycles of 98 °C 15 s, 67 °C 30 s, 72 °C 6 min. Finally, the cDNA was purified using 50 μL of SPRIselect beads. RNA-seq libraries were generated with the Nextera XT DNA Library Preparation kit (FC-131-1024, Illumina) and manufacturer’s protocol, using approximately 600 pg of purified cDNA from each sample. Samples were sequenced using an Illumina HiSeq 4000 with single-end 50 nt reads.
Data processing
Unless otherwise mentioned, all data analysis was performed with Bash scripts or with R (The R Foundation) scripts in RStudio. Sequencing reads were trimmed using default settings by Trim Galore! (Babraham Institute). Trimmed reads were aligned to the mm10 genome with Bowtie186. Peaks were called using MACS2 (q < 0.05)187. Blacklisted regions in mm10 as defined by ENCODE were removed to improve data quality188. Mapped reads from ChIP and input samples were extended by 100 bp on either side (250bp total) and a normalized signal was calculated using Eq. (1).
| 1 |
For Pearson’s correlation, the signal was calculated around the promoter region (TSS ± 2 kb) and plotted with the corr and levelplot functions. For visualization in IGV (Broad Institute), the signal was calculated in 100bp windows over the entire genome and output as a bigWig file. RNA-seq data was quantified using Salmon189 against the mm10 transcriptome using a full decoy and normalized counts were calculated with DESeq2190.
Enhancers analysis
To call enhancers, we considered H3K27achigh regions that did not intersect with promoter regions to be enhancer regions. First, consensus H3K27ac peak sets were generated for each of the experimental conditions. Peak widths were expanded to be 1000 bp long (summit ± 500 bp). Promoters were defined as TSS ± 2000 bp. Any H3K27ac 1 kb regions that intersected with a promoter region was removed and the remaining regions were designated as putative enhancers. The signal at each of the putative enhancers was then correlated (Spearman) to RNA-seq gene expression values within the same topological domain. Putative enhancers were linked to the gene with the highest correlation, however, links were only considered significant if the Spearman correlation coefficient (SCC) > 0.25 and if the correlation was considered significant if both empirical and quantitative p-values were less than 0.05. For both p-values, the SCC was calculated between the given putative enhancer and all genes on the same chromosome. The empirical p-value was then calculated as the fraction of genes on the same chromosome that has a higher correlation than the currently linked gene. The quantitative p-value was calculated by treating the calculated SCC values as a distribution and using the R function pnorm to calculate a significance. Enhancers were quantified using DiffBind191. Motif analysis was performed to determine enriched transcription factor binding motifs among the enhancer regions with HOMER192 (with options –size 1000 –mask –p 16 –nomotif).
RNA-seq analysis
Differential gene expression analysis was performed using DESeq2190, where genes with a fold-change > = 2 and FDR < 0.05 were considered to be significantly differentially expressed. Boxplots and MA plots were done in R using ggplot and ggpubr. Clustering was performed using clusterProfiler. Gene-set enrichment analysis was performed with GSEA193,194 using the Hallmark gene set and gene-set level permutation. Gene sets were considered significant if the FDR < 0.05. Significant differential transcript usage (p < 0.05, dIF > 0.1) was determined using IsoformSwitchAnalyzeR195,196 with the default DEXSeq197. Data output was then ran through CPAT198, PFAM199, SignalP200, NetSurfP-2.0201, and results were combined back into IsoformSwitchAnalyzeR to determine genes that might have functional consequences as a result of the DTUs. Genes were analyzed for gene ontologies with clusterProfiler.
Statistics and reproducibility
For each genotype (i.e. WT, Tram−/−, Irak-M−/−), cells from at least n = 6 mice were pooled and split into three experimental groups. Two technical replicates were analyzed from each genomic/experimental combination. Differences in genomic signal were calculated using unpaired t-tests and corrected for multiple testing using the Benjamini-Hochberg procedure. False discovery rates (FDR) were significant if FDR < 0.05. Unless specified otherwise, p-values were considered significant if p < 0.05. Flow cytometry was performed on n = 4 mice for each genotype and analyzed with a one-way ANOVA.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by US National Institutes of Health grants R01EB017235 (C.L.) and R01AI136386 (L.L.), and Virginia Tech ICTAS Center for Engineered Health seed grant (C.L.).
Author contributions
C.L. and L.L. designed the experiments and supervised the research. S.G. conducted the animal experiments and produced the cell samples. Y.P.H. and Z.Z. conducted the ChIP-seq and RNA-seq assays. L.B.N. analyzed the data. L.B.N., Y.P.H., S.G., L.L. and C.L. wrote the manuscript. All authors discussed the results and commented on the manuscript prior to submission.
Peer review
Peer review information
Communications Biology thanks Lian-Yun Li and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editors: Eirini Trompouki and George Inglis.
Data availability
The ChIP-seq and RNA-seq data sets are deposited in the Gene Expression Omnibus (GEO) repository with the following accession number: GSE168190. All other data is available from the corresponding author on reasonable request. Source values for each of the following figures are available in the corresponding Supplementary Data files: Fig. 1 – Supplementary Data 1; Fig. 2 – Supplementary Data 2; Fig. 3 – Supplementary Data 3; Supplementary Fig. 4 – Supplementary Data 4; Fig. 5 – Supplementary Data 5; Fig. 6 – Supplementary Data 6; Fig. 7 – Supplementary Data 7.
Code availability
We used the following software in the analysis: R v3.6.1 (The R Foundation), Trim Galore! v.0.4.1 (Babraham Institute), Bowtie v1.1.2, MACS2 v2.1.1.20160309 (q < 0.05), IGV v2.4.10 (Broad Institute), DiffBind v2.12.0, Salmon v1.2.1, DESeq2 v1.30.0, HOMER v4.10.3 (with options –size 1000 –mask –p 16 –nomotif), clusterProfiler v3.18.0, GSEA 4.1.0, IsoformSwitchAnalyzeR v3.12 (p < 0.05, dIF > 0.1), DEXSeq v1.35.1, CPAT v3.0.2, PFAM (HMMER) v2.41.1, SignalP v5.0, NetSurfP-2.0. All codes can be made available upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lynette B. Naler, Yuan-Pang Hsieh.
Contributor Information
Liwu Li, Email: lwli@vt.edu.
Chang Lu, Email: changlu@vt.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-03035-2.
References
- 1.Tao Q, et al. Association of chronic low-grade inflammation with risk of Alzheimer disease in ApoE4 carriers. JAMA Netw. Open. 2018;1:e183597. doi: 10.1001/jamanetworkopen.2018.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slavich GM, Irwin MR. From stress to inflammation and major depressive disorder: a social signal transduction theory of depression. Psychol. Bull. 2014;140:774–815. doi: 10.1037/a0035302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burhans MS, Hagman DK, Kuzma JN, Schmidt KA, Kratz M. Contribution of adipose tissue inflammation to the development of type 2 diabetes mellitus. Compr. Physiol. 2018;9:1–58. doi: 10.1002/cphy.c170040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, DuBois RN. Immunosuppression associated with chronic inflammation in the tumor microenvironment. Carcinogenesis. 2015;36:1085–1093. doi: 10.1093/carcin/bgv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrucci L, Fabbri E. Inflammageing: chronic inflammation in ageing, cardiovascular disease, and frailty. Nat. Rev. Cardiol. 2018;15:505–522. doi: 10.1038/s41569-018-0064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furman D, et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019;25:1822–1832. doi: 10.1038/s41591-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrisette-Thomas V, et al. Inflamm-aging does not simply reflect increases in pro-inflammatory markers. Mech Ageing Dev. 2014;139:49–57. doi: 10.1016/j.mad.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alpert A, et al. A clinically meaningful metric of immune age derived from high-dimensional longitudinal monitoring. Nat. Med. 2019;25:487–495. doi: 10.1038/s41591-019-0381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zitvogel L, Pietrocola F, Kroemer G. Nutrition, inflammation and cancer. Nat. Immunol. 2017;18:843–850. doi: 10.1038/ni.3754. [DOI] [PubMed] [Google Scholar]
- 10.Raichlen, D. A. et al. Physical activity patterns and biomarkers of cardiovascular disease risk in hunter-gatherers. Am. J. Hum. Biol.29, e22919 (2017). [DOI] [PubMed]
- 11.Burini RC, Anderson E, Durstine JL, Carson JA. Inflammation, physical activity, and chronic disease: an evolutionary perspective. Sports Med. Health Sci. 2020;2:1–6. doi: 10.1016/j.smhs.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll JE, et al. Partial sleep deprivation activates the DNA damage response (DDR) and the senescence-associated secretory phenotype (SASP) in aged adult humans. Brain Behav. Immun. 2016;51:223–229. doi: 10.1016/j.bbi.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Razzoli M, et al. Social stress shortens lifespan in mice. Aging Cell. 2018;17:e12778. doi: 10.1111/acel.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller GE, Chen E, Parker KJ. Psychological stress in childhood and susceptibility to the chronic diseases of aging: moving toward a model of behavioral and biological mechanisms. Psychol. Bull. 2011;137:959–997. doi: 10.1037/a0024768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brodin P, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc. Biol. 2007;81:1–5. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- 17.Chen L, et al. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget. 2018;9:7204–7218. doi: 10.18632/oncotarget.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mai J, Virtue A, Shen J, Wang H, Yang XF. An evolving new paradigm: endothelial cells-conditional innate immune cells. J. Hematol. Oncol. 2013;6:61. doi: 10.1186/1756-8722-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 20.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 21.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 22.Gasteiger G, et al. Cellular innate immunity: an old game with new players. J. Innate Immun. 2017;9:111–125. doi: 10.1159/000453397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Netea MG, et al. A guiding map for inflammation. Nat. Immunol. 2017;18:826–831. doi: 10.1038/ni.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Wang C-C. Inflammatory response of macrophages in infection. Hepatobiliary Pancreatic Dis. Int. 2014;13:138–152. doi: 10.1016/s1499-3872(14)60024-2. [DOI] [PubMed] [Google Scholar]
- 25.Fenyo IM, Gafencu AV. The involvement of the monocytes/macrophages in chronic inflammation associated with atherosclerosis. Immunobiology. 2013;218:1376–1384. doi: 10.1016/j.imbio.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Wan, X., Chowdhury, I. H., Jie, Z., Choudhuri, S. & Garg, N. J. Origin of monocytes/macrophages contributing to chronic inflammation in Chagas disease: SIRT1 inhibition of FAK-NFkappaB-dependent proliferation and proinflammatory activation of macrophages. Cells9, 80 (2019). [DOI] [PMC free article] [PubMed]
- 27.Finsterbusch M, et al. Patrolling monocytes promote intravascular neutrophil activation and glomerular injury in the acutely inflamed glomerulus. Proc. Natl Acad. Sci. USA. 2016;113:E5172–E5181. doi: 10.1073/pnas.1606253113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiesen S, et al. CD14(hi)HLA-DR(dim) macrophages, with a resemblance to classical blood monocytes, dominate inflamed mucosa in Crohn’s disease. J Leukoc Biol. 2014;95:531–541. doi: 10.1189/jlb.0113021. [DOI] [PubMed] [Google Scholar]
- 29.Sindrilaru A, et al. An unrestrained proinflammatory M1 macrophage population induced by iron impairs wound healing in humans and mice. J. Clin. Invest. 2011;121:985–997. doi: 10.1172/JCI44490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma WT, Gao F, Gu K, Chen DK. The role of monocytes and macrophages in autoimmune diseases: a comprehensive review. Front Immunol. 2019;10:1140. doi: 10.3389/fimmu.2019.01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erridge C, Attina T, Spickett CM, Webb DJ. A high-fat meal induces low-grade endotoxemia: evidence of a novel mechanism of postprandial inflammation. Am. J. Clin. Nutr. 2007;86:1286–1292. doi: 10.1093/ajcn/86.5.1286. [DOI] [PubMed] [Google Scholar]
- 32.Morris MC, Gilliam EA, Li L. Innate immune programing by endotoxin and its pathological consequences. Front Immunol. 2014;5:680. doi: 10.3389/fimmu.2014.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahtes A, Li L. Polarization of low-grade inflammatory monocytes through TRAM-mediated up-regulation of Keap1 by super-low dose endotoxin. Front Immunol. 2020;11:1478. doi: 10.3389/fimmu.2020.01478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng H, Maitra U, Morris M, Li L. Molecular mechanism responsible for the priming of macrophage activation. J. Biol. Chem. 2013;288:3897–3906. doi: 10.1074/jbc.M112.424390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maitra U, et al. Molecular mechanisms responsible for the selective and low-grade induction of proinflammatory mediators in murine macrophages by lipopolysaccharide. J. Immunol. 2012;189:1014–1023. doi: 10.4049/jimmunol.1200857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ullah MO, Sweet MJ, Mansell A, Kellie S, Kobe B. TRIF-dependent TLR signaling, its functions in host defense and inflammation, and its potential as a therapeutic target. J. Leukoc. Biol. 2016;100:27–45. doi: 10.1189/jlb.2RI1115-531R. [DOI] [PubMed] [Google Scholar]
- 37.Ciesielska, A., Matyjek, M. & Kwiatkowska, K. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell Mol. Life Sci.78, 1233–1261 (2021). [DOI] [PMC free article] [PubMed]
- 38.Sakai J, et al. Lipopolysaccharide-induced NF-kappaB nuclear translocation is primarily dependent on MyD88, but TNFalpha expression requires TRIF and MyD88. Sci. Rep. 2017;7:1428. doi: 10.1038/s41598-017-01600-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito T, Connett JM, Kunkel SL, Matsukawa A. The linkage of innate and adaptive immune response during granulomatous development. Front Immunol. 2013;4:10. doi: 10.3389/fimmu.2013.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lantz C, Radmanesh B, Liu E, Thorp EB, Lin J. Single-cell RNA sequencing uncovers heterogenous transcriptional signatures in macrophages during efferocytosis. Sci. Rep. 2020;10:14333. doi: 10.1038/s41598-020-70353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiariotti L, Coretti L, Pero R, Lembo F. Epigenetic Alterations Induced by Bacterial Lipopolysaccharides. Adv. Exp. Med. Biol. 2016;879:91–105. doi: 10.1007/978-3-319-24738-0_5. [DOI] [PubMed] [Google Scholar]
- 43.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 44.Patel U, et al. Macrophage polarization in response to epigenetic modifiers during infection and inflammation. Drug Discov. Today. 2017;22:186–193. doi: 10.1016/j.drudis.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrera-Uribe J, et al. Changes in H3K27ac at gene regulatory regions in porcine alveolar macrophages following LPS or PolyIC exposure. Front Genet. 2020;11:817. doi: 10.3389/fgene.2020.00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen S, Yang J, Wei Y, Wei X. Epigenetic regulation of macrophages: from homeostasis maintenance to host defense. Cell Mol. Immunol. 2020;17:36–49. doi: 10.1038/s41423-019-0315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruenjaiman V, et al. Profile of histone H3 Lysine 4 trimethylation and the effect of lipopolysaccharide/immune complex-activated macrophages on endotoxemia. Front Immunol. 2019;10:2956. doi: 10.3389/fimmu.2019.02956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Novakovic B, et al. beta-Glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell. 2016;167:1354–1368 e1314. doi: 10.1016/j.cell.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orecchioni M, Ghosheh Y, Pramod AB, Ley K. Macrophage polarization: different gene signatures in M1(LPS+) vs. classically and M2(LPS-) vs. alternatively activated macrophages. Front Immunol. 2019;10:1084. doi: 10.3389/fimmu.2019.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Das A, et al. High-resolution mapping and dynamics of the transcriptome, transcription factors, and transcription co-factor networks in classically and alternatively activated macrophages. Front Immunol. 2018;9:22. doi: 10.3389/fimmu.2018.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baillie JK, et al. Analysis of the human monocyte-derived macrophage transcriptome and response to lipopolysaccharide provides new insights into genetic aetiology of inflammatory bowel disease. PLoS Genet. 2017;13:e1006641. doi: 10.1371/journal.pgen.1006641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Das A, et al. Dual transcriptome sequencing reveals resistance of TLR4 ligand-activated bone marrow-derived macrophages to inflammation mediated by the BET inhibitor JQ1. Sci. Rep. 2015;5:16932. doi: 10.1038/srep16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu B, et al. MOWChIP-seq for low-input and multiplexed profiling of genome-wide histone modifications. Nat. Protoc. 2019;14:3366–3394. doi: 10.1038/s41596-019-0223-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao ZN, Chen CY, He B, Tan K, Lu C. A microfluidic device for epigenomic profiling using 100 cells. Nat. Methods. 2015;12:959–962. doi: 10.1038/nmeth.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 56.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 57.Lyroni K, et al. Epigenetic and Transcriptional Regulation of IRAK-M Expression in Macrophages. J. Immunol. 2017;198:1297–1307. doi: 10.4049/jimmunol.1600009. [DOI] [PubMed] [Google Scholar]
- 58.Geng S, et al. The persistence of low-grade inflammatory monocytes contributes to aggravated atherosclerosis. Nat. Commun. 2016;7:13436. doi: 10.1038/ncomms13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan R, et al. Low-grade inflammatory polarization of monocytes impairs wound healing. J. Pathol. 2016;238:571–583. doi: 10.1002/path.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geng, S., Zhang, Y., Yi, Z., Lu, R. & Li, L. Resolving monocytes generated through TRAM deletion attenuate atherosclerosis. JCI Insight6, e149651 (2021). [DOI] [PMC free article] [PubMed]
- 61.Peruzzi B, et al. Quantitative and qualitative alterations of circulating myeloid cells and plasmacytoid DC in SARS-CoV-2 infection. Immunology. 2020;161:345–353. doi: 10.1111/imm.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silvin A, et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1401–1418 e1418. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schulte-Schrepping J, et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440 e1423. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee, J. W. et al. Integrated analysis of plasma and single immune cells uncovers metabolic changes in individuals with COVID-19. Nat. Biotechnol.40, 110–120 (2022). [DOI] [PMC free article] [PubMed]
- 65.Pradhan, K., Geng, S., Zhang, Y., Lin, R. C. & Li, L. TRAM-Related TLR4 pathway antagonized by IRAK-M mediates the expression of adhesion/coactivating molecules on low-grade inflammatory monocytes. J. Immunol.206, 2980–2988 (2021). [DOI] [PMC free article] [PubMed]
- 66.Gates LA, Foulds CE, O’Malley BW. Histone marks in the ‘Driver’s Seat’: functional roles in steering the transcription cycle. Trends Biochem. Sci. 2017;42:977–989. doi: 10.1016/j.tibs.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muller WA. Getting leukocytes to the site of inflammation. Vet. Pathol. 2013;50:7–22. doi: 10.1177/0300985812469883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leick M, Azcutia V, Newton G, Luscinskas FW. Leukocyte recruitment in inflammation: basic concepts and new mechanistic insights based on new models and microscopic imaging technologies. Cell Tissue Res. 2014;355:647–656. doi: 10.1007/s00441-014-1809-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waltz P, et al. Lipopolysaccaride induces autophagic signaling in macrophages via a TLR4, heme oxygenase-1 dependent pathway. Autophagy. 2011;7:315–320. doi: 10.4161/auto.7.3.14044. [DOI] [PubMed] [Google Scholar]
- 70.Vural A, Kehrl JH. Autophagy in macrophages: impacting inflammation and bacterial infection. Scientifica (Cairo) 2014;2014:825463. doi: 10.1155/2014/825463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu K, et al. Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization. Autophagy. 2015;11:271–284. doi: 10.1080/15548627.2015.1009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong Z, et al. NF-kappaB restricts inflammasome activation via elimination of damaged mitochondria. Cell. 2016;164:896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu W, et al. Trem2 promotes anti-inflammatory responses in microglia and is suppressed under pro-inflammatory conditions. Hum. Mol. Genet. 2020;29:3224–3248. doi: 10.1093/hmg/ddaa209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keping Y, et al. Sestrin1 inhibits oxidized low-density lipoprotein-induced activation of NLRP3 inflammasome in macrophages in a murine atherosclerosis model. Eur. J. Immunol. 2020;50:1154–1166. doi: 10.1002/eji.201948427. [DOI] [PubMed] [Google Scholar]
- 75.Wu, M. Y. et al. PI3KC3 complex subunit NRBF2 is required for apoptotic cell clearance to restrict intestinal inflammation. Autophagy17, 1096–1111 (2021). [DOI] [PMC free article] [PubMed]
- 76.Tanaka T, Narazaki M, Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014;6:a016295. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cotton, J. A. et al. Interleukin-8 in gastrointestinal inflammation and malignancy: induction and clinical consequences. Int. J. Interferon, Cytokine Mediator Res.8, 13–34 (2016).
- 78.Rattigan KM, et al. Metabolomic profiling of macrophages determines the discrete metabolomic signature and metabolomic interactome triggered by polarising immune stimuli. PLoS ONE. 2018;13:e0194126. doi: 10.1371/journal.pone.0194126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nagy C, Haschemi A. Time and demand are two critical dimensions of immunometabolism: the process of macrophage activation and the pentose phosphate pathway. Front Immunol. 2015;6:164. doi: 10.3389/fimmu.2015.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lauterbach MA, et al. Toll-like receptor signaling rewires macrophage metabolism and promotes histone acetylation via ATP-citrate lyase. Immunity. 2019;51:997–1011 e1017. doi: 10.1016/j.immuni.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 81.Lee HT, Oh S, Ro DH, Yoo H, Kwon YW. The key role of DNA methylation and histone acetylation in epigenetics of atherosclerosis. J. Lipid Atheroscler. 2020;9:419–434. doi: 10.12997/jla.2020.9.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karami J, et al. Epigenetics in rheumatoid arthritis; fibroblast-like synoviocytes as an emerging paradigm in the pathogenesis of the disease. Immunol. Cell Biol. 2020;98:171–186. doi: 10.1111/imcb.12311. [DOI] [PubMed] [Google Scholar]
- 83.Angiolilli C, et al. Inflammatory cytokines epigenetically regulate rheumatoid arthritis fibroblast-like synoviocyte activation by suppressing HDAC5 expression. Ann. Rheum. Dis. 2016;75:430–438. doi: 10.1136/annrheumdis-2014-205635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Adukpo S, et al. Triggering receptor expressed on myeloid cells 1 (TREM-1) and cytokine gene variants in complicated and uncomplicated malaria. Trop. Med. Int. Health. 2016;21:1592–1601. doi: 10.1111/tmi.12787. [DOI] [PubMed] [Google Scholar]
- 85.Krausgruber T, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat. Immunol. 2011;12:231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 86.Lin W, et al. Function of CSF1 and IL34 in macrophage homeostasis, inflammation, and cancer. Front Immunol. 2019;10:2019. doi: 10.3389/fimmu.2019.02019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fontana MF, et al. JUNB is a key transcriptional modulator of macrophage activation. J. Immunol. 2015;194:177–186. doi: 10.4049/jimmunol.1401595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu X, et al. LPSinduced proinflammatory cytokine expression in human airway epithelial cells and macrophages via NFkappaB, STAT3 or AP1 activation. Mol. Med. Rep. 2018;17:5484–5491. doi: 10.3892/mmr.2018.8542. [DOI] [PubMed] [Google Scholar]
- 89.Liu W, et al. AP-1 activated by toll-like receptors regulates expression of IL-23 p19. J. Biol. Chem. 2009;284:24006–24016. doi: 10.1074/jbc.M109.025528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang C, et al. ATF4 is directly recruited by TLR4 signaling and positively regulates TLR4-trigged cytokine production in human monocytes. Cell Mol. Immunol. 2013;10:84–94. doi: 10.1038/cmi.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bush SJ, et al. Species-specificity of transcriptional regulation and the response to lipopolysaccharide in mammalian macrophages. Front Cell Dev. Biol. 2020;8:661. doi: 10.3389/fcell.2020.00661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kobayashi T, et al. NFIL3 is a regulator of IL-12 p40 in macrophages and mucosal immunity. J Immunol. 2011;186:4649–4655. doi: 10.4049/jimmunol.1003888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morgan R, Whiting K. Differential expression of HOX genes upon activation of leukocyte sub-populations. Int. J. Hematol. 2008;87:246–249. doi: 10.1007/s12185-008-0057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yin S, Cao W. Toll-like receptor signaling induces Nrf2 pathway activation through p62-triggered Keap1 degradation. Mol. Cell Biol. 2015;35:2673–2683. doi: 10.1128/MCB.00105-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hong S, Skaist AM, Wheelan SJ, Friedman AD. AP-1 protein induction during monopoiesis favors C/EBP: AP-1 heterodimers over C/EBP homodimerization and stimulates FosB transcription. J. Leukoc. Biol. 2011;90:643–651. doi: 10.1189/jlb.0111043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang D, Paz-Priel I, Friedman AD. NF-kappa B p50 regulates C/EBP alpha expression and inflammatory cytokine-induced neutrophil production. J. Immunol. 2009;182:5757–5762. doi: 10.4049/jimmunol.0803861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu SC, et al. A novel role for Oct-2 in the lipopolysaccharide-mediated induction of resistin gene expression in RAW264.7 cells. Biochem. J. 2007;402:387–395. doi: 10.1042/BJ20061096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lu SC, Wu HW, Lin YJ, Chang SF. The essential role of Oct-2 in LPS-induced expression of iNOS in RAW 264.7 macrophages and its regulation by trichostatin A. Am J Physiol Cell Physiol. 2009;296:C1133–C1139. doi: 10.1152/ajpcell.00031.2009. [DOI] [PubMed] [Google Scholar]
- 99.Cui H, Banerjee S, Guo S, Xie N, Liu G. IFN regulatory factor 2 inhibits expression of glycolytic genes and lipopolysaccharide-induced proinflammatory responses in macrophages. J. Immunol. 2018;200:3218–3230. doi: 10.4049/jimmunol.1701571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nhu QM, Cuesta N, Vogel SN. Transcriptional regulation of lipopolysaccharide (LPS)-induced Toll-like receptor (TLR) expression in murine macrophages: role of interferon regulatory factors 1 (IRF-1) and 2 (IRF-2) J. Endotoxin Res. 2006;12:285–295. doi: 10.1179/096805106X118834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blaszczyk K, et al. The unique role of STAT2 in constitutive and IFN-induced transcription and antiviral responses. Cytokine Growth Factor Rev. 2016;29:71–81. doi: 10.1016/j.cytogfr.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 102.Thygesen SJ, Stacey KJ. IRF1 and IRF2 regulate the non-canonical inflammasome. EMBO Rep. 2019;20:e48891. doi: 10.15252/embr.201948891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu, T., Zhang, L., Joo, D. & Sun, S. C. NF-kappaB signaling in inflammation. Signal Transduct Target Ther2, e17023 (2017). [DOI] [PMC free article] [PubMed]
- 104.Hajishengallis G, Genco RJ. Downregulation of the DNA-binding activity of nuclear factor-kappaB p65 subunit in Porphyromonas gingivalis fimbria-induced tolerance. Infect Immun. 2004;72:1188–1191. doi: 10.1128/IAI.72.2.1188-1191.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kang X, et al. Macrophage TCF-4 co-activates p65 to potentiate chronic inflammation and insulin resistance in mice. Clin. Sci. (Lond) 2016;130:1257–1268. doi: 10.1042/CS20160192. [DOI] [PubMed] [Google Scholar]
- 106.Riedlinger T, et al. NF-kappaB p65 dimerization and DNA-binding is important for inflammatory gene expression. FASEB J. 2019;33:4188–4202. doi: 10.1096/fj.201801638R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wolf Y, et al. Autonomous TNF is critical for in vivo monocyte survival in steady state and inflammation. J. Exp. Med. 2017;214:905–917. doi: 10.1084/jem.20160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Olejnik J, Hume AJ, Muhlberger E. Toll-like receptor 4 in acute viral infection: Too much of a good thing. PLoS Pathog. 2018;14:e1007390. doi: 10.1371/journal.ppat.1007390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Otero YF, et al. Enhanced glucose transport, but not phosphorylation capacity, ameliorates lipopolysaccharide-induced impairments in insulin-stimulated muscle glucose uptake. Shock. 2016;45:677–685. doi: 10.1097/SHK.0000000000000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Feingold KR, et al. Endotoxin rapidly induces changes in lipid metabolism that produce hypertriglyceridemia: low doses stimulate hepatic triglyceride production while high doses inhibit clearance. J. Lipid Res. 1992;33:1765–1776. [PubMed] [Google Scholar]
- 111.Roberts CA, Dickinson AK, Taams LS. The interplay between monocytes/macrophages and CD4(+) T cell subsets in rheumatoid arthritis. Front. Immunol. 2015;6:571. doi: 10.3389/fimmu.2015.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Imam T, Park S, Kaplan MH, Olson MR. Effector T helper cell subsets in inflammatory bowel diseases. Front Immunol. 2018;9:1212. doi: 10.3389/fimmu.2018.01212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tan C, et al. Decreased Histone Deacetylase 2 (HDAC2) in Peripheral Blood Monocytes (PBMCs) of COPD Patients. PLoS ONE. 2016;11:e0147380. doi: 10.1371/journal.pone.0147380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fang WF, et al. Histone deacetylase 2 (HDAC2) attenuates lipopolysaccharide (LPS)-induced inflammation by regulating PAI-1 expression. J. Inflamm. (Lond) 2018;15:3. doi: 10.1186/s12950-018-0179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu C, Li A, Hu J, Kang J. Histone deacetylase 2 is essential for LPS-induced inflammatory responses in macrophages. Immunol Cell Biol. 2019;97:72–84. doi: 10.1111/imcb.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang B, et al. Glycolysis-dependent histone deacetylase 4 degradation regulates inflammatory cytokine production. Mol. Biol. Cell. 2014;25:3300–3307. doi: 10.1091/mbc.E13-12-0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Park EJ, Kim YM, Kim HJ, Chang KC. Degradation of histone deacetylase 4 via the TLR4/JAK/STAT1 signaling pathway promotes the acetylation of high mobility group box 1 (HMGB1) in lipopolysaccharide-activated macrophages. FEBS Open Bio. 2018;8:1119–1126. doi: 10.1002/2211-5463.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Scarl RT, Lawrence CM, Gordon HM, Nunemaker CS. STEAP4: its emerging role in metabolism and homeostasis of cellular iron and copper. J. Endocrinol. 2017;234:R123–R134. doi: 10.1530/JOE-16-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Metais JY, et al. BCL2A1a over-expression in murine hematopoietic stem and progenitor cells decreases apoptosis and results in hematopoietic transformation. PLoS ONE. 2012;7:e48267. doi: 10.1371/journal.pone.0048267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chait A, et al. Presence of serum amyloid A3 in mouse plasma is dependent on the nature and extent of the inflammatory stimulus. Sci. Rep. 2020;10:10397. doi: 10.1038/s41598-020-66898-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.van der Laan LJ, et al. Regulation and functional involvement of macrophage scavenger receptor MARCO in clearance of bacteria in vivo. J. Immunol. 1999;162:939–947. [PubMed] [Google Scholar]
- 122.Luo C, Chen M, Madden A, Xu H. Expression of complement components and regulators by different subtypes of bone marrow-derived macrophages. Inflammation. 2012;35:1448–1461. doi: 10.1007/s10753-012-9458-1. [DOI] [PubMed] [Google Scholar]
- 123.Fensterl V, Sen GC. The ISG56/IFIT1 gene family. J. Interferon Cytokine Res. 2011;31:71–78. doi: 10.1089/jir.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wu C, et al. LRRC14 attenuates Toll-like receptor-mediated NF-kappaB signaling through disruption of IKK complex. Exp. Cell Res. 2016;347:65–73. doi: 10.1016/j.yexcr.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 125.Wammers M, et al. Reprogramming of pro-inflammatory human macrophages to an anti-inflammatory phenotype by bile acids. Sci. Rep. 2018;8:255. doi: 10.1038/s41598-017-18305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Marietta EV, Weis JJ, Weis JH. CD28 expression by mouse mast cells is modulated by lipopolysaccharide and outer surface protein A lipoprotein from Borrelia burgdorferi. J Immunol. 1997;159:2840–2848. [PubMed] [Google Scholar]
- 127.Introna M, Hamilton TA, Kaufman RE, Adams DO, Bast RC., Jr. Treatment of murine peritoneal macrophages with bacterial lipopolysaccharide alters expression of c-fos and c-myc oncogenes. J. Immunol. 1986;137:2711–2715. [PubMed] [Google Scholar]
- 128.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Blouin CC, Page EL, Soucy GM, Richard DE. Hypoxic gene activation by lipopolysaccharide in macrophages: implication of hypoxia-inducible factor 1alpha. Blood. 2004;103:1124–1130. doi: 10.1182/blood-2003-07-2427. [DOI] [PubMed] [Google Scholar]
- 130.Vijayan V, et al. Human and murine macrophages exhibit differential metabolic responses to lipopolysaccharide - A divergent role for glycolysis. Redox Biol. 2019;22:101147. doi: 10.1016/j.redox.2019.101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Soto-Pantoja DR, et al. Unfolded protein response signaling impacts macrophage polarity to modulate breast cancer cell clearance and melanoma immune checkpoint therapy responsiveness. Oncotarget. 2017;8:80545–80559. doi: 10.18632/oncotarget.19849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zuo, H. & Wan, Y. Metabolic reprogramming in mitochondria of myeloid cells. Cells9, 5 (2019). [DOI] [PMC free article] [PubMed]
- 133.O’Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Raulien N, et al. Fatty acid oxidation compensates for lipopolysaccharide-induced warburg effect in glucose-deprived monocytes. Front Immunol. 2017;8:609. doi: 10.3389/fimmu.2017.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang R, Wang L, Pan JH, Han J. A critical role of E2F transcription factor 2 in proinflammatory cytokines-dependent proliferation and invasiveness of fibroblast-like synoviocytes in rheumatoid Arthritis. Sci. Rep. 2018;8:2623. doi: 10.1038/s41598-018-20782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ribeiro MC, et al. LPS induces mTORC1 and mTORC2 activation during monocyte adhesion. Front Mol. Biosci. 2018;5:67. doi: 10.3389/fmolb.2018.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Greenhill CJ, et al. IL-6 trans-signaling modulates TLR4-dependent inflammatory responses via STAT3. J Immunol. 2011;186:1199–1208. doi: 10.4049/jimmunol.1002971. [DOI] [PubMed] [Google Scholar]
- 138.Cronin JG, Kanamarlapudi V, Thornton CA, Sheldon IM. Signal transducer and activator of transcription-3 licenses Toll-like receptor 4-dependent interleukin (IL)-6 and IL-8 production via IL-6 receptor-positive feedback in endometrial cells. Mucosal Immunol. 2016;9:1125–1136. doi: 10.1038/mi.2015.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hoareau L, et al. Signaling pathways involved in LPS induced TNFalpha production in human adipocytes. J. Inflamm. (Lond) 2010;7:1. doi: 10.1186/1476-9255-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.van der Bruggen T, Nijenhuis S, van Raaij E, Verhoef J, van Asbeck BS. Lipopolysaccharide-induced tumor necrosis factor alpha production by human monocytes involves the raf-1/MEK1-MEK2/ERK1-ERK2 pathway. Infect. Immun. 1999;67:3824–3829. doi: 10.1128/iai.67.8.3824-3829.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lee JY, Sullivan KE. Gamma interferon and lipopolysaccharide interact at the level of transcription to induce tumor necrosis factor alpha expression. Infect Immun. 2001;69:2847–2852. doi: 10.1128/IAI.69.5.2847-2852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dallagi A, et al. The activating effect of IFN-gamma on monocytes/macrophages is regulated by the LIF-trophoblast-IL-10 axis via Stat1 inhibition and Stat3 activation. Cell Mol Immunol. 2015;12:326–341. doi: 10.1038/cmi.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Akbar AN, Lord JM, Salmon M. IFN-α and IFN-β: a link between immune memory and chronic inflammation. Immunol. Today. 2000;21:337–342. doi: 10.1016/s0167-5699(00)01652-2. [DOI] [PubMed] [Google Scholar]
- 144.Kirou KA, et al. Coordinate overexpression of interferon-alpha-induced genes in systemic lupus erythematosus. Arthritis Rheum. 2004;50:3958–3967. doi: 10.1002/art.20798. [DOI] [PubMed] [Google Scholar]
- 145.Catalina MD, Bachali P, Geraci NS, Grammer AC, Lipsky PE. Gene expression analysis delineates the potential roles of multiple interferons in systemic lupus erythematosus. Commun. Biol. 2019;2:140. doi: 10.1038/s42003-019-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Molina JR, Adjei AA. The Ras/Raf/MAPK pathway. J. Thoracic Oncol. 2006;1:7–9. [PubMed] [Google Scholar]
- 147.Lin J, et al. Transcriptome-wide analysis reveals modulation of human macrophage inflammatory phenotype through alternative splicing. Arterioscler. Thromb Vasc. Biol. 2016;36:1434–1447. doi: 10.1161/ATVBAHA.116.307573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chunfa L, et al. The central role of IFI204 in IFN-beta release and autophagy activation during Mycobacterium bovis infection. Front Cell Infect Microbiol. 2017;7:169. doi: 10.3389/fcimb.2017.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Robbins CS, et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat. Med. 2013;19:1166–1172. doi: 10.1038/nm.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Griffin C, et al. TLR4, TRIF, and MyD88 are essential for myelopoiesis and CD11c(+) adipose tissue macrophage production in obese mice. J. Biol. Chem. 2018;293:8775–8786. doi: 10.1074/jbc.RA117.001526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.An H, et al. Up-regulation of TLR9 gene expression by LPS in mouse macrophages via activation of NF-κB, ERK and p38 MAPK signal pathways. Immunol. Letters. 2002;81:165–169. doi: 10.1016/s0165-2478(02)00010-x. [DOI] [PubMed] [Google Scholar]
- 153.Plitas G, Burt BM, Nguyen HM, Bamboat ZM, DeMatteo RP. Toll-like receptor 9 inhibition reduces mortality in polymicrobial sepsis. J. Exp. Med. 2008;205:1277–1283. doi: 10.1084/jem.20080162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tsuji N, et al. Role of mitochondrial DNA in septic AKI via toll-like receptor 9. J. Am. Soc. Nephrol. 2016;27:2009–2020. doi: 10.1681/ASN.2015040376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hu D, et al. Inhibition of Toll-like receptor 9 attenuates sepsis-induced mortality through suppressing excessive inflammatory response. Cell Immunol. 2015;295:92–98. doi: 10.1016/j.cellimm.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Celhar T, et al. TLR7 and TLR9 ligands regulate antigen presentation by macrophages. Int. Immunol. 2016;28:223–232. doi: 10.1093/intimm/dxv066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hoshino K, et al. IkappaB kinase-alpha is critical for interferon-alpha production induced by Toll-like receptors 7 and 9. Nature. 2006;440:949–953. doi: 10.1038/nature04641. [DOI] [PubMed] [Google Scholar]
- 158.De Nardo D, De Nardo CM, Nguyen T, Hamilton JA, Scholz GM. Signaling crosstalk during sequential TLR4 and TLR9 activation amplifies the inflammatory response of mouse macrophages. J. Immunol. 2009;183:8110–8118. doi: 10.4049/jimmunol.0901031. [DOI] [PubMed] [Google Scholar]
- 159.Jefferies CA. Regulating IRFs in IFN driven disease. Front Immunol. 2019;10:325. doi: 10.3389/fimmu.2019.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sin WX, et al. IRF-7 mediates type I IFN responses in endotoxin-challenged mice. Front Immunol. 2020;11:640. doi: 10.3389/fimmu.2020.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Krischuns T, et al. Phosphorylation of TRIM28 enhances the expression of IFN-beta and proinflammatory cytokines during HPAIV infection of human lung epithelial cells. Front Immunol. 2018;9:2229. doi: 10.3389/fimmu.2018.02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gong P, et al. Phosphorylation of mitogen- and stress-activated protein kinase-1 in astrocytic inflammation: a possible role in inhibiting production of inflammatory cytokines. PLoS ONE. 2013;8:e81747. doi: 10.1371/journal.pone.0081747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Litvak V, et al. A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature. 2012;490:421–425. doi: 10.1038/nature11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gates-Tanzer LT, Shisler JL. Cellular FLIP long isoform (cFLIPL)-IKKalpha interactions inhibit IRF7 activation, representing a new cellular strategy to inhibit IFNalpha expression. J. Biol. Chem. 2018;293:1745–1755. doi: 10.1074/jbc.RA117.000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Merchut-Maya, J. M. & Maya-Mendoza, A. The Contribution of Lysosomes to DNA Replication. Cells10, 1068 (2021). [DOI] [PMC free article] [PubMed]
- 167.Monguio-Tortajada M, Franquesa M, Sarrias MR, Borras FE. Low doses of LPS exacerbate the inflammatory response and trigger death on TLR3-primed human monocytes. Cell Death Dis. 2018;9:499. doi: 10.1038/s41419-018-0520-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Fontaine M, et al. S100A8/A9 mRNA induction in an ex vivo model of endotoxin tolerance: roles of IL-10 and IFNgamma. PLoS ONE. 2014;9:e100909. doi: 10.1371/journal.pone.0100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Denstaedt SJ, et al. S100A8/A9 drives neuroinflammatory priming and protects against anxiety-like behavior after sepsis. J. Immunol. 2018;200:3188–3200. doi: 10.4049/jimmunol.1700834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Guo Q, et al. Induction of alarmin S100A8/A9 mediates activation of aberrant neutrophils in the pathogenesis of COVID-19. Cell Host Microbe. 2021;29:222–235 e224. doi: 10.1016/j.chom.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Puthia M, et al. IRF7 inhibition prevents destructive innate immunity-A target for nonantibiotic therapy of bacterial infections. Sci. Transl. Med. 2016;8:336ra359. doi: 10.1126/scitranslmed.aaf1156. [DOI] [PubMed] [Google Scholar]
- 172.Li L, McCall C, Hu X. Editorial: innate immunity programming and memory in resolving and non-resolving inflammation. Front Immunol. 2020;11:177. doi: 10.3389/fimmu.2020.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Saravia J, Raynor JL, Chapman NM, Lim SA, Chi H. Signaling networks in immunometabolism. Cell Res. 2020;30:328–342. doi: 10.1038/s41422-020-0301-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bhattacharyya ND, Feng CG. Regulation of T helper cell fate by TCR signal strength. Front Immunol. 2020;11:624. doi: 10.3389/fimmu.2020.00624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Glaros TG, et al. Causes and consequences of low grade endotoxemia and inflammatory diseases. Front Biosci. (Schol Ed.) 2013;5:754–765. doi: 10.2741/s405. [DOI] [PubMed] [Google Scholar]
- 176.Rimmele T, et al. Immune cell phenotype and function in sepsis. Shock. 2016;45:282–291. doi: 10.1097/SHK.0000000000000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Boomer JS, Green JM, Hotchkiss RS. The changing immune system in sepsis: is individualized immuno-modulatory therapy the answer? Virulence. 2014;5:45–56. doi: 10.4161/viru.26516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chen, H. J., Tas, S. W. & de Winther, M. P. J. Type-I interferons in atherosclerosis. J. Exp. Med.217, e20190459 (2020). [DOI] [PMC free article] [PubMed]
- 179.Voloshyna I, Littlefield MJ, Reiss AB. Atherosclerosis and interferon-gamma: new insights and therapeutic targets. Trends Cardiovasc. Med. 2014;24:45–51. doi: 10.1016/j.tcm.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Feng Y, et al. MyD88 and Trif signaling play distinct roles in cardiac dysfunction and mortality during endotoxin shock and polymicrobial sepsis. Anesthesiology. 2011;115:555–567. doi: 10.1097/ALN.0b013e31822a22f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cuenca AG, et al. TRIF-dependent innate immune activation is critical for survival to neonatal gram-negative sepsis. J. Immunol. 2015;194:1169–1177. doi: 10.4049/jimmunol.1302676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Totura AL, et al. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. mBio. 2015;6:e00638–15. doi: 10.1128/mBio.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hong T, Xing J, Li L, Tyson JJ. A simple theoretical framework for understanding heterogeneous differentiation of CD4+ T cells. BMC Syst. Biol. 2012;6:66. doi: 10.1186/1752-0509-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Geng, S., Pradhan, K. & Li, L. Signal-strength and history-dependent innate immune memory dynamics in health and disease. Handb Exp Pharmacol, 10.1007/164_2021_485 (2021). [DOI] [PubMed]
- 185.Cao Z, Chen C, He B, Tan K, Lu C. A microfluidic device for epigenomic profiling using 100 cells. Nat Methods. 2015;12:959–962. doi: 10.1038/nmeth.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Amemiya HM, Kundaje A, Boyle AP. The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep. 2019;9:9354. doi: 10.1038/s41598-019-45839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Stark, R. & Brown, G. D. DiffBind: differential binding analysis of ChIP-seq peak data. http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf (2022).
- 192.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 195.Vitting-Seerup K, Sandelin A. The landscape of isoform switches in human cancers. Mol Cancer Res. 2017;15:1206–1220. doi: 10.1158/1541-7786.MCR-16-0459. [DOI] [PubMed] [Google Scholar]
- 196.Vitting-Seerup K, Sandelin A. IsoformSwitchAnalyzeR: analysis of changes in genome-wide patterns of alternative splicing and its functional consequences. Bioinformatics. 2019;35:4469–4471. doi: 10.1093/bioinformatics/btz247. [DOI] [PubMed] [Google Scholar]
- 197.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang L, et al. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Almagro Armenteros JJ, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 201.Klausen MS, et al. NetSurfP-2.0: Improved prediction of protein structural features by integrated deep learning. Proteins. 2019;87:520–527. doi: 10.1002/prot.25674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The ChIP-seq and RNA-seq data sets are deposited in the Gene Expression Omnibus (GEO) repository with the following accession number: GSE168190. All other data is available from the corresponding author on reasonable request. Source values for each of the following figures are available in the corresponding Supplementary Data files: Fig. 1 – Supplementary Data 1; Fig. 2 – Supplementary Data 2; Fig. 3 – Supplementary Data 3; Supplementary Fig. 4 – Supplementary Data 4; Fig. 5 – Supplementary Data 5; Fig. 6 – Supplementary Data 6; Fig. 7 – Supplementary Data 7.
We used the following software in the analysis: R v3.6.1 (The R Foundation), Trim Galore! v.0.4.1 (Babraham Institute), Bowtie v1.1.2, MACS2 v2.1.1.20160309 (q < 0.05), IGV v2.4.10 (Broad Institute), DiffBind v2.12.0, Salmon v1.2.1, DESeq2 v1.30.0, HOMER v4.10.3 (with options –size 1000 –mask –p 16 –nomotif), clusterProfiler v3.18.0, GSEA 4.1.0, IsoformSwitchAnalyzeR v3.12 (p < 0.05, dIF > 0.1), DEXSeq v1.35.1, CPAT v3.0.2, PFAM (HMMER) v2.41.1, SignalP v5.0, NetSurfP-2.0. All codes can be made available upon reasonable request.