Abstract
DNA is a biological polymer that encodes and stores genetic information in all living organism. Particularly, the precise nucleobase pairing inside DNA is exploited for the self-assembling of nanostructures with defined size, shape and functionality. These DNA nanostructures are known as framework nucleic acids (FNAs) for their skeleton-like features. Recently, FNAs have been explored in various fields ranging from physics, chemistry to biology. In this review, we mainly focus on the recent progress of FNAs in a pharmaceutical perspective. We summarize the advantages and applications of FNAs for drug discovery, drug delivery and drug analysis. We further discuss the drawbacks of FNAs and provide an outlook on the pharmaceutical research direction of FNAs in the future.
KEY WORDS: DNA nanotechnology, Self-assembly, Framework nucleic acids, DNA nanostructures, Drug discovery, Therapeutic molecules, Drug delivery, Drug analysis
Graphical abstract
DNA can be self-assembled into skeleton-like nanostructures known as framework nucleic acids (FNAs) with defined geometry. This review describes the pharmaceutical application of FNAs covering drug discovery, delivery and analysis.
1. Introduction
Framework nucleic acids (FNAs) are nanostructures assembled from DNA molecules with specific shapes and size in one to three dimensions. The biological function of DNA has been studied for more than a century. DNA is one of the fundamental materials of all living organisms. DNA consists of four basic building blocks, including adenine (A), thymine (T), guanine (G) and cytosine (C). The sequence composed of these building blocks encodes the genetic information of life1. The assembly of DNA strictly follows the principles of Watson‒Crick base pairing2. From a chemical perspective, nucleic acids can be regarded as a functional polymer3, with the capability of self-assembly at nanoscale with single base-pair precision. In recent decades, studies on DNA molecules for chemical engineering have only emerged. Since the 1980s, researchers of DNA have changed their attention from biology to a new interdisciplinary combining biology, physics, chemistry, materials science and medicine4. For the first time, Seeman et al.5,6 reported the synthetic DNA branched junction, pioneering the early development of DNA nanotechnology. The early stage of constructing DNA materials was based on stoichiometric self-assembly of DNA tile. Nevertheless, the reaction conditions are strict and the efficiency is relatively low7,8. In 2006, the emergence of DNA origami greatly increased the size and complexity of nanostructures, bringing DNA nanotechnology to a new area9. The basic method of DNA origami is to fold a long strand of scaffold DNA into a specific shape by hybridizing DNA with a large number of short, single-stranded staple DNA9. In the experiment by Rothemund et al.10, the scaffold strand is single stranded, circular viral DNA from M13mp18 phage with 7249 nucleotides. By designing a large number of DNA staples, the DNA scaffold can hybridize with the staples in specific regions to form FNAs of customized shapes.
FNAs have various applications in physics, chemistry and biology4,11. These functions, come from the special properties of FNAs at nanoscale. First of all, FNAs have shown controlled self-assembly, excellent programmability and addressability. Inside each DNA strands, all the atom and chemical bonds have definite location and number. DNA also has nanometer-sized helix of precise diameters (2 nm) and helix pitches (3.4 nm and about 10.5 base pairs per turn). Therefore, the geometry and location parameters of various FNAs are predictable. In this regard, FNAs can be incorporated with addressable molecules by the facile synthesis of oligonucleotide, which can provide excellent and precise regulation of chemical and biochemical reactions at single molecular level12,13. Secondly, FNAs have shown different properties from linear nucleic acids, such as unique interaction with cells14,15. In contrast to ssDNA as well as dsDNA, some FNAs have good cell membrane penetrability and can efficiently load cargo molecules by virtue of their own structure16,17. Furthermore, FNAs are made of nucleic acids and therefore have good biocompatibility18, low immunogenicity19, and good structural stability in the physiological environment20, which greatly promotes the applications of FNAs in biomedical field.
In this review, we aim to summarize the recent progress of FNAs in pharmaceutical science. Pharmaceutical science is a scientific field focusing on the discovery and development of new drugs and therapies. Pharmaceutical science covers the secondary research areas including but not limited to drug discovery, drug delivery and drug analysis. In the following sections, we highlight the applications of FNAs around these secondary fields.
2. FNAs for drug discovery
2.1. FNAs as tools for biological study
Unlike other nanoscale materials, the customized structure and site-specific functionalization of FNAs render these nanostructures as a useful tool for biological research involved in drug discovery, such as imaging, ligand screening, and study of structure–activity relationship.
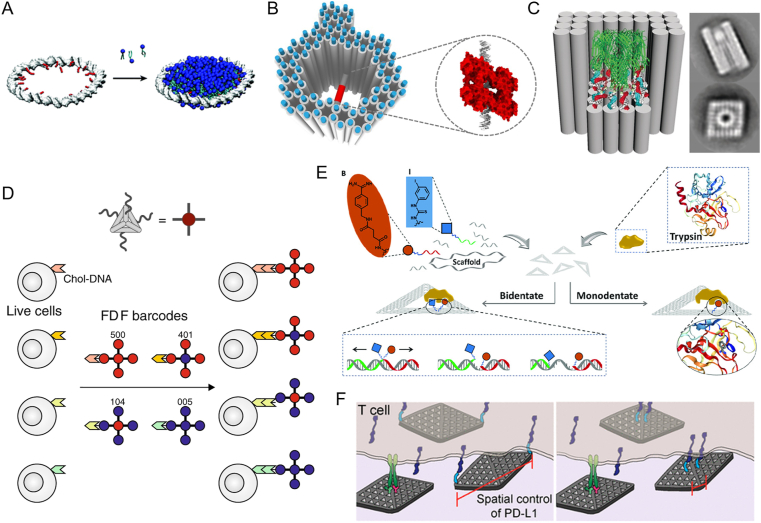
The first step of drug discovery includes identification and analysis of biological targets related to disease. During this process, a broad range of techniques were used to deals with target analysis, such as Cryo electron microscopy (cryo-EM)21. Among many drug targets, membrane proteins are primary. In contrast to soluble proteins, studying membrane proteins is difficult as their poor solubility and loss of native conformation without the cell-membrane environment. The fabrication of nanodiscs by membrane scaffolding proteins has provided a solution to host membrane proteins22. With the advantages of DNA nanotechnology, Iric et al.23 introduced a toroidal DNA ring with alkyl modification on the inner side to prepare nanodiscs (Fig. 1A). Apart from the intrinsic properties of drug targets, many samples of macromolecular complexes are often destroyed by harmful conditions during the preparation of cryo-EM samples. To protect these biological complexes, Martin et al.24 developed a hollow DNA origami to support and enclose the complex from harmful forces (Fig. 1B). The cavity of the designed DNA origami spans a protein-binding molecule for the anchoring of target proteins. This supporting structure provided control over the protein orientations and alleviated protein aggregation or effects during sample preparation. Similarly, Dong et al.25 described a DNA nanobarrel with lipid environment for precise support of membrane proteins (Fig. 1C). Additionally, the significant contrast of phosphate backbone renders the possibility of FNAs to tag individual protein in electron microscopy. Leveraging this advantage, Silvester et al.26 proposed an asymmetry DNA origami as signposts for electron cryotomography to identify molecules of interest located on crowded biological surfaces.
Figure 1.
Examples of framework nucleic acids (FNAs) for drug discovery. (A) DNA-scaffolded nanodiscs. Reprinted with permission from Ref. 23. Copyright © 2018 The Royal Society of Chemistry. (B) Three-dimensional structure based on hollow DNA origami for the support and encapsulation of protein samples during cryo-EM. Reprinted with permission from Ref. 24. Copyright © 2016 National Academy of Sciences. (C) Lipid-functionalized DNA nanobarrel as substitution of nanodiscs for membrane protein support. Reprinted with permission from Ref. 25. Copyright © 2018 John Wiley and Sons. (D) Fluorescence probe by FNAs with multiplex signal encoding. Reprinted with permission from Ref. 28. Copyright © 2020 Springer Nature. (E) Pharmacophore nanoarrays on DNA origami substrates for fragment-based drug discovery. Reprinted with permission from Ref. 31. Copyright © 2020 John Wiley and Sons. (F) DNA nanosheets for probing the relationship between receptor activity and the spatial organization of ligands. Reprinted with permission from Ref. 35. Copyright © 2021 American Chemical Society.
Optical techniques have also shown great impact on drug discovery. From molecular assays to cell imaging, fluorescently labeled molecules can be applied. However, continuous imaging of fluorescent samples is restricted by photobleaching. In this context, Niekamp et al.27 presented a compact DNA Cube labeled with multiple dyes for continuous fluorescence emission. The DNA cube provides precise control over the distance between multiple dyes to eliminate self-quenching. The DNA FluoroCubes can be further attached to targets of interests. The authors demonstrated higher photobleaching lifetime and photons emission than single organic dyes. By site-precisely modification of DNA nanostructure with distinct fluorophores, Li et al.28 developed fractal DNA frameworks for multiplex fluorescence encoding (Fig. 1D). The site specificity enabled by the fractal structure can minimize interference between fluorophore and allows the construction of up to 36 color-encoded probes. These barcodes based on DNA frameworks were applied to multiplexed cell imaging and discrimination, which is an advantage during drug screening.
Besides, the precise patterning of molecules by FNAs was also applied to fragment-based drug discovery (FBDD). FBDD is an important and lengthy process involved in drug screening29. On the DNA origami substrates, pharmacophores are displayed as individuals or in pairs30,31 (Fig. 1E). The single-molecule binding of target to the displayed ligands is detected by atomic force microscopy (AFM), which screens the strong binder. With this strategy, Huang et al.31 have discovered a bidentate trypsin binder based on the pair of benzamidine and aromatic fragments.
The development of potential therapy mainly focuses on cell signal transduction pathways32. Most receptors are membrane protein that can trigger intracellular signaling cascades when they bind to their extracellular ligands. Drugs play their therapeutic role by promotion or inhibition of the receptor33. Understanding the relevance between the nanoscale topology of ligands and the interaction with its own receptor may guide rational design of future therapies. Shaw et al.34 utilized DNA nanocalipers with precise modification of ephrin-A5 to prove that the spatial distribution of ephrin-A5 has an effect on the activation levels of EphA2 receptor and the invasiveness of breast cancer cell. The approach based on DNA origami provides accurate nanoscale distance manipulation, which can be regulated independently of ligand concentration. The same group35 also fabricated DNA nanosheets with various organization of PD-L1 ligands to study the inhibitory effect on the activation of T cell in vitro (Fig. 1F). Similarly, Veneziano et al.36 designed a serious of DNA origami nanoparticles with distinct pattern of HIV-1 glycoprotein-120 to investigate the roles of antigen spacing or number on B-cell triggering. The authors demonstrated that B-cell signaling is maximized when displaying as few as five antigens with approximately 25–30 nm spacing. According to the above researches, we can draw a conclusion that the FNAs can independently control over antigen stoichiometry, inter-antigen distance, as well as the spatial dimensionality of ligand display. Such strategy may provide new insights for the design principles of ligand-based therapies by revealing the structure activity relationships.
2.2. FNAs as potential therapeutic molecules
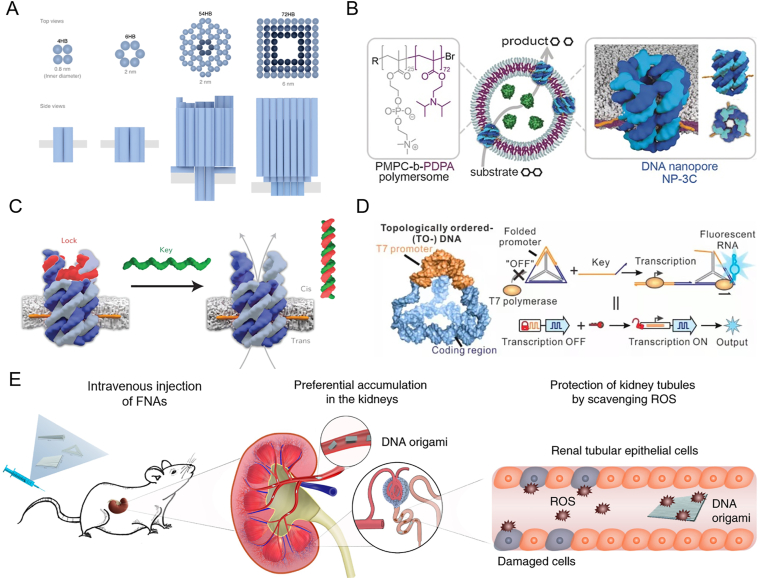
The defined shape fabricated by DNA nanotechnology in nanoscale endows the FNAs with the ability to mimic the structure of natural proteins. The most common biomimetic applications of DNA frameworks belong to membrane channel, which greatly relied on their pore structure to transport molecules across cell membrane37. A transmembrane channel created by DNA origami was first proposed by using α-hemolysin as a model38. The synthetic DNA channels can successfully span the lipid bilayers via precise attachment of hydrophobic moieties to the nanostructures. The membrane-spanning nanopores made of DNA origami are reported to show stable electrical properties and allow the translocation of small molecules or folded proteins across the lipid bilayer39,40. These biomimetic DNA nanopores can be further downsized through DNA tile-based assembly41,42 (Fig. 2A). Therefore, the yield of DNA nanopores is increased in contrast to those made of DNA origami, which makes DNA nanopores available in cell studies for therapeutics.
Figure 2.
Potential therapeutic molecules of FNAs. (A) Biomimetic DNA nanopores assembled through DNA tiles (the two on the left) or DNA origami (the two on the right). Reprinted with permission from Ref. 41. Copyright © 2021 Springer Nature. (B) The functionalization of vehicle by DNA nanopores. Reprinted with permission from Ref. 47. Copyright © 2016 John Wiley and Sons. (C) Oligonucleotide-gated DNA nanopores. Reprinted with permission from Ref. 48. Copyright © 2016 Springer Nature. (D) Gene regulation on topologically switchable FNAs. Reprinted with permission from Ref. 54. Copyright © 2020 American Chemical Society. (E) Alleviation of acute kidney injury by rectangular DNA origami. Reprinted with permission from Ref. 15. Copyright © 2018 Springer Nature.
Initially, Burns et al.43 reported that the sized-reduced DNA nanopores showed cytotoxicity by perforating the plasma membrane. To improve the incorporation of nanopores into lipid bilayers, our group44 reported a fusogenic liposome-incorporated transmembrane DNA nanopores. The developed strategy can bypass the direct insertion process through membrane fusion, during which the embedded DNA nanopores directly transfer from the liposomal bilayer to the plasma membrane. The incorporation process can further have targeting ability via an on-command cleavable steric shield on the liposomal surface. We showed that our fusogenic DNA nanopores can induce an unexpected pyroptosis-like cell death and inhibit tumor growth in xenograft models. Additionally, based on the transport capacity of these synthetic channels, application of DNA nanopores has been proposed to fight against doxorubicin resistance caused by drug efflux45. Recently, a DNA nanopores which can modulate immune response of white blood cells is also reported46. The cholesterol moiety of the structures renders the nanopores selectively adhere on white blood cells, forming a steric block for toll-like receptors against pro-inflammatory endotoxin lipopolysaccharide.
Furthermore, DNA nanopores are proposed to be applied in the functionalization of synthetic vesicles47 (Fig. 2B). The functionalized vesicles feature size-selected permeability that the enzyme substrates can diffuse across the nanopore-embedded membrane while larger enzymes are retained. With the programmability of DNA hybridization reactions, the DNA nanopores can be gated by oligonucleotides48,49 (Fig. 2C), temperature50, small molecules51 and proteins52. As such, control of drug release can be achieved in this kind of nanopore-functionalized vesicles52. Besides DNA nanopores, some membrane-spanning DNA nanostructures can induce a toroidal pore on lipid bilayer and mimic the function of scramblase53. The synthetic scramblase can catalyze the transport of phospholipids between the bilayer leaflets and exposed phosphatidylserine lipids on the outer membrane of human cells.
The programmable and reconfigurable topology of nucleic acid structures inside FNAs can be also explored to mimic some other function in biology. For example, the topology of genes regulates the translational process. Recently, Jiao et al.54 reported the development of topologically switchable FNAs to program the transcription of prokaryotic genes (Fig. 2D). The DNA templates are integrated on the DNA framework where the position of T7 promoter is topologically constrained. The promoter can be activated after topology stress is released by reconfiguration of the DNA framework upon stimuli. The switchable transcription of DNA framework is applied in genetic engineering of bacterial cells. Likewise, the switchable function of protein based on confirmation change can be achieved in framework nucleic acids. Tian et al.55 proposed the switchable activity of melittin by encapsulation of melittin in conformationally changeable FNAs.
In some instances, FNAs can exhibit therapeutic efficacy without structural mimicking. Jiang et al.15 investigated the protection against acute kidney injury by FNAs with diverse shapes, and observed that the rectangular DNA origami showed similar efficacy compared with clinical standard treatment (Fig. 2E). The authors indicated that this therapeutic effect was due to the shape-dependent accumulation of FNAs to the kidneys. Such results could promote drug discovery and delivery for the treatment of kidney diseases. Likewise, Ma et al.56 proved that tetrahedral FNAs combined with neural stem cells transplantation have better therapeutic achievement including higher cell viability, stronger capacity of differentiation and proliferation. Although the results showed potential application on nerve regeneration, there are still some limitations like short of animal models and physiological details during the treatment progress.
3. FNAs for drug delivery
Compared with most organic or inorganic nanoparticles, FNAs are considered as a natural biomacromolecule with excellent monodispersity, biodegradability and biocompatibility, which have rendered FNAs great promise as drug carriers. Besides, many molecules for therapeutics, targeting and stimuli-responsiveness can be easily integrated onto FNAs via base pairing or simple chemical ligation. Owing to the addressability of FNAs, these modifications on FNAs can be quantified and allow tunable loading efficiency. Particularly, FNAs can also exhibit some unique properties. First, as one of the FNAs, tetrahedral DNA nanostructures57 (TDNs) could rapidly enter the cells without the presence of transfection agents while remaining their structural integrity58. Studies by single-particle tracking have indicated that the endocytosis was mediated by lipid-raft-mediated pathway with minimum electrostatic repulsion59 (Fig. 3A) in a caveolin-dependent manner, followed by lysosomes transport60. Apart from cellular uptake, FNAs have been reported as topical transdermal drug delivery carriers61. Secondly, structures with cavities and DNA logic gate can be achieved on FNAs for cargo protection and responsiveness, respectively. In this section, we make an in-depth discussion about the applications of FNAs in drug delivery. The following summaries span from FNAs-based delivery carriers (for small molecules, macromolecules and inorganic nanoparticles), to templates for preparation of drug carriers.
Figure 3.
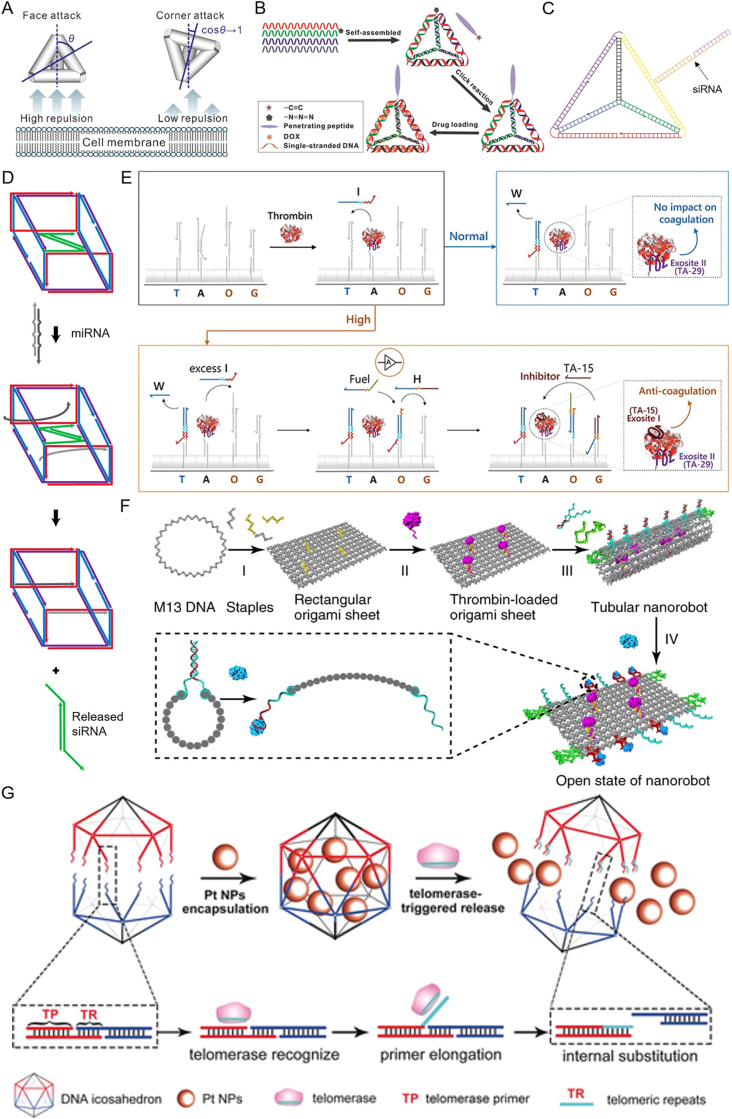
Examples of FNAs for drug delivery. (A) The cell-entry mode of TDNs. Reprinted with permission from Ref. 59. Copyright © 2018 American Chemical Society. (B) TPP-conjugated FNAs as delivery carrier for DOX. Reprinted with permission from Ref. 62. Copyright © 2016 American Chemical Society. (C) siRNA delivered by TNDs with folate ligands. Reprinted with permission from Ref. 83. Copyright © 2012 Springer Nature. (D) siRNA-encapsulated DNA nanocube that can release cargo upon intracellular trigger. Reprinted with permission from Ref. 86. Copyright © 2016 American Chemical Society. (E) DNA nanorobot that can intelligently regulate thrombin functions in human plasma. Reproduced with permission from Ref. 102. Copyright © 2020 John Wiley and Sons. (F) DNA nanorobot by origami for the targeted delivery of thrombin. Reprinted with permission from Ref. 106. Copyright © 2018 Springer Nature. (G) Platinum-nanoparticle-caged DNA icosahedron and unpacked in the presence of telomerase. Reprinted with permission from Ref. 112. Copyright © 2018 John Wiley and Sons.
3.1. Small molecule delivery
Small molecules drugs are considered to be the most widely used pharmaceutical ingredients. For instance, doxorubicin (DOX) is a type of non-specific, broad-spectrum chemotherapeutic agent, which produces strong cytotoxicity by means of intercalating into double-stranded DNA to inhibit biosynthesis of nucleic acid. Nevertheless, various drawbacks have emerged during the clinical applications of DOX alone, including poor selectivity, adverse side effects, multidrug resistance.
To address the above challenges, researchers have made considerable efforts to achieve DOX targeted delivery. Inspired by the interaction between DOX and DNA, FNAs have been exploited as alternative carriers. DOX can be loaded on tetrahedral DNA nanostructures (TNDs) functionalized with tumor-penetrating peptide (TPP)62 (Fig. 3B), SL2B aptamer or folic acid63. Likewise, 3D DNA icosahedrons with aptamer conjugation were applied to deliver DOX64. The high payload and ligand decoration enabled by FNAs resulted in targeted delivery, increased cellular uptake and sufficient anticancer efficacy at a much lower DOX concentration. Besides, Jiang et al.65 reported a DOX-loaded triangular DNA origami being able to circumvent resistance of DOX mediated by tumor intrinsic efflux-pump. The Origami-based FNAs were demonstrated to enhance passive targeting and tumor accumulation properties of the loaded DOX in vivo66. Furthermore, the origami structures could be designed with varying degrees of global twist and different amounts of relaxation in the DNA double-helix structure to rationally tune the encapsulation efficiency and release kinetics of DOX67.
Besides DOX, other small molecule drugs also have shown to be delivered by FNAs by noncovalent absorption. Cisplatin is a therapeutic drug against cancer mainly via covalent and non-covalent interactions with DNA base pairs. Wu et al.68 developed a double-bundle TDNs with nanobody conjugation for targeted cisplatin delivery. Paclitaxel (PTX) is another effective chemotherapeutic molecule against cancer. Xie et al.69 reported a PTX-loaded TDNs for the treatment of PTX-resistant non-small cell lung cancer (NSCLC), which exerted strong lethality on A549/T cell lines. The PTX-loaded TDNs could exert in vitro and in vivo targeted apoptosis in glioma cells when further modified with GMT8 and Gint4. T aptamers70. Moreover, Shi et al.71 constructed wogonin-loaded TDNs for osteoarthritis therapy, which has offered new prospective for effective relief of inflammatory reactions.
Small molecule drugs can also be covalently grafted on the backbone of FNAs. Mou et al.72 proposed floxuridine-integrated DNA polyhedral for constructing trojan-horse-like anticancer drug delivery system. Later, the same group73 developed camptothecin (CPT)-linked TDNs through reaction between the phosphorothioate in the TDNs and carbonethyl bromide in the CPT derivative. A cleavable disulfide was introduced in the linkage to endow the CPT-TDNs with glutathione-responsiveness. Zhong et al.74 synthesized DNA-conjugated cisplatin prodrug and further assembled into DNA nanostructure for prodrug delivery. Wang et al.75 assembled a DNA octahedral framework composed of Sgc8c aptamers functionalized with combretastatin A-4 (a broad-spectrum microtubule inhibitor) for targeted cancer therapy.
3.2. Oligonucleotide delivery
During or following the assembly of FNAs, oligonucleotides can be integrated into FNAs. These integrations are mainly based on strand hybridization between oligonucleotide and the protruded scaffold of FNAs76. Since oligonucleotides and FNAs are same kinds of material, oligonucleotides can also be organically combined with FNAs during the solid-phase synthesis of scaffold strands77. Therefore, FNAs are capable of loading therapeutic nucleic acids.
RNA interference is a highly conserved biological reaction during evolution. Upon induction by double-stranded RNA, RNA interference can specifically degrade or inhibit the expression of homologous mRNAs78. Small interfering RNAs (siRNAs) are 21–25-nucleotide small RNAs produced by enzyme Dicer, which plays an important role in RNAi pathway in eukaryotic cells79. Therefore, synthetic siRNAs have become a class of potential effective nucleic acid drug candidates for treatment by disrupting pathogenic or disease-promoting genes. Many drug delivery systems based on polymeric nanoparticles and liposomes have been developed for in vivo delivery of siRNA80,81. However, most of the carriers are still limited by weak tissue specificity, potential toxicity, and poor drug release82.
By taking the advantage of DNA nanostructures, FNAs have recently become new carriers for siRNA delivery. In 2012, Lee et al.83 synthesized TDNs to efficiently deliver siRNA into tumor cells and induce target gene silencing in vivo. The authors also found that the spatial orientation and density of folate ligands may affect the efficiency of TDNs in uptake and gene silencing (Fig. 3C). Rahman et al.84 used the modular DNA brick method to assemble many rectangular and tubular DNA nanostructures with varied sizes for delivery of Bcl 2 siRNA. To our knowledge, BCL2 or P-glycoprotein (P-gp) overexpression is always associated with tumors. They confirmed the optimized size of structures for the effective uptake and tumor growth inhibition in mice. Furthermore, Wang et al.85 used DNA origami technology to construct a glutathione (GSH)-responsive tubular DNA nanodevice for co-delivery of Bcl2 siRNA, P-gp siRNA, and DOX. The nanodevice was further modified with multiple functional moieties, including cell-penetrating peptides and disulfide bond-containing DNA locks, so as to improve the cell uptake efficiency of the carrier and the stability of the drug.
FNAs not only serve as delivery vectors for siRNA, but also help the cargo resist nuclease degradation. Bujold et al.86 designed a trigger-responsive DNA "nanosuitcase” that encapsulates siRNA (Fig. 3D). The locked siRNA survived longer in the progress of nuclease degradation than free siRNA. This three-dimensional DNA prism structure used two gating strands which can identify selected genetic markers and unwind by chain displacement to release the siRNAs from the cavity.
The "lock and key” function enable by DNA nanotechnology overcomes the problems of non-specific adsorption and off-target effects, and can be also used in accurate siRNA delivery. Ren et al.87 designed a self-assembled oligonucleotide nanocarrier (ONV) with a double lock and key system to achieve precise delivery of siRNA. Through the sequential binding of the two aptamers sgc8c and sgc4f16 on the cell surface, the nanostructures could achieve specific recognition of cell subtypes, greatly reducing systemic toxicity.
Tumor suppressive microRNA (miRNA) is another effective molecule that may cure cancer, which have similarities with siRNAs both in size and function. Qian et al.88 reported a shuriken-shape DNA wireframe, which was formed by hybridizing three copies of miR-145 with a DNA star motif. Compared with ordinary liposomal transfection agents, this multi-pronged configuration increased the cell uptake rate and showed a significant therapeutic effect on human colon cancer cells. As a type of inhibitor of apoptosis protein (IAP), Survivin has been reported to be overexpressed in tumor cells and is a potential candidate protein for tumor therapy. Li et al.89 designed TDNs for delivery of miRNA-214-3p (miR-214-3p), which could combine with Survivin mRNA to reduce its expression in cells, activate the mitochondrial apoptotic pathway, and ultimately cause tumor cell apoptosis. Nahar et al.90 demonstrated DNA nanostructure containing three anti-miRNAs that can target tumor oncomiRNA-27A, 96, and 182. These miRNAs synergistically down-regulated the expression of the transcription factor FOXO1a, which contributed to malignant transformation and carcinogenic status.
Antisense oligonucleotide (ASO) is a small single-stranded DNA complementary to the target mRNA sequence. It is involved in the regulation of gene expression and can become a targeted therapeutic gene with great potential91. In 2018, Yang et al.92 developed a double-beam TDN that could bind nuclear targeting peptides and antisense oligonucleotides that silence the proto-oncogene c-Raf. Experimental results proved the increased delivery of antisense oligonucleotides to the nucleus, resulting in down-regulation of target mRNA and inhibition of cell proliferation. The same research group93 integrated antisense nucleic acid drugs for gene therapy with KillerRed protein for photodynamic therapy on TDN. Likewise, Zhang et al.94 designed TDNs for transporting antisense peptide nucleic acids into methicillin-resistant Staphylococcus aureus (MRSA) cells. The complex effectively reduced the expression of the target gene and significantly inhibited cell growth in a concentration-dependent manner.
FNAs can also be nanocarriers for therapeutic genes. In 2018, Ding and co-workers95 constructed a DNA triangle origami containing tumor suppressor gene P53 and chemotherapeutic DOX as co-delivery system for the treatment of multidrug resistant tumors.
CpG (unmethylated cytosine guanosine phosphate) is found to be ubiquitous in natural viral and bacterial DNA96. Synthetic CpG oligonucleotide is a class of therapeutic nucleic acid with powerful immunostimulatory effects97. CpG-containing sequences can be easily modified into DNA framework and delivered as anti-cancer immunotherapeutic drug. In 2011, Schuller et al.19 constructed a 30-helix DNA nanotube grafted with up to 62 CpG sequences, as non-cytotoxic immunostimulants. In the same year, Li et al.98 combined CpG motifs on different vertices of the TDNs. They found that DNA nanostructures can effectively resist nuclease degradation in fetal bovine serum and cells, and the modified multivalent CpG motif greatly enhanced the immunostimulatory activities of CpG. Further, CpG also has the potential to become a vaccine or immunomodulator. Liu et al.99 used tetrahedral DNA as a scaffold to assemble streptavidin (STV) and CpG oligodeoxynucleotide adjuvant into a vaccine complex. Experimental results of immunized mice showed that this antigen adjuvant-DNA nanostructure was similar to natural virus particles and can induce a strong and long-lasting antibody response. In the study of Qu et al.100, DNA dendrimers are programmable and safe carriers that can be used to deliver immunomodulators loop-CpG and TAT peptide to activate immune responses.
In addition to the above-mentioned oligonucleotide drugs, Ma et al.101 explored the TDNs in delivering an anti-HER2 aptamer for enhancing lysosomal degradation of HER2 protein on breast cancer cells. The HER2 degradation further arrested cell growth and induced cell apoptosis through inhibition of downstream PI3K/AKT signaling pathway. Moreover, DNA logic gates can be integrated into FNAs as potential intelligent personalized nanomedicine. For autonomous blood anticoagulation in human plasma, Yang et al.102 designed a DNA nanorobot with thrombin-triggered strand hybridization cascades embedded on DNA barrel (Fig. 3E). The nanorobot provided a logic loop to sense the thrombin concentration and released thrombin aptamer when exceed the tunable threshold. The released aptamer further inhibited thrombin functions and thus achieved autonomous and precise control over thrombin dosage. Together, these advanced researches have proven that the DNA framework is a powerful tool for oligonucleotides delivery.
3.3. Peptide and protein delivery
Peptides and Proteins are important components of human body and play a vital function in life. In recent years, intracellular protein delivery is developing for novel therapeutics103,104. Thrombin, converts fibrinogen into fibrin, activates platelets and aggregates them, eventually leading to thrombus formation105. Using thrombin to selectively block tumor blood vessels and cause tumor cell death is an attractive anti-cancer strategy. Li et al.106 assembled a DNA nanorobot using DNA origami technology, with nucleolar targeting aptamers on the outside, which could bind to the nucleolin on the surface of tumor cells and expose the encapsulated thrombin (Fig. 3F). After intravenous injection of DNA nanorobot into tumor-bearing mouse models, thrombin would specifically reach the tumor blood vessels and induce thrombosis, thereby achieving tumor necrosis and inhibiting tumor growth.
New peptide-based antibacterial drugs have become potential substitutes in reducing the probability of bacterial resistance. Liu et al.107 used TDNs as a carrier with the conjugation of antibacterial peptide GL13K to study the inhibitory effect on Escherichia coli and Porphyromonas gingivalis. They found that TDNs not only enhanced the antimicrobial effect of GL13K, but also protected it from degradation by proteases.
Peptides can also be used for cancer treatment by regulating immune responses. Liu et al.108 designed a tubular DNA origami as a cancer vaccine to transport antigen peptides and molecular adjuvants. This structure had inner cavity wrapped with antigen peptides and two types of Toll-like receptor (TLR) agonists. A pH-responsive DNA strands locked the cavity for gating the exposure of cargos. They found that the nano-vaccine can produce long-term T cell responses in mice, effectively preventing tumor metastasis and recurrence.
3.4. Delivering inorganic nanoparticle-FNAs complex
Inorganic nanoparticles are submicroscopic particles with sizes ranging from 1 to 100 nm, which have the advantages of well-defined shape, plasma and magnetic properties, and controllable surface chemical and physical properties109. In recent years, FNAs have been integrated during the delivery of inorganic nanoparticles. Jiang et al.110 constructed gold nanorods on the surface of triangular DNA origami as a nanotheranostics. Experimental outcomes showed that the complexes significantly enhanced tumor cell accumulation and improved photothermal decomposition ability. On this basis, Du et al.111 confirmed that the gold-nanorods-triangular-origami could be both an effective diagnostic tool and a therapeutic agent to inhibit tumor growth.
In addition, Ma et al.112 designed a telomerase-responsive DNA icosahedron linked by two pyramid-shaped DNA cages for encapsulation of platinum nanoparticles (Fig. 3G). The nanoplatform achieved precise nanoparticle delivery and treated cisplatin-resistant tumor cells. According to a relative research, Wang et al.113 designed DNA origami structures of different geometry labeled with gold nanoparticles, and observed their internalization process in multiple human cancer cell lines. These researches may provide new insight for the delivery of nanoparticles with enhanced therapeutic efficacy.
3.5. Synthetic templates for nanocarrier
The customized and precise geometry of FNAs possess tremendous promise for template-based preparation of organic and inorganic nanoparticles. Significantly, the FNAs-templated strategy offers a near-homogeneous size distribution in comparison to conventional methods. For instance, Yang et al.114 presented a DNA origami-enabled strategy to generate artificial small unilamellar vesicles with controlled shape, size, and outstanding monodispersity (Fig. 4A). DNA origami with lipid molecule decoration served as the templates or “exoskeleton” and precisely guided liposome formation of different morphology. Inspired by the highly ordered architecture and self-assembly of clathrin, Baumann et al.115 applied DNA triskelion for liposome coating to enhance liposome mechanical stability.
Figure 4.
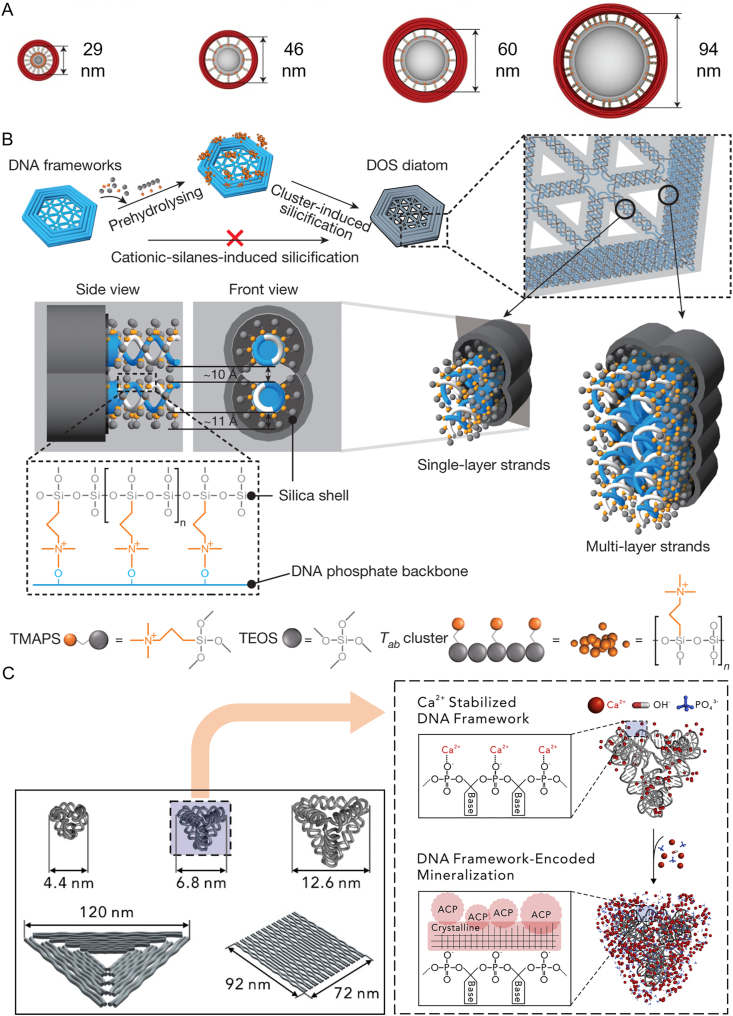
FNAs-based templates for the preparation of drug carriers. (A) Size-controlled generation of liposomes on DNA origami rings. Reprinted with permission from Ref. 114. Copyright © 2016 Springer Nature. (B) Silicification on DNA origami to produce silica nanoparticles with complex geometry. Reprinted with permission from Ref. 116. Copyright © 2018 Springer Nature. (C) FNA-templated crystallization of calcium phosphate. Reprinted with permission from Ref. 118. Copyright © 2019 Elsevier.
Recently, by integration of DNA origami and Stöber method, Liu et al.116 established a promising approach for template-fabrication of silica nanostructures with various size and shapes (Fig. 4B). These silica nanostructures showed higher toughness than the DNA template while maintaining softness, which was a unique property other than existing silica nanoparticles. Of note, the softness inherited in these nanoparticles may play an important role in their delivery or cellular uptake117. The same group118 also reported a FNAs-templated strategy for calcium phosphate (CaP) crystallization (Fig. 4C). The fabricated CaP exhibited user-defined geometry with nanoscale precision. Cellular experiments confirmed that template CaP nanoparticles could serve as a sustainable agent for live-cell delivery of CpG with enhanced bioactivities. Furthermore, CaP nanoparticles with site-specific protein modification can be obtained by the FNAs-templated synthesis119.
4. FNAs for drug analysis
Drug analysis is an important component of pharmaceutical sciences. With the development of DNA nanotechnology, FNAs are widely used to construct a variety of sensing platforms during sample analysis120. There have been tremendous researches in FNAs-based biosensors for the detection of DNA, protein and nucleic acids121, 122, 123, 124, 125, 126. One of the major obstacles for the biosensor design depends on the restricted target accessibility at the solid water interface. For electrochemical sensor, recognition ligands especially aptamers are disordered on the interface of the modified probes127,128, which limits the accessibility of the target and lowers the detection sensitivity of the target. FNAs offer a convenient approach to improve the order of aptamer at the interface, alleviate the interference to aptamer-target recognition, and thus improve the detection sensitivity121,129. In this section, we provide an overview mainly on the involvement of FNAs in drug analysis.
FNAs, especially TDNs, have been widely used in the fabrication of electrochemical sensors130, 131, 132. Li et al.133 grafted an aptamer-conjugated TNDs on a gold electrode of micronano structure for metabolite detection in whole blood. The conformational change of the aptamer upon ATP binding enabled MB proximal to the electrode surface and led a strong electrochemical signal, which realized rapid ATP detection (Fig. 5A). The limit of ATP detection was demonstrated as 50 μmol/L in rabbit whole blood.
Figure 5.
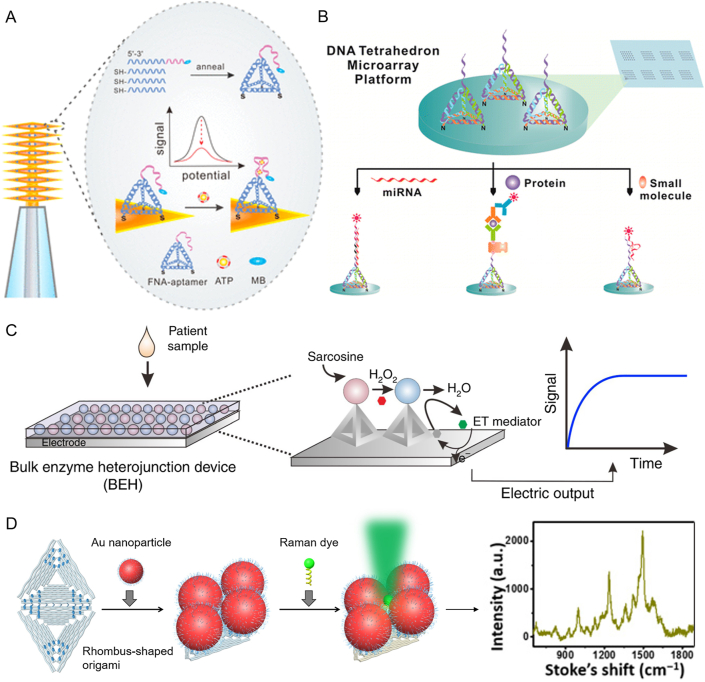
Examples of FNAs for drug delivery. (A) TDNs-functionalized electrode for ATP analysis in human blood. Reprinted with permission from Ref. 133. Copyright © 2020 American Chemical Society. (B) A microarray platform based on TNDs for multiple analysis of various bioactive molecules such as cocaine. Reprinted with permission from Ref. 135. Copyright © 2014 American Chemical Society. (C) TNDs programmed enzyme heterojunctions on the electrode for sarcosine sensing. Reprinted with permission from Ref. 137. Copyright © 2020 Springer Nature. (D) DNA origami-anchored metal particles for single molecule Raman imaging. Reprinted with permission from Ref. 138. Copyright © 2019 American Association for the Advancement of Science.
For some addictive drugs, such as cocaine, illegal use can lead to social unrest. Analytical techniques for rapid detection of cocaine are important in border controls or in drug investigation and tracking. Several electrochemical sensors have been developed to detect cocaine based on cocaine aptamer. Wen et al.134 constructed a sensing platform based on FNAs to develop an ultra-sensitive, highly selective electrochemical cocaine sensor. The electrode surface was modified with a suspended aptamer-TNDs structure. An anti-cocaine aptamer was split into two segments. One of the split aptamers extended from the top of the TNDs to improve the cocaine recognition. Another split aptamer was labeled with biotin and brought the avidin-horseradicadric-peroxidase to the TDNs upon cocaine binding. The peroxidase converted the aptamer binding event into an electrochemical signal, achieving the detection limit of 33 nmol/L. In addition, the tetrahedral modified electrode surface has anti-fouling properties and reduces the interference of background noise, leading highly selectivity during the analysis of complex samples. The same strategy was also introduced to a microarray platform to realize the fluorescent detection of cocaine and achieve the detection limit of 100 nmol/L135 (Fig. 5B).
In addition, TNDs-based sensors can also be used to detect theophylline. Theophylline is a “star drug” for the treatment of respiratory diseases. The toxicity and side effects of theophylline narrow the safe range in clinical administration. Wang et al.136 proposed an electrochemical method for theophylline detection based on the cascade assembly of aptamer on the TDNs-coated electrodes. In the presence of theophylline, the RNA aptamer subsequently self-assembled into nanowires on the DNA tetrahedron and further adsorbed silver nanoparticles. Therefore, the concentration of theophylline can be measured via the silver stripping current. This method achieved the detection limit of 50 nmol/L.
The TDNs can also regulate the enzyme distance on the electrode interface to improve the detection efficiency. Song et al.137 reported bulk enzyme heterojunction (BEH) based on TDNs to program the multi-enzyme catalytic cascade on the electrode for sarcosine sensing (Fig. 5C). The TNDs achieved delicate regulation of the distance between enzyme pairs within the critical coupling length of about 10 nm. As a result, the overall catalytic efficiency is increased by approximately 10 times. On this basis, sarcosine was efficiently oxidized in the enzyme cascade, which resulted in an electrocatalytic current in the electrode.
Besides electrodes, continuous exploration of FNAs has been achieved in the field of Raman detection. In a recent publication, Fang et al.138 reported the precise anchoring of gold nanoparticles on a DNA origami to enable Fano resonances for quantified surface-enhanced Raman scattering (Fig. 5D). This proof-of-concept nanoplatform has provided an insight to ultrasensitive sensing of pharmaceutically-related molecules.
5. Conclusions and future perspectives
To sum up, we highlight the advantages of FNAs and review the pharmaceutical applications of FNAs (Table 1). The precise base pairing makes oligonucleotides assemble into FNAs with customized structures and narrow particle size distribution. FNAs are natural biomacromolecules which show biodegradability and biocompatibility. FNAs are easily subjected to chemical modification through introducing reactive function groups during oligonucleotide synthesis. Since each oligonucleotide inside the FNAs is localizable at nanoscale, site-specific or quantified functionalization on FNAs could be achieved. Moreover, FNAs could naturally blend in with logical DNA hybridization reaction to exhibit programmable functions and response to external stimuli. These advantages of FNAs have been proved to meet certain requirements in pharmaceutical research.
Table 1.
Summary on the application and advantages of framework nucleic acids (FNAs).
| Research area | Application | Example | Advantage |
|---|---|---|---|
| Drug discovery | Tools for biological study | Supporting membrane protein for structural analysis As labels in electron cryotomography Long-term or multiplexed fluorescence imaging Fragment-based drug discovery by atom force microscopy Spatial organization of ligands |
Customized structure Narrow particle size distribution Easy of chemical modification Site-specific functionalization Quantified cargo loading Logical DNA strand displacement reactions for programmable functions Biodegradability and biocompatibility Unique cellular uptake |
| Potential therapeutic molecules | Structural or functional mimicking of their biological counterparts Alleviation of acute kidney injury and other therapeutic efficacy |
||
| Drug delivery | Small molecule delivery | Doxorubicin, cisplatin, paclitaxel, wogonin, camptothecin, cisplatin prodrug, combretastatin A-4 | |
| Oligonucleotide delivery | Small interfering RNAs, microRNA, antisense oligonucleotide, unmethylated cytosine guanosine phosphate, aptamer | ||
| Peptide and protein delivery | Thrombin, antibacterial peptide, antigen peptides | ||
| Co-delivery with inorganic nanoparticles | Gold nanorods, platinum nanoparticles, gold nanoparticles | ||
| Synthetic templates for nanocarrier | Small unilamellar liposomes, silica nanoparticles, calcium phosphate nanoparticles | ||
| Drug analysis | Electrochemical detection of metabolite in whole blood Quantified surface-enhanced Raman scattering |
||
Despite significance advances achieved in biomedicine, several drawbacks of FNAs still exist. Firstly, the yield of FNAs is limited by the quantity of scaffold strands purchased from the company. Generally, the production of FNAs is only sufficient to reach effective concentration in cell experiments or animal models. Besides, high cost of the scaffold strands is another obstacle factor for clinical applications of FNAs. Secondly, the complex folding kinetics of oligonucleotides could play a critical role in the successful assembly of FNAs. In fact, many factors could cut off the assemble process such as high strand concentration, similarity between scaffold sequences, preferred conformation or secondary structure of each strands, dimers, and adverse intermolecular forces from chemical modification. Lastly, FNAs are susceptible to physiological conditions and may show structural disassembly. As is known, DNA contains multiple phosphate groups with negative charge, which could bring electrostatic repulsion inside the FNAs in low-salt solutions. Even though the introduction of divalent ions could block the negative charges, but the action of these ions in vivo should be taken into consideration. FNAs are also unstable in high temperature since the number of base pair for effective FNAs assembly is limited (generally 7–24 base pairs). Moreover, the nucleic-acid nature of FNAs makes it susceptible to nuclease digestion in vivo. Of note, the enzymatic stability of FNAs is able to be enhanced by costly modification of scaffold strands such as phosphorothioate or non-natural nucleotides.
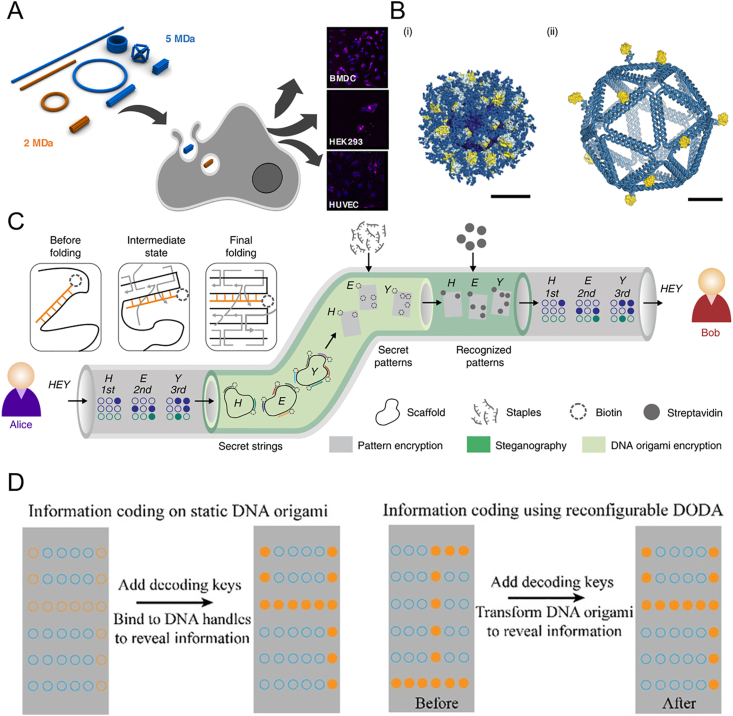
Beyond the summarized applications in this reviews, the usage of FNAs in biomedicine still have a lot to be explored in the future. Importantly, previous researches have indicated the role of size and mechanical properties in nanoparticle delivery117,139. FNAs have various properties including size, shape and stiffness. Therefore, how the properties of FNAs influence the immunogenicity, biodistribution and clearance of FNAs in vivo remains to be illuminated for better translation of research into practical applications. In fact, structures of FNAs are demonstrated to modulate the uptake or immunorecognition by cells140,141 (Fig. 6A). Besides, the quantity and site-specific organization of ligands on FNAs are relative to ligand–target interaction and the corresponding drug actions. This principle could also be elucidated to optimize the regulation of biological process such as increasing vaccination efficacy36 and viral inhibition142 (Fig. 6B). To our knowledge, the biocompatible and the encoding nature of nucleic acids render DNA capable of in-drug labeling for pharmaceutical anti-counterfeiting143. Currently, the applications of DNA in authentication mainly focus on the unique sequence buried in oligonucleotides144,145, or a unique DNA marker formed from the fragmentation of double strand DNA146. With the development of DNA nanotechnology, DNA can be encoded in a different way by folding single strands into numerous DNA framework with pre-designed size, shape and ligand patterning. Recently, Zhang et al.147 develop DNA-origami-based cryptography (Fig. 6C). The technique exploits folding of a M13mp18 single-stranded DNA into a customized nanostructure with a specific set of staple strands. Some of the staple strands are modified with biotin so that encrypted molecular patterns are placed on the DNA origami. The decryption can be accomplished upon streptavidin binding.
Figure 6.
Future applications of FNAs in pharmaceutical research. (A) Shape-dependent and cell-dependent cellular uptake of FNAs. Reprinted with permission from Ref. 140. Copyright © 2018 American Chemical Society. (B) Spatial organization of HIV immunogens on FNAs to enhance B-cell activation during vaccination. Reprinted with permission from Ref. 36. Copyright © 2020 Springer Nature. (C) Information coding in DNA origami for message confidentiality. Reprinted with permission from Ref. 147. Copyright © 2019 Springer Nature. (D) Secondary message coding by reconfigurable DNA origami. Reprinted with permission from Ref. 148. Copyright © 2020 John Wiley and Sons.
Besides, the molecule patterns on DNA origami can also be dynamic. Fan et al.148 proposed a fantastic cryptography based on reconfigurable DNA origami domino array (DODA, Fig. 6D). The nanoscale patterns on DNA origami can be transformed after DNA-programmed structure reconfiguration of the DNA framework, allowing the information coding. The confidentiality of this strategy can be further enhanced by implementation of reconfiguration-mediated toehold strand displacement reaction on the dynamic DNA origami149. The units engaged in strand displacement cascades are assembled on the DNA origami. Upon conformational transformation, the reaction units were brought into close proximity and triggered the cascades. The strand displacement reaction thus changed the molecular pattern on the DNA origami, which served as a secondary coding. It is foreseeable that DNA framework may be potentially applied to combat counterfeiting of pharmaceuticals.
Acknowledgments
This study was supported by National Natural Science Foundation (No.82072087, China), Key Technologies Research and Development Program (No. 2016YFA0201200, China), and the Guangdong Natural Science Fund for Distinguished Young Scholars (No. 2017A030306016, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Contributor Information
Yuanqing Zhang, Email: zhangyq65@mail.sysu.edu.cn.
Chunhai Fan, Email: fanchunhai@sjtu.edu.cn.
Author contributions
Liang Chen generated the conception of the review. Ziyan Zhang drafted the Introduction. Liang Chen drafted the section of drug discovery and conclusion. Jie Zhang and Miao Mao drafted the section of drug delivery. Zhun Lin and Jiacheng Wu drafted the section of drug analysis. Liang Chen edited the manuscript. Yuanqing Zhang and Chunhai Fan supervised the manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
References
- 1.Sessler J.L., Lawrence C.M., Jayawickramarajah J. Molecular recognition via base-pairing. Chem Soc Rev. 2007;36:314–325. doi: 10.1039/b604119c. [DOI] [PubMed] [Google Scholar]
- 2.Watson J.D., Crick F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y., Tu J., Wang D., Zhu H., Maity S.K., Qu X., et al. Programmable and multifunctional DNA-based materials for biomedical applications. Adv Mater. 2018;30 doi: 10.1002/adma.201703658. [DOI] [PubMed] [Google Scholar]
- 4.Seeman N.C., Sleiman H.F. DNA nanotechnology. Nat Rev Mater. 2017;3:17068. [Google Scholar]
- 5.Seeman N.C. Nucleic acid junctions and lattices. J Theor Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 6.Seeman N.C., Kallenbach N.R. Design of immobile nucleic acid junctions. Biophys J. 1983;44:201–209. doi: 10.1016/S0006-3495(83)84292-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seeman N.C. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 8.Lin C., Liu Y., Rinker S., Yan H. DNA tile based self-assembly: building complex nanoarchitectures. ChemPhysChem. 2006;7:1641–1647. doi: 10.1002/cphc.200600260. [DOI] [PubMed] [Google Scholar]
- 9.Hong F., Zhang F., Liu Y., Yan H. DNA origami: scaffolds for creating higher order structures. Chem Rev. 2017;117:12584–12640. doi: 10.1021/acs.chemrev.6b00825. [DOI] [PubMed] [Google Scholar]
- 10.Rothemund P.W. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 11.Dey S., Fan C., Gothelf K.V., Li J., Lin C., Liu L., et al. DNA origami. Nat Rev Methods Prim. 2021;1:13. [Google Scholar]
- 12.Ge Z., Gu H., Li Q., Fan C. Concept and development of framework nucleic acids. J Am Chem Soc. 2018;140:17808–17819. doi: 10.1021/jacs.8b10529. [DOI] [PubMed] [Google Scholar]
- 13.Ramezani H., Dietz H. Building machines with DNA molecules. Nat Rev Genet. 2020;21:5–26. doi: 10.1038/s41576-019-0175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He L., Lu D., Liang H., Xie S., Zhang X., Liu Q., et al. mRNA-initiated, three-dimensional DNA amplifier able to function inside living cells. J Am Chem Soc. 2018;140:258–263. doi: 10.1021/jacs.7b09789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang D., Ge Z., Im H.J., England C.G., Ni D., Hou J., et al. DNA origami nanostructures can exhibit preferential renal uptake and alleviate acute kidney injury. Nat Biomed Eng. 2018;2:865–877. doi: 10.1038/s41551-018-0317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang T., Tian T., Zhou R., Li S., Ma W., Zhang Y., et al. Design, fabrication and applications of tetrahedral DNA nanostructure-based multifunctional complexes in drug delivery and biomedical treatment. Nat Protoc. 2020;15:2728–2757. doi: 10.1038/s41596-020-0355-z. [DOI] [PubMed] [Google Scholar]
- 17.Madhanagopal B.R., Zhang S., Demirel E., Wady H., Chandrasekaran A.R. DNA nanocarriers: programmed to deliver. Trends Biochem Sci. 2018;43:997–1013. doi: 10.1016/j.tibs.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Xia K., Kong H., Cui Y., Ren N., Li Q., Ma J., et al. Systematic study in mammalian cells showing No adverse response to tetrahedral DNA nanostructure. ACS Appl Mater Interfac. 2018;10:15442–15448. doi: 10.1021/acsami.8b02626. [DOI] [PubMed] [Google Scholar]
- 19.Schuller V.J., Heidegger S., Sandholzer N., Nickels P.C., Suhartha N.A., Endres S., et al. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano. 2011;5:9696–9702. doi: 10.1021/nn203161y. [DOI] [PubMed] [Google Scholar]
- 20.Shen X., Jiang Q., Wang J., Dai L., Zou G., Wang Z.G., et al. Visualization of the intracellular location and stability of DNA origami with a label-free fluorescent probe. Chem Commun. 2012;48:11301–11303. doi: 10.1039/c2cc36185j. [DOI] [PubMed] [Google Scholar]
- 21.Renaud J.P., Chari A., Ciferri C., Liu W.T., Remigy H.W., Stark H., et al. Cryo-EM in drug discovery: achievements, limitations and prospects. Nat Rev Drug Discov. 2018;17:471–492. doi: 10.1038/nrd.2018.77. [DOI] [PubMed] [Google Scholar]
- 22.Denisov I.G., Sligar S.G. Nanodiscs in membrane biochemistry and biophysics. Chem Rev. 2017;117:4669–4713. doi: 10.1021/acs.chemrev.6b00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iric K., Subramanian M., Oertel J., Agarwal N.P., Matthies M., Periole X., et al. DNA-encircled lipid bilayers. Nanoscale. 2018;10:18463–18467. doi: 10.1039/c8nr06505e. [DOI] [PubMed] [Google Scholar]
- 24.Martin T.G., Bharat T.A., Joerger A.C., Bai X.C., Praetorius F., Fersht A.R., et al. Design of a molecular support for cryo-EM structure determination. Proc Natl Acad Sci U S A. 2016;113:E7456–E7463. doi: 10.1073/pnas.1612720113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong Y., Chen S., Zhang S., Sodroski J., Yang Z., Liu D., et al. Folding DNA into a lipid-conjugated nanobarrel for controlled reconstitution of membrane proteins. Angew Chem Int Ed Engl. 2018;57:2072–2076. doi: 10.1002/anie.201710147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silvester E., Vollmer B., Prazak V., Vasishtan D., Machala E.A., Whittle C., et al. DNA origami signposts for identifying proteins on cell membranes by electron cryotomography. Cell. 2021;184 doi: 10.1016/j.cell.2021.01.033. 1110-21.e1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niekamp S., Stuurman N., Vale R.D. A 6-nm ultra-photostable DNA FluoroCube for fluorescence imaging. Nat Methods. 2020;17:437–441. doi: 10.1038/s41592-020-0782-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J., Dai J., Jiang S., Xie M., Zhai T., Guo L., et al. Encoding quantized fluorescence states with fractal DNA frameworks. Nat Commun. 2020;11:2185. doi: 10.1038/s41467-020-16112-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erlanson D.A., Fesik S.W., Hubbard R.E., Jahnke W., Jhoti H. Twenty years on: the impact of fragments on drug discovery. Nat Rev Drug Discov. 2016;15:605–619. doi: 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- 30.Kielar C., Reddavide F.V., Tubbenhauer S., Cui M., Xu X., Grundmeier G., et al. Pharmacophore nanoarrays on DNA origami substrates as a single-molecule assay for fragment-based drug discovery. Angew Chem Int Ed Engl. 2018;57:14873–14877. doi: 10.1002/anie.201806778. [DOI] [PubMed] [Google Scholar]
- 31.Huang J., Suma A., Cui M., Grundmeier G., Carnevale V., Zhang Y., et al. Arranging small molecules with subnanometer precision on DNA origami substrates for the single-molecule investigation of protein–ligand interactions. Small Struct. 2020;1:2000038. [Google Scholar]
- 32.Attwood M.M., Jonsson J., Rask-Andersen M., Schioth H.B. Soluble ligands as drug targets. Nat Rev Drug Discov. 2020;19:695–710. doi: 10.1038/s41573-020-0078-4. [DOI] [PubMed] [Google Scholar]
- 33.Rosenbaum M.I., Clemmensen L.S., Bredt D.S., Bettler B., Stromgaard K. Targeting receptor complexes: a new dimension in drug discovery. Nat Rev Drug Discov. 2020;19:884–901. doi: 10.1038/s41573-020-0086-4. [DOI] [PubMed] [Google Scholar]
- 34.Shaw A., Lundin V., Petrova E., Fordos F., Benson E., Al-Amin A., et al. Spatial control of membrane receptor function using ligand nanocalipers. Nat Methods. 2014;11:841–846. doi: 10.1038/nmeth.3025. [DOI] [PubMed] [Google Scholar]
- 35.Fang T., Alvelid J., Spratt J., Ambrosetti E., Testa I., Teixeira A.I. Spatial regulation of T-cell signaling by programmed death-ligand 1 on wireframe DNA origami flat sheets. ACS Nano. 2021;15:3441–3452. doi: 10.1021/acsnano.0c10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veneziano R., Moyer T.J., Stone M.B., Wamhoff E.C., Read B.J., Mukherjee S., et al. Role of nanoscale antigen organization on B-cell activation probed using DNA origami. Nat Nanotechnol. 2020;15:716–723. doi: 10.1038/s41565-020-0719-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song L., Hobaugh M.R., Shustak C., Cheley S., Bayley H., Gouaux J.E. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 38.Langecker M., Arnaut V., Martin T.G., List J., Renner S., Mayer M., et al. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science. 2012;338:932–936. doi: 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krishnan S., Ziegler D., Arnaut V., Martin T.G., Kapsner K., Henneberg K., et al. Molecular transport through large-diameter DNA nanopores. Nat Commun. 2016;7:12787. doi: 10.1038/ncomms12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diederichs T., Pugh G., Dorey A., Xing Y., Burns J.R., Hung Nguyen Q., et al. Synthetic protein-conductive membrane nanopores built with DNA. Nat Commun. 2019;10:5018. doi: 10.1038/s41467-019-12639-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanphere C., Offenbartl-Stiegert D., Dorey A., Pugh G., Georgiou E., Xing Y., et al. Design, assembly, and characterization of membrane-spanning DNA nanopores. Nat Protoc. 2021;16:86–130. doi: 10.1038/s41596-020-0331-7. [DOI] [PubMed] [Google Scholar]
- 42.Chidchob P., Offenbartl-Stiegert D., McCarthy D., Luo X., Li J., Howorka S., et al. Spatial presentation of cholesterol units on a DNA cube as a determinant of membrane protein-mimicking functions. J Am Chem Soc. 2019;141:1100–1108. doi: 10.1021/jacs.8b11898. [DOI] [PubMed] [Google Scholar]
- 43.Burns J.R., Al-Juffali N., Janes S.M., Howorka S. Membrane-spanning DNA nanopores with cytotoxic effect. Angew Chem Int Ed Engl. 2014;53:12466–12470. doi: 10.1002/anie.201405719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen L., Liang S., Chen Y., Wu M., Zhang Y. Destructing the plasma membrane with activatable vesicular DNA nanopores. ACS Appl Mater Interfac. 2020;12:96–105. doi: 10.1021/acsami.9b14746. [DOI] [PubMed] [Google Scholar]
- 45.Lv C., Gu X., Li H., Zhao Y., Yang D., Yu W., et al. Molecular transport through a biomimetic DNA channel on live cell membranes. ACS Nano. 2020;14:14616–14626. doi: 10.1021/acsnano.0c03105. [DOI] [PubMed] [Google Scholar]
- 46.Arulkumaran N., Lanphere C., Gaupp C., Burns J.R., Singer M., Howorka S. DNA nanodevices with selective immune cell interaction and function. ACS Nano. 2021;15:4394–4404. doi: 10.1021/acsnano.0c07915. [DOI] [PubMed] [Google Scholar]
- 47.Messager L., Burns J.R., Kim J., Cecchin D., Hindley J., Pyne A.L., et al. Biomimetic hybrid nanocontainers with selective permeability. Angew Chem Int Ed Engl. 2016;55:11106–11109. doi: 10.1002/anie.201604677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burns J.R., Seifert A., Fertig N., Howorka S. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat Nanotechnol. 2016;11:152–156. doi: 10.1038/nnano.2015.279. [DOI] [PubMed] [Google Scholar]
- 49.Yang Q., Guo Z., Liu H., Peng R., Xu L., Bi C., et al. A cascade signaling network between artificial cells switching activity of synthetic transmembrane channels. J Am Chem Soc. 2021;143:232–240. doi: 10.1021/jacs.0c09558. [DOI] [PubMed] [Google Scholar]
- 50.Arnott P.M., Howorka S. A temperature-gated nanovalve self-assembled from DNA to control molecular transport across membranes. ACS Nano. 2019;13:3334–3340. doi: 10.1021/acsnano.8b09200. [DOI] [PubMed] [Google Scholar]
- 51.Peng R., Xu L., Wang H., Lyu Y., Wang D., Bi C., et al. DNA-based artificial molecular signaling system that mimics basic elements of reception and response. Nat Commun. 2020;11:978. doi: 10.1038/s41467-020-14739-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lanphere C., Arnott P.M., Jones S.F., Korlova K., Howorka S. A biomimetic DNA-based membrane gate for protein-controlled transport of cytotoxic drugs. Angew Chem Int Ed Engl. 2021;60:1903–1908. doi: 10.1002/anie.202011583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohmann A., Li C.Y., Maffeo C., Al Nahas K., Baumann K.N., Gopfrich K., et al. A synthetic enzyme built from DNA flips 10(7) lipids per second in biological membranes. Nat Commun. 2018;9:2426. doi: 10.1038/s41467-018-04821-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiao K., Zhu B., Guo L., Zhou H., Wang F., Zhang X., et al. Programming switchable transcription of topologically constrained DNA. J Am Chem Soc. 2020;142:10739–10746. doi: 10.1021/jacs.0c01962. [DOI] [PubMed] [Google Scholar]
- 55.Tian T.R., Xiao D.X., Zhang T., Li Y.J., Shi S.R., Zhong W.Y., et al. A framework nucleic acid based robotic nanobee for active targeting therapy. Adv Funct Mater. 2021:31. [Google Scholar]
- 56.Ma W., Zhan Y., Zhang Y., Xie X., Mao C., Lin Y. Enhanced neural regeneration with a concomitant treatment of framework nucleic acid and stem cells in spinal cord injury. ACS Appl Mater Interfac. 2020;12:2095–2106. doi: 10.1021/acsami.9b19079. [DOI] [PubMed] [Google Scholar]
- 57.Goodman R.P., Schaap I.A., Tardin C.F., Erben C.M., Berry R.M., Schmidt C.F., et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 58.Walsh A.S., Yin H., Erben C.M., Wood M.J., Turberfield A.J. DNA cage delivery to mammalian cells. ACS Nano. 2011;5:5427–5432. doi: 10.1021/nn2005574. [DOI] [PubMed] [Google Scholar]
- 59.Ding H., Li J., Chen N., Hu X., Yang X., Guo L., et al. DNA nanostructure-programmed like-charge attraction at the cell-membrane interface. ACS Cent Sci. 2018;4:1344–1351. doi: 10.1021/acscentsci.8b00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang L., Li J., Li Q., Huang Q., Shi J., Yan H., et al. Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew Chem Int Ed Engl. 2014;53:7745–7750. doi: 10.1002/anie.201403236. [DOI] [PubMed] [Google Scholar]
- 61.Wiraja C., Zhu Y., Lio D.C.S., Yeo D.C., Xie M., Fang W., et al. Framework nucleic acids as programmable carrier for transdermal drug delivery. Nat Commun. 2019;10:1147. doi: 10.1038/s41467-019-09029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia Z., Wang P., Liu X., Liu T., Yan Y., Yan J., et al. Tumor-penetrating peptide-modified DNA tetrahedron for targeting drug delivery. Biochemistry. 2016;55:1326–1331. doi: 10.1021/acs.biochem.5b01181. [DOI] [PubMed] [Google Scholar]
- 63.Sun P., Zhang N., Tang Y., Yang Y., Chu X., Zhao Y. SL2B aptamer and folic acid dual-targeting DNA nanostructures for synergic biological effect with chemotherapy to combat colorectal cancer. Int J Nanomed. 2017;12:2657–2672. doi: 10.2147/IJN.S132929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang M., Yang C.S., Huang D.M. Aptamer-conjugated DNA icosahedral nanoparticles as a carrier of doxorubicin for cancer therapy. ACS Nano. 2011;5:6156–6163. doi: 10.1021/nn200693a. [DOI] [PubMed] [Google Scholar]
- 65.Jiang Q., Song C., Nangreave J., Liu X., Lin L., Qiu D., et al. DNA origami as a carrier for circumvention of drug resistance. J Am Chem Soc. 2012;134:13396–13403. doi: 10.1021/ja304263n. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Q., Jiang Q., Li N., Dai L., Liu Q., Song L., et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano. 2014;8:6633–6643. doi: 10.1021/nn502058j. [DOI] [PubMed] [Google Scholar]
- 67.Zhao Y.X., Shaw A., Zeng X., Benson E., Nystrom A.M., Hogberg B. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano. 2012;6:8684–8691. doi: 10.1021/nn3022662. [DOI] [PubMed] [Google Scholar]
- 68.Wu T., Liu J., Liu M., Liu S., Zhao S., Tian R., et al. A nanobody-conjugated DNA nanoplatform for targeted platinum-drug delivery. Angew Chem Int Ed Engl. 2019;58:14224–14228. doi: 10.1002/anie.201909345. [DOI] [PubMed] [Google Scholar]
- 69.Xie X., Shao X., Ma W., Zhao D., Shi S., Li Q., et al. Overcoming drug-resistant lung cancer by paclitaxel loaded tetrahedral DNA nanostructures. Nanoscale. 2018;10:5457–5465. doi: 10.1039/c7nr09692e. [DOI] [PubMed] [Google Scholar]
- 70.Shi S., Fu W., Lin S., Tian T., Li S., Shao X., et al. Targeted and effective glioblastoma therapy via aptamer-modified tetrahedral framework nucleic acid-paclitaxel nanoconjugates that can pass the blood brain barrier. Nanomedicine. 2019;21:102061. doi: 10.1016/j.nano.2019.102061. [DOI] [PubMed] [Google Scholar]
- 71.Shi S., Chen Y., Tian T., Li S., Lin S., Zhang Y., et al. Effects of tetrahedral framework nucleic acid/wogonin complexes on osteoarthritis. Bone Res. 2020;8:6. doi: 10.1038/s41413-019-0077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mou Q., Ma Y., Pan G., Xue B., Yan D., Zhang C., et al. DNA trojan horses: self-assembled floxuridine-containing DNA polyhedra for cancer therapy. Angew Chem Int Ed Engl. 2017;56:12528–12532. doi: 10.1002/anie.201706301. [DOI] [PubMed] [Google Scholar]
- 73.Zhang J., Guo Y., Ding F., Pan G., Zhu X., Zhang C. A camptothecin-grafted DNA tetrahedron as a precise nanomedicine to inhibit tumor growth. Angew Chem Int Ed Engl. 2019;58:13794–13798. doi: 10.1002/anie.201907380. [DOI] [PubMed] [Google Scholar]
- 74.Zhong Y.F., Cheng J., Liu Y., Luo T., Wang Y., Jiang K., et al. DNA nanostructures as Pt(IV) prodrug delivery systems to combat chemoresistance. Small. 2020;16 doi: 10.1002/smll.202003646. [DOI] [PubMed] [Google Scholar]
- 75.Wang D., Peng R.Z., Peng Y.B., Deng Z.Y., Xu F.Y., Su Y.Y., et al. Hierarchical fabrication of DNA wireframe nanoarchitectures for efficient cancer imaging and targeted therapy. ACS Nano. 2020;14:17365–17375. doi: 10.1021/acsnano.0c07495. [DOI] [PubMed] [Google Scholar]
- 76.Huang Z., Qiu L., Zhang T., Tan W. Integrating DNA nanotechnology with aptamers for biological and biomedical applications. Matter. 2021;4:461–489. [Google Scholar]
- 77.Madsen M., Gothelf K.V. Chemistries for DNA nanotechnology. Chem Rev. 2019;119:6384–6458. doi: 10.1021/acs.chemrev.8b00570. [DOI] [PubMed] [Google Scholar]
- 78.Zamore P.D. RNA interference: listening to the sound of silence. Nat Struct Biol. 2001;8:746–750. doi: 10.1038/nsb0901-746. [DOI] [PubMed] [Google Scholar]
- 79.Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 80.Charbe N.B., Amnerkar N.D., Ramesh B., Tambuwala M.M., Bakshi H.A., Aljabali A.A.A., et al. Small interfering RNA for cancer treatment: overcoming hurdles in delivery. Acta Pharm Sin B. 2020;10:2075–2109. doi: 10.1016/j.apsb.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo Q., Jiang C. Delivery strategies for macromolecular drugs in cancer therapy. Acta Pharm Sin B. 2020;10:979–986. doi: 10.1016/j.apsb.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kanasty R., Dorkin J.R., Vegas A., Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 83.Lee H., Lytton-Jean A.K., Chen Y., Love K.T., Park A.I., Karagiannis E.D., et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rahman M.A., Wang P., Zhao Z., Wang D., Nannapaneni S., Zhang C., et al. Systemic delivery of bc12-targeting siRNA by DNA nanoparticles suppresses cancer cell growth. Angew Chem Int Ed Engl. 2017;56:16023–16027. doi: 10.1002/anie.201709485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Z., Song L., Liu Q., Tian R., Shang Y., Liu F., et al. A tubular DNA nanodevice as a siRNA/chemo-drug Co-delivery vehicle for combined cancer therapy. Angew Chem Int Ed Engl. 2021;60:2594–2598. doi: 10.1002/anie.202009842. [DOI] [PubMed] [Google Scholar]
- 86.Bujold K.E., Hsu J.C.C., Sleiman H.F. Optimized DNA "nanosuitcases" for encapsulation and conditional release of siRNA. J Am Chem Soc. 2016;138:14030–14038. doi: 10.1021/jacs.6b08369. [DOI] [PubMed] [Google Scholar]
- 87.Ren K., Liu Y., Wu J., Zhang Y., Zhu J., Yang M., et al. A DNA dual lock-and-key strategy for cell-subtype-specific siRNA delivery. Nat Commun. 2016;7:13580. doi: 10.1038/ncomms13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qian H., Tay C.Y., Setyawati M.I., Chia S.L., Lee D.S., Leong D.T. Protecting microRNAs from RNase degradation with steric DNA nanostructures. Chem Sci. 2017;8:1062–1067. doi: 10.1039/c6sc01829g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li S., Sun Y., Tian T., Qin X., Lin S., Zhang T., et al. MicroRNA-214-3p modified tetrahedral framework nucleic acids target survivin to induce tumour cell apoptosis. Cell Prolif. 2020;53 doi: 10.1111/cpr.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nahar S., Nayak A.K., Ghosh A., Subudhi U., Maiti S. Enhanced and synergistic downregulation of oncogenic miRNAs by self-assembled branched DNA. Nanoscale. 2017;10:195–202. doi: 10.1039/c7nr06601e. [DOI] [PubMed] [Google Scholar]
- 91.Ming X., Laing B. Bioconjugates for targeted delivery of therapeutic oligonucleotides. Adv Drug Deliv Rev. 2015;87:81–89. doi: 10.1016/j.addr.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang J., Jiang Q., He L., Zhan P., Liu Q., Liu S., et al. Self-assembled double-bundle DNA tetrahedron for efficient antisense delivery. ACS Appl Mater Interfac. 2018;10:23693–23699. doi: 10.1021/acsami.8b07889. [DOI] [PubMed] [Google Scholar]
- 93.Wu T., Liu Q., Cao Y., Tian R., Liu J., Ding B. Multifunctional double-bundle DNA tetrahedron for efficient regulation of gene expression. ACS Appl Mater Interfac. 2020;12:32461–32467. doi: 10.1021/acsami.0c08886. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Y., Ma W., Zhu Y., Shi S., Li Q., Mao C., et al. Inhibiting methicillin-resistant Staphylococcus aureus by tetrahedral DNA nanostructure-enabled antisense peptide nucleic acid delivery. Nano Lett. 2018;18:5652–5659. doi: 10.1021/acs.nanolett.8b02166. [DOI] [PubMed] [Google Scholar]
- 95.Liu J., Song L., Liu S., Jiang Q., Liu Q., Li N., et al. A DNA-based nanocarrier for efficient gene delivery and combined cancer therapy. Nano Lett. 2018;18:3328–3334. doi: 10.1021/acs.nanolett.7b04812. [DOI] [PubMed] [Google Scholar]
- 96.Krieg A.M., Yi A.K., Matson S., Waldschmidt T.J., Bishop G.A., Teasdale R., et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 97.Klinman D.M. Immunotherapeutic uses of CpG oligodeoxynucleotides. Nat Rev Immunol. 2004;4:249–258. doi: 10.1038/nri1329. [DOI] [PubMed] [Google Scholar]
- 98.Li J., Pei H., Zhu B., Liang L., Wei M., He Y., et al. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano. 2011;5:8783–8789. doi: 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- 99.Liu X., Xu Y., Yu T., Clifford C., Liu Y., Yan H., et al. A DNA nanostructure platform for directed assembly of synthetic vaccines. Nano Lett. 2012;12:4254–4259. doi: 10.1021/nl301877k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qu Y., Yang J., Zhan P., Liu S., Zhang K., Jiang Q., et al. Self-assembled DNA dendrimer nanoparticle for efficient delivery of immunostimulatory CpG motifs. ACS Appl Mater Interfac. 2017;9:20324–20329. doi: 10.1021/acsami.7b05890. [DOI] [PubMed] [Google Scholar]
- 101.Ma W., Zhan Y., Zhang Y., Shao X., Xie X., Mao C., et al. An intelligent DNA nanorobot with in vitro enhanced protein lysosomal degradation of HER2. Nano Lett. 2019;19:4505–4517. doi: 10.1021/acs.nanolett.9b01320. [DOI] [PubMed] [Google Scholar]
- 102.Yang L., Zhao Y., Xu X., Xu K., Zhang M., Huang K., et al. An intelligent DNA nanorobot for autonomous anticoagulation. Angew Chem Int Ed Engl. 2020;59:17697–17704. doi: 10.1002/anie.202007962. [DOI] [PubMed] [Google Scholar]
- 103.Gu Z., Biswas A., Zhao M., Tang Y. Tailoring nanocarriers for intracellular protein delivery. Chem Soc Rev. 2011;40:3638–3655. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]
- 104.Leader B., Baca Q.J., Golan D.E. Protein therapeutics: a summary and pharmacological classification. Nat Rev Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 105.Sambrano G.R., Weiss E.J., Zheng Y.W., Huang W., Coughlin S.R. Role of thrombin signalling in platelets in haemostasis and thrombosis. Nature. 2001;413:74–78. doi: 10.1038/35092573. [DOI] [PubMed] [Google Scholar]
- 106.Li S., Jiang Q., Liu S., Zhang Y., Tian Y., Song C., et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat Biotechnol. 2018;36:258–264. doi: 10.1038/nbt.4071. [DOI] [PubMed] [Google Scholar]
- 107.Liu Y., Sun Y., Li S., Liu M., Qin X., Chen X., et al. Tetrahedral framework nucleic acids deliver antimicrobial peptides with improved effects and less susceptibility to bacterial degradation. Nano Lett. 2020;20:3602–3610. doi: 10.1021/acs.nanolett.0c00529. [DOI] [PubMed] [Google Scholar]
- 108.Liu S., Jiang Q., Zhao X., Zhao R., Wang Y., Wang Y., et al. A DNA nanodevice-based vaccine for cancer immunotherapy. Nat Mater. 2021;20:421–430. doi: 10.1038/s41563-020-0793-6. [DOI] [PubMed] [Google Scholar]
- 109.Lucky S.S., Soo K.C., Zhang Y. Nanoparticles in photodynamic therapy. Chem Rev. 2015;115:1990–2042. doi: 10.1021/cr5004198. [DOI] [PubMed] [Google Scholar]
- 110.Jiang Q., Shi Y., Zhang Q., Li N., Zhan P., Song L., et al. A self-assembled DNA origami-gold nanorod complex for cancer theranostics. Small. 2015;11:5134–5141. doi: 10.1002/smll.201501266. [DOI] [PubMed] [Google Scholar]
- 111.Du Y., Jiang Q., Beziere N., Song L., Zhang Q., Peng D., et al. DNA-Nanostructure-Gold-Nanorod hybrids for enhanced in vivo optoacoustic imaging and photothermal therapy. Adv Mater. 2016;28:10000–10007. doi: 10.1002/adma.201601710. [DOI] [PubMed] [Google Scholar]
- 112.Ma Y., Wang Z., Ma Y., Han Z., Zhang M., Chen H., et al. A telomerase-responsive DNA icosahedron for precise delivery of platinum nanodrugs to cisplatin-resistant cancer. Angew Chem Int Ed Engl. 2018;57:5389–5393. doi: 10.1002/anie.201801195. [DOI] [PubMed] [Google Scholar]
- 113.Wang P., Rahman M.A., Zhao Z., Weiss K., Zhang C., Chen Z., et al. Visualization of the cellular uptake and trafficking of DNA origami nanostructures in cancer cells. J Am Chem Soc. 2018;140:2478–2484. doi: 10.1021/jacs.7b09024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang Y., Wang J., Shigematsu H., Xu W., Shih W.M., Rothman J.E., et al. Self-assembly of size-controlled liposomes on DNA nanotemplates. Nat Chem. 2016;8:476–483. doi: 10.1038/nchem.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Baumann K.N., Piantanida L., Garcia-Nafria J., Sobota D., Voitchovsky K., Knowles T.P.J., et al. Coating and stabilization of liposomes by clathrin-inspired DNA self-assembly. ACS Nano. 2020;14:2316–2323. doi: 10.1021/acsnano.9b09453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu X., Zhang F., Jing X., Pan M., Liu P., Li W., et al. Complex silica composite nanomaterials templated with DNA origami. Nature. 2018;559:593–598. doi: 10.1038/s41586-018-0332-7. [DOI] [PubMed] [Google Scholar]
- 117.Hui Y., Yi X., Hou F., Wibowo D., Zhang F., Zhao D., et al. Role of nanoparticle mechanical properties in cancer drug delivery. ACS Nano. 2019;13:7410–7424. doi: 10.1021/acsnano.9b03924. [DOI] [PubMed] [Google Scholar]
- 118.Liu X.G., Jing X.X., Liu P., Pan M.C., Liu Z., Dai X.P., et al. DNA framework-encoded mineralization of calcium phosphate. Chem-Us. 2020;6:472–485. [Google Scholar]
- 119.Wu S., Zhang M., Song J., Weber S., Liu X., Fan C., et al. Fine customization of calcium phosphate nanostructures with site-specific modification by DNA templated mineralization. ACS Nano. 2021;15:1555–1565. doi: 10.1021/acsnano.0c08998. [DOI] [PubMed] [Google Scholar]
- 120.Yang F., Li Q., Wang L., Zhang G.J., Fan C. Framework-nucleic-acid-enabled biosensor development. ACS Sens. 2018;3:903–919. doi: 10.1021/acssensors.8b00257. [DOI] [PubMed] [Google Scholar]
- 121.Sundah N.R., Ho N.R.Y., Lim G.S., Natalia A., Ding X., Liu Y., et al. Barcoded DNA nanostructures for the multiplexed profiling of subcellular protein distribution. Nat Biomed Eng. 2019;3:684–694. doi: 10.1038/s41551-019-0417-0. [DOI] [PubMed] [Google Scholar]
- 122.Wu W., Yu X., Wu J., Wu T., Fan Y., Chen W., et al. Surface plasmon resonance imaging-based biosensor for multiplex and ultrasensitive detection of NSCLC-associated exosomal miRNAs using DNA programmed heterostructure of Au-on-Ag. Biosens Bioelectron. 2021;175:112835. doi: 10.1016/j.bios.2020.112835. [DOI] [PubMed] [Google Scholar]
- 123.Liu Y.B., Chen D.B., Zhang W.Q., Zhang Y.Q. Mobile DNA tetrahedron on ultra-low adsorption lipid membrane for directional control of cell sensing. Sensor Actuator B Chem. 2020;307 [Google Scholar]
- 124.Chen D., Sun D., Wang Z., Qin W., Chen L., Zhou L., et al. A DNA nanostructured aptasensor for the sensitive electrochemical detection of HepG2 cells based on multibranched hybridization chain reaction amplification strategy. Biosens Bioelectron. 2018;117:416–421. doi: 10.1016/j.bios.2018.06.041. [DOI] [PubMed] [Google Scholar]
- 125.Qu X., Zhang H., Chen H., Aldalbahi A., Li L., Tian Y., et al. Convection-driven pull-down assays in nanoliter droplets using scaffolded aptamers. Anal Chem. 2017;89:3468–3473. doi: 10.1021/acs.analchem.6b04475. [DOI] [PubMed] [Google Scholar]
- 126.Lin M., Song P., Zhou G., Zuo X., Aldalbahi A., Lou X., et al. Electrochemical detection of nucleic acids, proteins, small molecules and cells using a DNA-nanostructure-based universal biosensing platform. Nat Protoc. 2016;11:1244–1263. doi: 10.1038/nprot.2016.071. [DOI] [PubMed] [Google Scholar]
- 127.Wong E.L., Chow E., Gooding J.J. DNA recognition interfaces: the influence of interfacial design on the efficiency and kinetics of hybridization. Langmuir. 2005;21:6957–6965. doi: 10.1021/la050725m. [DOI] [PubMed] [Google Scholar]
- 128.Irving D., Gong P., Levicky R. DNA surface hybridization: comparison of theory and experiment. J Phys Chem B. 2010;114:7631–7640. doi: 10.1021/jp100860z. [DOI] [PubMed] [Google Scholar]
- 129.Pei H., Zuo X., Zhu D., Huang Q., Fan C. Functional DNA nanostructures for theranostic applications. Acc Chem Res. 2014;47:550–559. doi: 10.1021/ar400195t. [DOI] [PubMed] [Google Scholar]
- 130.Chen X., Zhou G., Song P., Wang J., Gao J., Lu J., et al. Ultrasensitive electrochemical detection of prostate-specific antigen by using antibodies anchored on a DNA nanostructural scaffold. Anal Chem. 2014;86:7337–7342. doi: 10.1021/ac500054x. [DOI] [PubMed] [Google Scholar]
- 131.Ge Z., Lin M., Wang P., Pei H., Yan J., Shi J., et al. Hybridization chain reaction amplification of microRNA detection with a tetrahedral DNA nanostructure-based electrochemical biosensor. Anal Chem. 2014;86:2124–2130. doi: 10.1021/ac4037262. [DOI] [PubMed] [Google Scholar]
- 132.Dong S., Zhao R., Zhu J., Lu X., Li Y., Qiu S., et al. Electrochemical DNA biosensor based on a tetrahedral nanostructure probe for the detection of avian influenza A (H7N9) virus. ACS Appl Mater Interfac. 2015;7:8834–8842. doi: 10.1021/acsami.5b01438. [DOI] [PubMed] [Google Scholar]
- 133.Li M., Liu J.B., Zhao H.P., Song L., Mao X.H., Ge Z.L., et al. DNA framework-programmed micronano hierarchy sensor interface for metabolite analysis in whole blood. Acs Appl Bio Mater. 2020;3:53–58. doi: 10.1021/acsabm.9b00841. [DOI] [PubMed] [Google Scholar]
- 134.Wen Y., Pei H., Wan Y., Su Y., Huang Q., Song S., et al. DNA nanostructure-decorated surfaces for enhanced aptamer-target binding and electrochemical cocaine sensors. Anal Chem. 2011;83:7418–7423. doi: 10.1021/ac201491p. [DOI] [PubMed] [Google Scholar]
- 135.Li Z., Zhao B., Wang D., Wen Y., Liu G., Dong H., et al. DNA nanostructure-based universal microarray platform for high-efficiency multiplex bioanalysis in biofluids. ACS Appl Mater Interfac. 2014;6:17944–17953. doi: 10.1021/am5047735. [DOI] [PubMed] [Google Scholar]
- 136.Wang J., Cheng W., Meng F., Yang M., Pan Y., Miao P. Hand-in-hand RNA nanowire-based aptasensor for the detection of theophylline. Biosens Bioelectron. 2018;101:153–158. doi: 10.1016/j.bios.2017.10.025. [DOI] [PubMed] [Google Scholar]
- 137.Song P., Shen J., Ye D., Dong B., Wang F., Pei H., et al. Programming bulk enzyme heterojunctions for biosensor development with tetrahedral DNA framework. Nat Commun. 2020;11:838. doi: 10.1038/s41467-020-14664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fang W., Jia S., Chao J., Wang L., Duan X., Liu H., et al. Quantizing single-molecule surface-enhanced Raman scattering with DNA origami metamolecules. Sci Adv. 2019;5 doi: 10.1126/sciadv.aau4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yu W., Liu R., Zhou Y., Gao H. Size-tunable strategies for a tumor targeted drug delivery system. ACS Cent Sci. 2020;6:100–116. doi: 10.1021/acscentsci.9b01139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bastings M.M.C., Anastassacos F.M., Ponnuswamy N., Leifer F.G., Cuneo G., Lin C., et al. Modulation of the cellular uptake of DNA origami through control over mass and shape. Nano Lett. 2018;18:3557–3564. doi: 10.1021/acs.nanolett.8b00660. [DOI] [PubMed] [Google Scholar]
- 141.Hong E., Halman J.R., Shah A.B., Khisamutdinov E.F., Dobrovolskaia M.A., Afonin K.A. Structure and composition define immunorecognition of nucleic acid nanoparticles. Nano Lett. 2018;18:4309–4321. doi: 10.1021/acs.nanolett.8b01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kwon P.S., Ren S., Kwon S.J., Kizer M.E., Kuo L., Xie M., et al. Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition. Nat Chem. 2020;12:26–35. doi: 10.1038/s41557-019-0369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang H., Hua D., Huang C., Samal S.K., Xiong R., Sauvage F., et al. Materials and Technologies to combat counterfeiting of pharmaceuticals: current and future problem tackling. Adv Mater. 2020;32 doi: 10.1002/adma.201905486. [DOI] [PubMed] [Google Scholar]
- 144.Paunescu D., Fuhrer R., Grass R.N. Protection and deprotection of DNA--high-temperature stability of nucleic acid barcodes for polymer labeling. Angew Chem Int Ed Engl. 2013;52:4269–4272. doi: 10.1002/anie.201208135. [DOI] [PubMed] [Google Scholar]
- 145.Paunescu D., Puddu M., Soellner J.O., Stoessel P.R., Grass R.N. Reversible DNA encapsulation in silica to produce ROS-resistant and heat-resistant synthetic DNA 'fossils'. Nat Protoc. 2013;8:2440–2448. doi: 10.1038/nprot.2013.154. [DOI] [PubMed] [Google Scholar]
- 146.Gooch J., Daniel B., Abbate V., Frascione N. Taggant materials in forensic science: a review. Trac Trends Anal Chem. 2016;83:49–54. [Google Scholar]
- 147.Zhang Y., Wang F., Chao J., Xie M., Liu H., Pan M., et al. DNA origami cryptography for secure communication. Nat Commun. 2019;10:5469. doi: 10.1038/s41467-019-13517-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fan S., Wang D., Cheng J., Liu Y., Luo T., Cui D., et al. Information coding in a reconfigurable DNA origami domino array. Angew Chem Int Ed Engl. 2020;59:12991–12997. doi: 10.1002/anie.202003823. [DOI] [PubMed] [Google Scholar]
- 149.Fan S., Cheng J., Liu Y., Wang D., Luo T., Dai B., et al. Proximity-induced pattern operations in reconfigurable DNA origami domino array. J Am Chem Soc. 2020;142:14566–14573. doi: 10.1021/jacs.0c06061. [DOI] [PubMed] [Google Scholar]