Abstract
The first rate-limiting enzyme of the serine synthesis pathway (SSP), phosphoglycerate dehydrogenase (PHGDH), is hyperactive in multiple tumors, which leads to the activation of SSP and promotes tumorigenesis. However, only a few inhibitors of PHGDH have been discovered to date, especially the covalent inhibitors of PHGDH. Here, we identified withangulatin A (WA), a natural small molecule, as a novel covalent inhibitor of PHGDH. Affinity-based protein profiling identified that WA could directly bind to PHGDH and inactivate the enzyme activity of PHGDH. Biolayer interferometry and LC–MS/MS analysis further demonstrated the selective covalent binding of WA to the cysteine 295 residue (Cys295) of PHGDH. With the covalent modification of Cys295, WA blocked the substrate-binding domain (SBD) of PHGDH and exerted an allosteric effect to induce PHGDH inactivation. Further studies revealed that with the inhibition of PHGDH mediated by WA, the glutathione synthesis was decreased and intracellular levels of reactive oxygen species (ROS) were elevated, leading to the inhibition of tumor proliferation. This study indicates WA as a novel PHGDH covalent inhibitor, which identifies Cys295 as a novel allosteric regulatory site of PHGDH and holds great potential in developing anti-tumor agents for targeting PHGDH.
KEY WORDS: Serine synthesis pathway, Phosphoglycerate dehydrogenase, Colon cancer, Withanolides, Withangulatin A, Chemical proteomics, Covalent inhibitor, Oxidative stress
Abbreviations: 3-PG, 3-phosphoglycerate; 3-PHP, 3-phosphohydroxypyruvate; ABPP, affinity-based protein profiling; BLI, biolayer interferometry assay; CETSA, cellular thermal shift assay; CuAAC, copper-catalyzed alkyne–azide cycloaddition; DARTS, drug affinity responsive target stability; GSH, glutathione; MD, molecular dynamics; NADPH, nicotinamide adenine dinucleotide phosphate; PHGDH, phosphoglycerate dehydrogenase; PSAT, phosphoserine aminotransferase; RMSD, root mean square deviation; RMSF, root mean square fluctuations; ROS, reactive oxygen species; SBD, substrate-binding domain; SSP, serine synthesis pathway; TBTA, tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine; TCEP, tris(2-carboxyethyl) phosphine
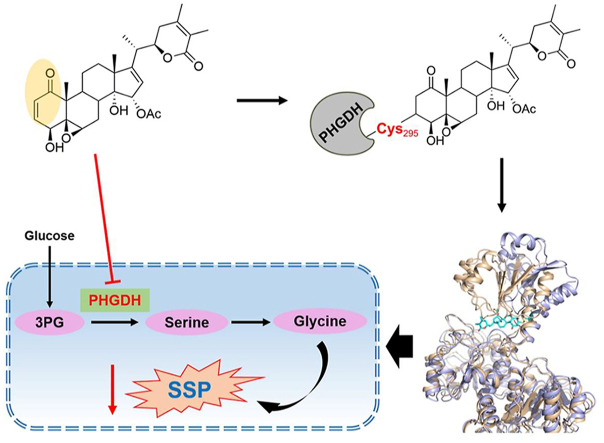
Graphical abstract
WA is a novel covalent-allosteric inhibitor of phosphoglycerate dehydrogenase (PHGDH), and inhibits the serine synthesis pathway and proliferation of colon cancer cells with overexpression of PHGDH.
1. Introduction
Tumor cells reprogram their metabolism to satisfy their biosynthetic requirements and support rapid proliferation1, 2, 3. Serine, a major source of one-carbon units, is a crucial precursor for the biosynthesis of amino acids, nucleotides, and lipids, which maintain the biosynthetic needs and redox homeostasis of tumor cells4, 5, 6. Serine can be de novo synthesized from glucose via the serine synthesis pathway (SSP)7. The SSP becomes overactive and makes a substantial contribution to serine availability in tumor cells2. The upregulation of SSP is increasingly found in serine-deprived circumstances, which reveals the significance of serine in the proliferation of tumor cells6, 7, 8. Therefore, targeting the SSP provides a promising strategy for cancer therapy5.
Phosphoglycerate dehydrogenase (PHGDH), as the first rate-limiting enzyme of the SSP, catalyzes the conversion of 3-phosphoglycerate (3-PG) into 3-phosphohydroxypyruvate (3-PHP)9, 10, 11. Subsequently, 3-PHP is converted to serine via phosphoserine aminotransferase (PSAT) and phosphoserine phosphatase (PSPH)4. PHGDH is also overexpressed and contributes to cell proliferation in multiple cancers12, 13, 14. The genetic depletion of PHGDH significantly suppresses the growth and SSP of the tumor cells with elevated expression of PHGDH, while tumor cells with low expression of PHGDH are insensitive to the deficiency of PHGDH4,7. The SSP provides essential precursors for the synthesis of glutathione (GSH) and the production of nicotinamide adenine dinucleotide phosphate (NADPH)6,10. Accordingly, inhibition of PHGDH induced oxidative damage by decreasing the GSH and NADPH synthesis15,16. Meanwhile, the reported selective inhibitors of PHGDH, such as CBR-5884 and NCT-503, effectively downregulate SSP and inhibit the proliferation of tumor cells with high expression of PHGDH, while having no significant inhibition on the tumor cells with low expression of PHGDH17,18. Thus, selective inhibition of PHGDH provides a potential therapeutic strategy for certain cancers with overexpressed PHGDH. Nonetheless, only a few inhibitors of PHGDH have been discovered to date, which is limited to the development of targeting PHGDH therapeutic strategy in cancer.
Notably, most of the PHGDH inhibitors are competitive inhibitors. Only a few synthetic compounds, such as CBR-5884 and disulfiram, have been discovered as covalent inhibitors of PHGDH11,19. Herein, we first report that withangulatin A (WA), a natural small molecule, is a novel covalent inhibitor of PHGDH. In the present study, we found that WA significantly inhibited the proliferation of HCT-116 colon cancer cells which have a high expression of PHGDH20, and suppressed the enzyme activity of PHGDH. Different from other PHGDH competitive inhibitors, we demonstrated that WA selective covalently modified to the Cys295 residue (Cys295) of PHGDH by the α,β-unsaturated ketone moiety and exhibited an allosteric effect, which blocked the substrate-binding domain (SBD) of PHGDH and inactivated the PHGDH. Further studies discovered that WA increased the intracellular reactive oxygen species (ROS) and decreased the generation of GSH, which was dependent on the inhibition of PHGDH. These findings not only reveal that the Cys295 is a novel allosteric site for targeting PHGDH, but also indicate that WA is a novel PHGDH covalent inhibitor, which could be used as a promising lead compound in the development of PHGDH inhibitor. Besides that, WA is also applied as a novel probe to explore the functions of PHGDH and SSP.
2. Materials and methods
2.1. Compounds and cell lines
Withangulatin A (WA) was isolated from Physalis angulata by our laboratory. ZR-75-1 and HCT-116 cells were purchased from the Cell Bank of Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). All cells were negative for Mycoplasma during this investigation. ZR-75-1 and HCT-116 cells were cultured in DMEM (GIBCO) supplemented with 10% FBS (GIBCO) at 37 °C with 5% CO2. Normal colon epithelial cell line NCM460 was obtained from Corues Biotechnology Co., Ltd. (Nanjing, China). NCM460 cells were cultured in PRMI-1640 (GIBCO) supplemented with 10% FBS (GIBCO) at 37 °C with 5% CO2.
2.2. Cell viability and proliferation assay
The CCK-8 assay was applied to test the cell viability. Cells (7 × 103/well) were seeded into a 96-well culture plate (Corning) for 24 h. Then cells were incubated with or without serial dilutions of WA for 24 h. Then the detection reagent was added and incubated for 4 h. The absorbance at 450 nm was measured by a microplate reader (Molecular Devices, CA, USA). For the colony formation assay, cells were seeded into 35 mm plates (Corning) at a density of 1 × 103/well and incubated overnight. Then, the cells were treated with various concentrations of WA for 24 h. The growth medium was refreshed every three days. After 14 days, the cells were fixed and stained with a crystal violet solution (Sigma–Aldrich) for 15 min, and images of colonies were taken manually.
2.3. The PHGDH enzyme assay
The enzyme assay was performed as previously reported21. Briefly, PHGDH activity was measured in 96-well plates at 37 °C by monitoring resorufin fluorescence (Ex 550 nm/Em 580 nm) over time with a FLUOstar Omega (BMG Labtech). PSAT1 was included to prevent product inhibition of PHGDH. Assays were performed in the PHGDH assay buffer (50 mmol/L Tris, pH 8.5, and 1 mmol/L EDTA). Substrates and enzyme concentrations were as follows: 3-PG (Sigma–Aldrich), 240 μmol/L; NAD+ (Sigma–Aldrich), 120 μmol/L; glutamate (Yuanye Bio-Technology), 30 mmol/L; resazurin (Yuanye Bio-Technology), 0.1 mmol/L; PHGDH (Zoonbio Biotechnology), 15 ng/μL; PSAT1 (Zoonbio Biotechnology), 20 ng/μL; diaphorase (Yuanye Bio-Technology), 0.18 μg/μL. For inhibition assay, compounds and enzyme were preincubated for 30 min before initiating the enzyme reaction with substrate mix.
2.4. In situ pull-down experiment and target validation
The general pull-down experiments were based on previously reported procedures22. HCT-116 cells were grown in culture medium until 70%–80% confluence. The medium was removed and the cells were preincubated with or without WA for 2 h, then the cells were treated with WP for 4 h. After incubation, the cells were lysed and the soluble fractions were diluted to 1.5 mg/mL with lysis buffer. Next 100 μmol/L of Biotin-N3 (from 100 mmol/L stock solution in DMSO, Aladdin Biochemical Technology), 1 mmol/L of CuSO4 (from 50 mmol/L stock solution in deionized water, Sigma–Aldrich), 100 μmol/L of tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA, from 100 mmol/L stock solution in deionized water, Sigma–Aldrich) and 1 mmol/L of tris(2-carboxyethyl)phosphine (TCEP, from 100 mmol/L stock solution in deionized water, Sigma–Aldrich) were added to the lysates. The reaction was further incubated for 2 h with gentle mixing at room temperature. Next, tagged proteins were precipitated with prechilled acetone. The precipitated proteins were subsequently collected by centrifugation at 15,000 rpm (ST16R, ThermoFisher Scientific, Waltham, MA, USA) for 10 min at 4 °C. Then the precipitated proteins were redissolved with 200 μL of lysis buffer by sonication. Upon incubation with streptavidin beads (ThermoFisher Scientific) for 2 h at room temperature, the beads were washed with PBS. The enriched proteins were suspended by 1×loading buffer and boiled for 15 min, then separated by SDS-PAGE for target validation and analyzed by LC–MS/MS.
2.5. Drug affinity responsive target stability (DARTS)
DARTS was performed as previously reported23. Briefly, the cells were collected and total protein was isolated by using lysis buffer. The cell lysates were centrifuged at 15,000 rpm (ST16R, ThermoFisher Scientific) for 15 min at 4 °C. The supernatants were 1:10 diluted with 10× TNC buffer (500 mmol/L Tris-HCl, pH 8.0, 500 mmol/L NaCl, 100 mmol/L CaCl2) and treated with different concentrations of WA or DMSO. After incubation for 1 h at room temperature, pronase (5 μg/mL, Roche) was added into the lysates for a further 15 min at 37 °C. Reactions were stopped by adding protease inhibitor cocktail (Selleck) and SDS-PAGE loading buffer followed by immunoblotting.
2.6. Cellular thermal shift assay (CETSA)
CETSA was performed as previously reported23. Briefly, HCT-116 cells were treated with WA (5 μmol/L) or DMSO and incubated for 4 h. Cells were collected and the pelleted washed with PBS, then resuspended to a density of 5 × 106 cells/mL in PBS supplemented with protease inhibitor. 100 μL of each cell suspension was then dispensed into PCR tubes and heated at 44–60 °C by a thermal cycler for 3 min and immediately lysed by freeze–thaw in liquid nitrogen. The cell lysates were clarified by centrifugation at 15,000 rpm (ST16R, ThermoFisher Scientific) for 15 min at 4 °C. Then supernatants were analyzed by immunoblotting.
2.7. Biolayer interferometry assay (BLI)
The binding affinities of compounds for WT or mutant PHGDH were determined by a biolayer interferometry assay using Octet RED96 (ForteBio, CA, USA). Ni-NTA biosensor tips (ForteBio, CA, USA) were used to immobilize the his-labeled proteins after prewetting with kinetic buffer (PBS, 0.05% bovine serum albumin, 0.01% Tween 20). The equilibrated Ni-NTA biosensors were loaded with WT or mutant PHGDH (100 μg/mL). Background binding controls used a duplicate set of sensors that incubated in buffer without proteins. All assays were performed by a standard protocol in 96-well black plates with a total volume of 250 μL/well at 30 °C. All the data were analyzed by Octet data analysis software. The signals were analyzed by a double reference subtraction protocol to deduce nonspecific and background signals and signal drifts caused by biosensor variability. Equilibrium dissociation constant (Kd) values were calculated from the ratio of Koff to Kon.
2.8. U-13C-glucose stable isotope labeling
The U-13C-glucose stable isotope labeling experiments were based on previously reported procedures17. HCT-116 cells (5 × 105 cells) were plated in 6-mm culture dishes and grown until 70%–80% confluence. Cells were then replaced with DMEM medium containing U-13C-glucose (Cambridge Isotope Laboratories) and incubated for 24 h. For U-13C-glucose tracing with drug treatments, cells were preincubated with corresponding compounds for 2 h, followed by medium replacement with U-13C-glucose and corresponding drug treatment for 6 h. Metabolites were then extracted. The cell samples containing 50% aqueous methanol collected by customer was processed by 5 cycles of 1 min ultra-sonication and 1 min interval in ice-water bath and stood for 30 min at −20 °C. After centrifugation at 15,000 rpm (ST16R, ThermoFisher Scientific) for 15 min at 4 °C, all supernatant was evaporated to dryness. The residues were reconstituted in 100 μL of 50% aqueous acetonitrile (1:1, v/v) prior to UHPLC–HRMS/MS analysis.
2.9. Generation of PHGDH KO cell lines by CRISPR/Cas9 system
To obtain single clones of PHGDH KO HCT-116 cells, HCT-116 cells were transfected with the pLentiCRISPR (Addgene, plasmid#49535) plasmid containing each target sgRNA sequence or empty vector as described previously24. After 48 h transfection, cells were selected with puromycin for two weeks, then single clones were picked and validated by immunoblotting analysis. The sgRNA sequences were showed in Supporting Information Table S1.
2.10. Molecular dynamics (MD) simulation
The crystal structure of PHGDH (PDB entry: 2G76) was used to generate initial binding complexes. Then the classic MD simulation was performed using sander. MPI of AmberTools19 simulation program using the AMBER ff14SB and gaff2 force field. The system was first minimized by 5000 steps of steepest and 5000 steps of conjugate gradient. After minimization, the system was heated to the target temperature of 300 K for a period of 20 ps with a time step of 1.0 fs in constant pressure periodic boundary conditions (NVT). The system was equilibrated by 50 ns under constant pressure of 1 bar and temperature of 300 K with time step of 1 fs, which was followed by 50 ns of production simulation performed in the same conditions. All the simulations used the Berenden barostat and Langevin thermostat with a time constant of 2 ps. A cutoff of 10 Å was used for non-bonded interactions and long-range electrostatic interactions were treated by means of the Particle Mesh Ewald (PME) method. Two independent simulations were carried out. The MD simulation results were analyzed using the pytraj and PyMol.
2.11. Measurement of ROS
The intracellular ROS levels were measured as previously described25,26. Briefly, the HCT-116 cells were incubated with serial dilutions of WA for 24 h. Then incubated in the dark with 10 mmol/L oxidation-sensitive fluorescent probe DCFH-DA (Beyotime Biotechnology) for 20 min at 37 °C. The fluorescence intensity was then measured with a C6 flow cytometer (BD Biosciences, NJ, USA) or an imageXpress confocal microscope (Molecular Devices, CA, USA).
2.12. Measurement of GSH levels
Intracellular GSH concentration was measured using a GSSG/GSH assay kit (Beyotime Biotechnology). The detailed measures were carried out with reference of the kit instructions.
2.13. Measurement of NADPH levels
Intracellular NADPH levels were measured using a NADP+/NADPH assay kit (Beyotime Biotechnology). The detailed measures were carried out with reference of the kit instructions.
2.14. Xenograft tumor model
Five-week-old male BALB/c nude mice were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China). For the tumor xenograft assay, 3 × 106 HCT-116 cells were suspended in 200 μL PBS and subcutaneously injected into the right flank of the mice. When the tumors reached approximately 100 mm3, the tumor-bearing mice were randomly divided into four groups. Then, WA (10 or 20 mg/kg) and NCT-503 (40 mg/kg) were intraperitoneally administered every three days. After four weeks, all of the mice were euthanized. Then, the tumors and visceral organs of each group were collected and fixed in 4% paraformaldehyde. All animal experimental procedures followed the National Institutes of Health guide for the care and use of laboratory animals and were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of China Pharmaceutical University Experimental Animal Center.
2.15. Induction and treatment of colitis-associated colon cancer
To induce colitis-associated colon cancer, mice were injected intraperitoneally with a single dose (10 mg/kg) of AOM. One week later, mice were provided with the 2.5% DSS (MP Biomedicals) in drinking water for 7 days followed by 14 days of normal water, and repeated another two cycles. WA was administered through injected intraperitoneally every 3 days at the indicated dose during the interval between DSS cycles. Mice were sacrificed after 17 weeks induction. The colon tissues and visceral organs of each group were collected and fixed in 4% paraformaldehyde. All animal experimental procedures followed the National Institutes of Health guide for the care and use of laboratory animals and were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of China Pharmaceutical University Experimental Animal Center.
2.16. Statistical analysis
Statistical significance was determined using GraphPad Prism Software 8. Data are represented as mean ± standard deviation (SD). Student's t test and one-way ANOVA test were used for statistical analyses of the data. P values <0.05, <0.01 or <0.001 were considered statistically significant.
3. Results
3.1. Syntheses and biological evaluations of WA-based activity probe
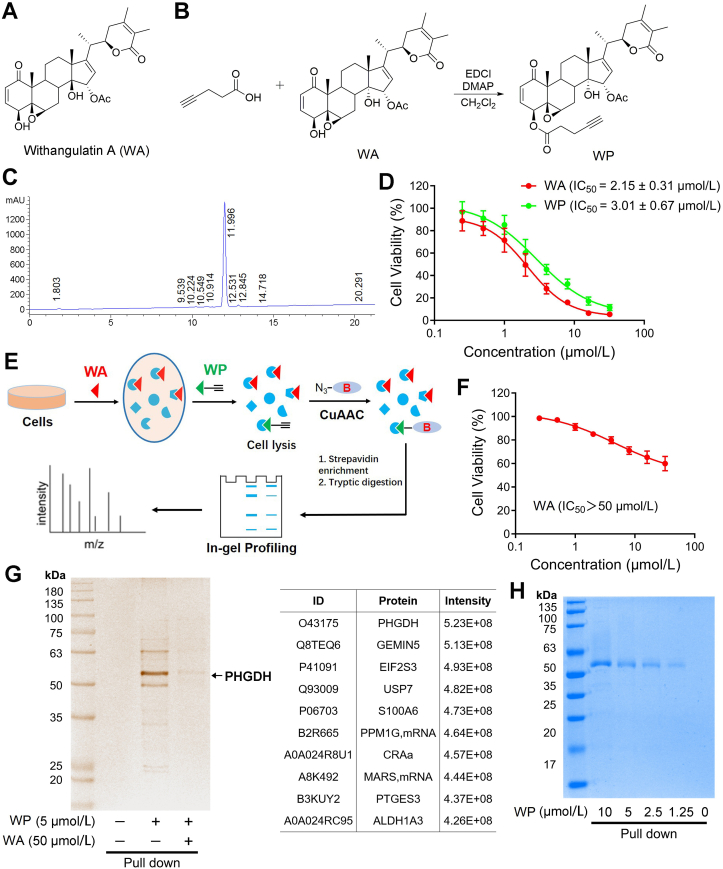
To explore and identify the cellular targets of WA, an alkyne-tagged WA-based probe WP was designed and synthesized (Fig. 1A–C and Supporting Information Scheme S1). The cytotoxic experiment showed that WA significantly suppressed the proliferation of HCT-116 colon cells (Fig. 1D and Supporting Information Fig. S1A). WP had similar cytotoxicity in comparison with WA (Fig. 1D), suggesting that the addition of alkyne tag had no significant effect on the cytotoxic activity. The alkyne tag can be further appended with an azide-biotin tag through click chemistry, allowing the cellular targets of WA to be affinity purified for mass spectrum identification (Fig. 1E)27. Immunoblotting results showed that PHGDH was highly expressed in colon cancer cells, while lower in normal colonic epithelial NCM460 cells (Fig. S1B). Moreover, WA had lower cytotoxicity in the NCM460 cells (Fig. 1F), suggesting that WA showed selective cytotoxicity in colon cancer cells.
Figure 1.
The chemical proteomics approach for identifying the targets of withangulatin A. (A) Chemical structures of withangulatin A (WA). (B) Synthesis of withangulatin A-alkyne probe (WP). (C) HPLC of WP. (D) The cell viability of HCT-116 cells treated by WA and WP were measured by CCK-8 assay. Data are presented as the mean ± SD, n = 6. (E) Schematic workflow of chemical proteomic platform to profile protein targets labelled by the probe WP in situ. (F) The cell viability of NCM460 cells treated by WA was measured by CCK-8 assay. Data are presented as the mean ± SD, n = 6. (G) Proteome reactivity profiles of HCT-116 cells treated with WP (5 μmol/L) in the presence or absence of WA (50 μmol/L). Protein affinity pull-down assay was performed, then the whole cell lysate (WCL) was pull-down with streptavidin beads and the precipitated proteins were separated by SDS-PAGE and processed by silver staining and LC–MS/MS. (H) Labeling of recombinant PHGDH with different concentrations of WP. Protein affinity pull-down assay was performed and the precipitated proteins were processed by Coomassie blue staining.
3.2. PHGDH is identified as a direct target of WA
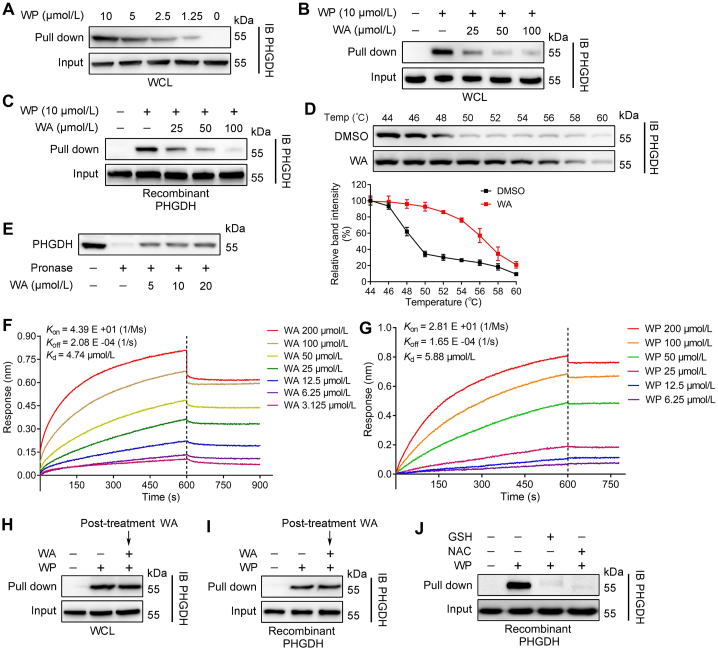
Affinity-based protein profiling (ABPP), as a chemical proteomics strategy, is usually employed to identify the direct intracellular molecular targets28, 29, 30. Thus, we applied WP for identifying the cellular targets of WA through ABPP experiments in situ. After WP treatment, the HCT-116 cells were lysed, and the WP-labeled proteins were conjugated with biotin-azide through copper-catalyzed alkyne–azide cycloaddition (CuAAC)31, 32, 33. The WP-labeled proteins were affinity purified and further identified by LC–MS/MS analysis (Fig. 1E). The click reaction in vitro also confirmed that WP could be appended with an azide-biotin tag through CuAAC (Supporting Information Scheme S2). The silver stain analysis showed that a visible band between 55 and 60 kDa was precipitated by WP, and the protein band was remarkably competed by excessive WA (Fig. 1G). LC–MS/MS analysis indicated that the protein intensity of PHGDH was the highest (56.5 kDa, Fig. 1G and Supporting Information Table S2). Therefore, we speculated that PHGDH was the major target protein of WA. Furthermore, the results of the immunoblotting assay revealed that PHGDH was pulled down by WP both in HCT-116 cells and recombinant PHGDH protein in a dose-dependent manner (Figure 1, Figure 2A). Notably, the binding of WP to PHGDH was blocked by preincubating with WA both in HCT-116 cells and recombinant PHGDH protein (Fig. 2B and C). Together, these data indicate that PHGDH is a direct target of WA in HCT-116 cells.
Figure 2.
WA directly binds to PHGDH. (A) Protein affinity pull-down assay was performed in HCT-116 cells by different concentrations of WP, and immunoblotting by PHGDH antibody. (B) Immunoblotting analysis of HCT-116 cells or (C) the recombinant PHGDH protein treated with WP in the absence or presence of WA for the competitive binding, and followed by protein affinity pull-down assay. The PHGDH bound to the WP were detected by immunoblotting. (D) CETSA assay was used to evaluate the binding between WA (5 μmol/L) and PHGDH in thermodynamic levels. The expression of PHGDH was detected by immunoblotting. Data are presented as the mean ± SD, n = 3. (E) HCT-116 cells lysates were treated with different concentrations of WA, and then incubated with pronase (5 μg/mL). The expression of PHGDH was detected by immunoblotting. (F) Interaction between WA and recombinant PHGDH protein was analysed by BLI assay. (G) Interaction between WP and recombinant PHGDH protein was analyzed by BLI assay. (H) HCT-116 cells were preincubated with WP (5 μmol/L) for 4 h and then further incubated with or without WA (50 μmol/L) for 2 h, then protein affinity pull-down assay was performed. (I) The recombinant PHGDH protein were preincubated with WP (5 μmol/L) for 2 h and then further incubated with or without WA (50 μmol/L) for 2 h, then protein affinity pull-down assay was performed. (J) WP was preincubated with glutathione (GSH, 1 mmol/L) and N-acetyl-cysteine (NAC, 1 mmol/L) for 2 h, then further incubated with recombinant PHGDH protein for 2 h, then protein affinity pull-down assay was performed.
To further validate the interaction of WA with PHGDH, we performed the DARTS and CETSA in HCT-116 cells34. The thermal stability of PHGDH was increased after the treatment of WA in CETSA, suggesting the interaction of WA with PHGDH (Fig. 2D). Meanwhile, WA also significantly restrained the proteolysis of PHGDH by pronase, suggesting WA could bind to PHGDH (Fig. 2E). More importantly, BLI assay further confirmed that there was a strong direct interaction between WA and PHGDH, and the Kd value of WA binding to PHGDH was 4.74 μmol/L (Fig. 2F). Furthermore, there was similar direct interaction between WP and PHGDH compared with WA (Fig. 2G). Collectively, these results indicate that WA could directly bind to PHGDH.
3.3. WA covalently binds to PHGDH
We further investigated whether WA could irreversibly bind to PHGDH. Interestingly, posttreatment with excessive WA could not prevent PHGDH binding to WP both in HCT-116 cells and recombinant PHGDH protein (Fig. 2H and I), revealing the existence of an irreversible bond between WA and PHGDH. Due to the α,β-unsaturated ketone moiety is considered as a Michael acceptor that could covalently bind to the thiol of cysteine in protein35, which are essential for the biological activity of multiple compounds36. We speculated whether WA is bound to PHGDH by the α,β-unsaturated ketone moiety. As shown in Fig. 2J, the binding of WP to PHGDH was reversed by preincubating with N-acetyl-cysteine (NAC) or glutathione (GSH) which having a sulphydryl, suggesting that WA might covalently bind to the thiol of cysteine in PHGDH protein.
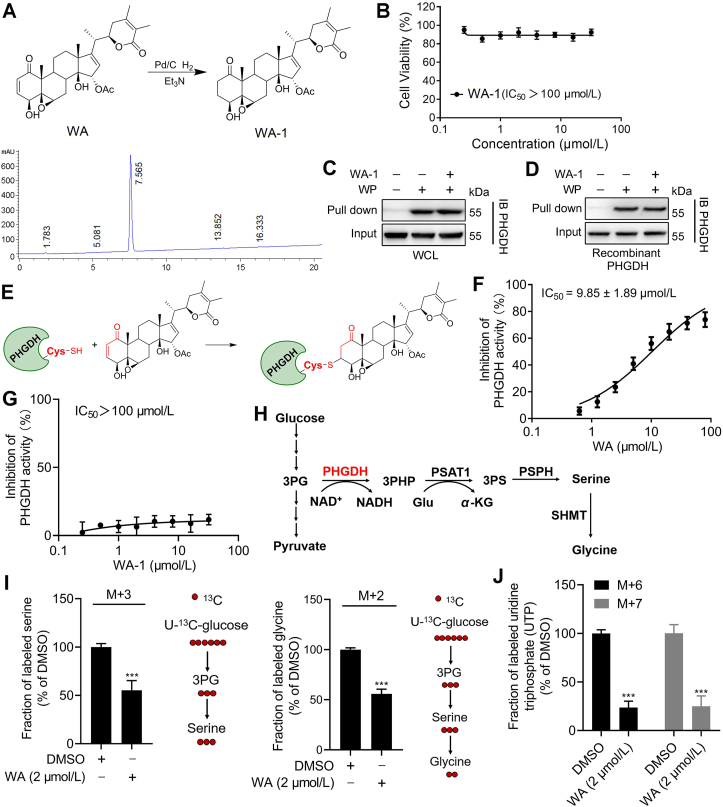
To further certify that WA could covalently bind to PHGDH by the α,β-unsaturated ketone moiety, we synthesized WA-1 by the reduction reaction of WA to destroy the α,β-unsaturated ketone moiety (Fig. 3A and Supporting Information Scheme S3). Strikingly, WA-1 had no obvious influence on the proliferation of HCT-116 cells (Fig. 3B), suggesting that the cytotoxicity of WA depended on the α,β-unsaturated ketone moiety. It was noteworthy that preincubation of WA-1 could not prevent PHGDH binding to WP both in HCT-116 cells and recombinant PHGDH protein (Fig. 3C and D), further indicating that WA covalently bound to PHGDH by the α,β-unsaturated ketone moiety (Fig. 3E). The further LC–MS/MS confirmed that WA could form a complex with GSH by Michael addition reaction in vitro (Supporting Information Fig. S2). Taken together, these data suggest that WA covalently binds to PHGDH by the α,β-unsaturated ketone moiety.
Figure 3.
WA covalently binds to PHGDH and inhibits the activity of PHGDH. (A) Synthesis of WA analog WA-1 (with the reduced α,β-unsaturated ketone moiety) and HPLC of WA-1. (B) The cell viability of HCT-116 cells treated by WA-1 was measured by CCK-8 assay. Data are presented as the mean ± SD, n = 6. (C) HCT-116 cells were preincubated with or without WA-1 (50 μmol/L) for 4 h and then further incubated with WP (5 μmol/L) for 2 h, then protein affinity pull-down assay was performed. (D) The recombinant PHGDH protein were preincubated with or without WA-1 (50 μmol/L) for 2 h and then further incubated with WP (5 μmol/L) for 2 h, then protein affinity pull-down assay was performed. (E) Proposed mechanism of interaction between WA and PHGDH. (F) Dose–response inhibition curve of PHGDH enzyme activity by WA and (G) WA-1 were measured by PHGDH enzyme assay. Data are presented as the mean ± SD, n = 6. (H) Serine synthesis from glucose via the serine synthetic pathway. (I) The 13C-serine, 13C-glycine and (J) UTP labeling from U-13C-glucose in HCT-116 cells after 6 h treatment with WA were measured by UHPLC–HRMS/MS analysis. Data are presented as the mean ± SD, n = 6, ∗∗∗P < 0.001 versus DMSO group.
3.4. WA inhibits the activity of PHGDH and SSP
With the covalent binding of WA to PHGDH, we examined the effect of WA on the function of PHGDH via an enzyme activity assay21. As shown in Fig. 3F and Supporting Information Fig. S3, the enzyme activity of PHGDH was significantly inhibited by WA and WP. In contrast, WA-1 had a slight inhibitory effect on the enzyme activity of PHGDH (Fig. 3G), further revealing that WA might inactivate PHGDH protein by the covalent binding with the α,β-unsaturated ketone moiety. Since the inhibition or deficiency of PHGDH remarkably inhibits the SSP of tumor cells7,8,37. Therefore, we investigated the effects of WA on the SSP in HCT-116 cells. As shown in Fig. 3H and I, WA remarkably decreased the production of M+3-serine and its product M+2-glycine from 13C-glucose, indicating that the SSP of HCT-116 cells was inhibited by WA. Besides, the M+6 and M+7 glucose labeling of UTP were significantly reduced after treatment with WA (Fig. 3J), suggesting a decrease of nucleotide synthesis upon the inhibition of PHGDH. These results demonstrate that WA could inhibit the SSP of HCT-116 cells.
3.5. Cysteine 295 residue of PHGDH is covalently modified by WA
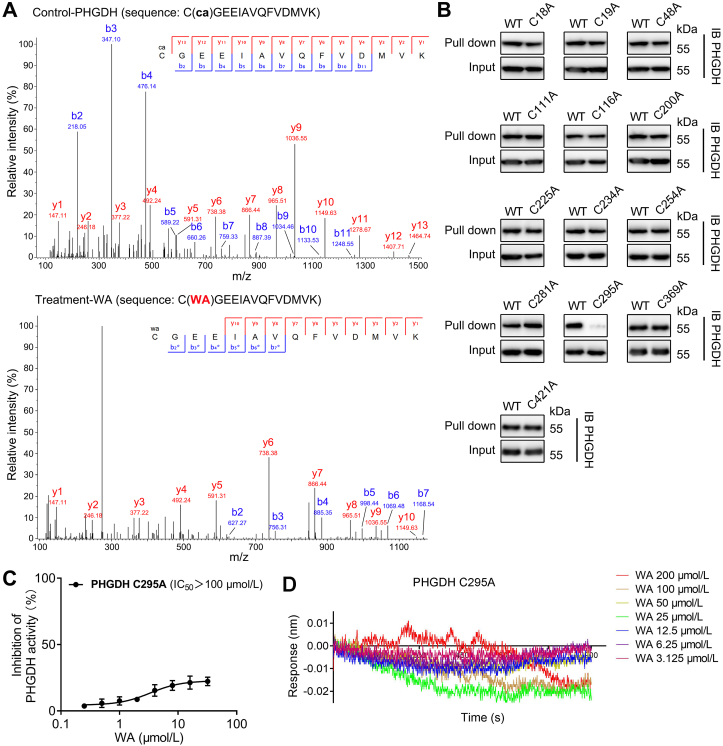
Since the α,β-unsaturated ketone moiety of WA is most likely a reactive Michael acceptor, which might form a covalent bond with cysteine residues of proteins. Therefore, we speculated that the cysteine residues in PHGDH might be the binding sites of WA. We scanned the full-length human PHGDH protein sequence and identified thirteen cysteine residues (Supporting Information Fig. S4). To confirm which of these residues was modified by WA, we incubated recombinant PHGDH protein with or without WA followed by LC–MS/MS analysis. The MS analysis showed that the Cys295-containing peptide C295GEEIAVQFVDMVK had a 526.26 Da mass shift in the presence of WA, which exactly matched the molecular weight of one WA molecule (Supporting Information Fig. S5). Further MS/MS analysis showed that no ion information was available to identify the amino acid residue bound to WA (Supporting Information Fig. S6). Therefore, we speculated whether WA would break into fragments during the LC–MS/MS analysis. As expected, the MS/MS spectra of WA show a sodiated ion at m/z 489.2255, which might be obtained via neutral losses of CH3COOH (Supporting Information Fig. S7A). Thus, we performed the protein MS/MS database search by using the mass of 466.24 Da. The further MS/MS analysis displayed that the Cys295-containing peptide C295GEEIAVQFVDMVK (from b2 to b7 fragment ions) had a 466.24 Da mass shift in the peptide spectra of the WA treatment group (Fig. 4A and Supporting Information Table S3), which was consistent with the addition of one molecular unit of WA-residue (466.24 Da). These data indicate that the Cys295 residue of PHGDH is covalently modified by WA.
Figure 4.
WA selective covalently binds to the Cys295 residue of PHGDH. (A) The recombinant PHGDH incubated without (up) or with (down) WA, then the peptide fragments were identified by MS/MS. ca, carbamidomethylation, 57.02 Da. (B) The recombinant PHGDH WT and PHGDH mutant proteins were incubated with WP, and followed by protein affinity pull-down assay. The PHGDH bound to the WP were detected by immunoblotting. (C) Dose–response inhibition curve of PHGDH enzyme activity by WA with the recombinant Cys295A PHGDH protein. Data are presented as the mean ± SD, n = 6. (D) BLI analysis of interaction between WA and recombinant Cys295A PHGDH protein.
To further prove the binding of WA to the Cys295 residue of PHGDH, we mutated each cysteine residue of PHGDH into alanine. Protein affinity pull-down assay further demonstrated that only Cys295 mutation abolished the capacity of PHGDH binding to WP (Fig. 4B), further supporting that the Cys295 residue of PHGDH was covalently modified by WA. We further investigated whether the Cys295 mutation affected the inhibition of WA on the activity of PHGDH. As shown in Fig. 4C, the Cys295 mutation observably decreased the inhibition of WA on PHGDH activity. Significantly, the BLI assay showed that WA had no interaction with the recombinant Cys295A PHGDH proteins (Fig. 4D). Taken together, these results demonstrate that WA selective covalently binds to the Cys295 residue of PHGDH.
3.6. WA exerts an allosteric regulation on PHGDH
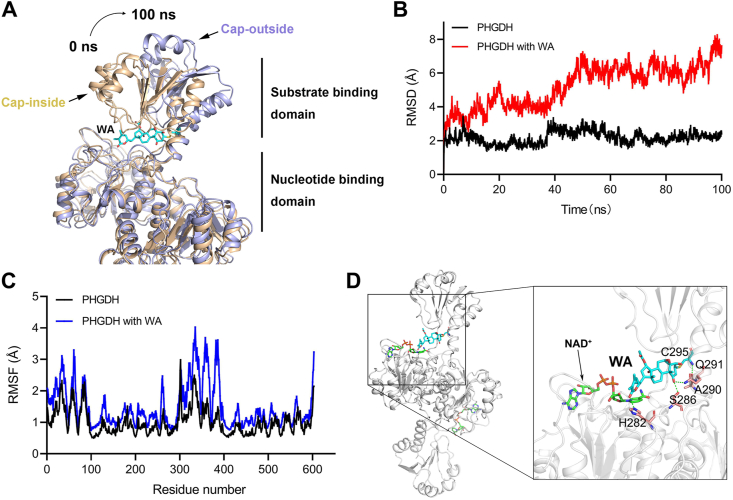
The Cys295 residue is located in the SBD of PHGDH, which is identified as an allosteric binding pocket and connected with the nucleotide-binding domain (NBD) via short flexible hinges24. Therefore, we performed a 100 ns molecular dynamics (MD) simulation to investigate whether WA exerted an allosteric regulation on PHGDH. Based on the 100 ns MD simulation analysis, WA occupied the SBD of PHGDH, which might block the capacity of the substrate-binding (Fig. 5A). Meanwhile, WA facilitated the curving of the SBD of PHGDH outwards, which resulted in the allosteric effect of PHGDH (Fig. 5A). The B-factor of the SBD was elevated after WA binding to PHGDH in the MD simulation, indicating an allosteric effect of PHGDH when bound to WA (Fig. S7B). As shown in Fig. 5B, in comparison with the PHGDH alone, the root mean square deviation (RMSD) of PHGDH was more unstable and increased when bound to WA. The root mean square fluctuations (RMSF) of SBD in PHGDH also revealed more flexibility when bound to WA (Fig. 5C). Moreover, the radius of gyration (radgyr) of PHGDH was also elevated when bound to WA, further revealing an allosteric effect of PHGDH after binding to WA (Fig. S7C). In addition, hydrogen-bonding interactions were also found between WA and the binding pocket in the substrate-binding domain (Fig. 5D), which promoted the interaction between PHGDH and WA. Collectively, these data indicate that WA has an allosteric regulation effect on PHGDH, and the Cys295 residue likely acts as a novel allosteric site of PHGDH.
Figure 5.
Cys295 residue is an allosteric regulatory site of PHGDH. (A) A representative conformation of the WA-mediated allosteric effect of PHGDH protein (PDB: 2G76) after a 100 ns molecular dynamics (MD) simulation. PHGDH protein alone (Cap-inside) is shown by the yellow ribbon model, WA–PHGDH complex (Cap-outside) is shown by the gray ribbon model. (B) RMSD values (Å) of PHGDH alone and WA-PHGDH complex in 100 ns MD simulation. (C) Calculation RMSF (Å) for free PHGDH protein and WA–PHGDH complex. (D) Hydrogen bonds between WA and the binding pocket in the substrate-binding domain were identified by the MD simulation.
3.7. WA induces the oxidative stress in HCT-116 cells
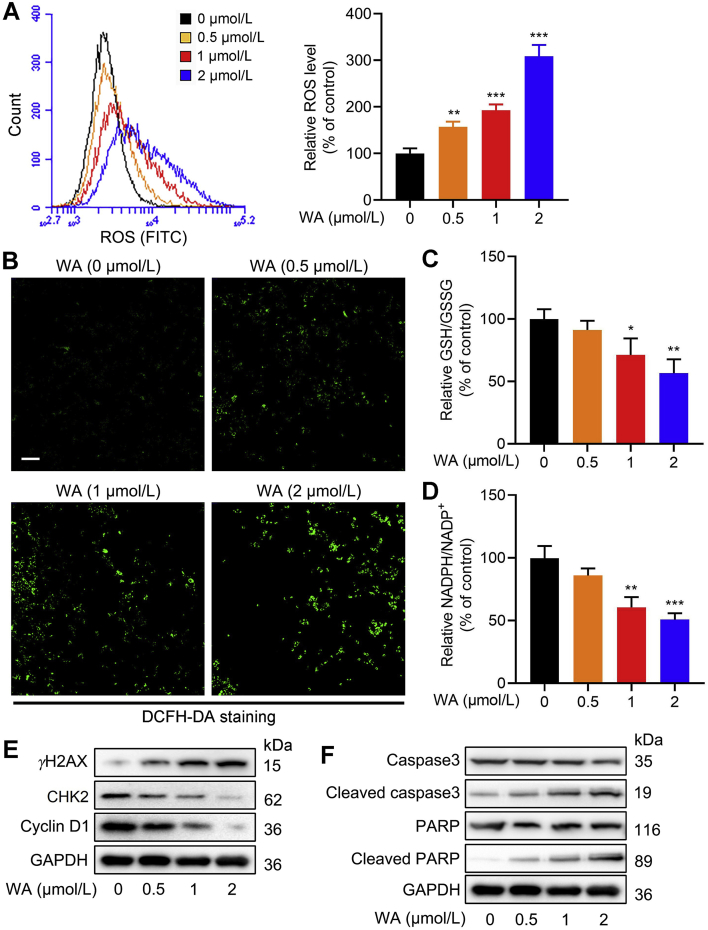
The SSP is pivotal in maintaining the cellular redox homeostasis4,6. Especially, the SSP provides essential precursors for the synthesis of GSH, which is one of the important intracellular antioxidants37. The SSP also contributes to the production of NADPH, a key functional metabolite that provides reducing power for redox reactions6. Therefore, we further evaluated the effect of WA on the redox balance in HCT-116 cells. As shown in Fig. 6A and B, WA increased the ROS levels of HCT-116 cells. Meanwhile, WA significantly decreased the cellular ratio of GSH/GSSG and NADPH/NADP+ (Fig. 6C and D), which represented the increased oxidative stress. Elevated ROS contributes to the apoptosis of tumor cells. Further immunoblotting analysis showed that the expression of γH2AX (Fig. 6E), cleaved caspase3 and cleaved PARP (Fig. 6F) were elevated (Supporting Information Fig. S8A and S8B), while the expression of CHK2 and cyclin D1 were reduced after treatment of WA (Fig. 6E and Fig. S8A). Together, these results reveal that WA increases the production of ROS in HCT-116 cells.
Figure 6.
WA increases intracellular ROS levels of HCT-116 cells. (A) The ROS levels in HCT-116 cells after treatment with various concentrations of WA were measured by flow cytometry. Data are presented as the mean ± SD, n = 3, ∗∗P < 0.01, ∗∗∗P < 0.001 versus control group (0 μmol/L). (B) The ROS levels in HCT-116 cells after treatment with various concentrations of WA were measured by fluorescence microscope (DCFH-DA staining). Scale bar = 50 μm. (C) GSH/GSSG ratio of HCT-116 cells after treatment with various concentrations of WA were measured by a glutathione assay kit. Data are presented as the mean ± SD, n = 3, ∗P < 0.05, ∗∗P < 0.01 versus control group (0 μmol/L). (D) Analysis of NADPH/NADP+ ratio was performed by a NADPH Assay Kit. Data are presented as the mean ± SD, n = 3, ∗∗P < 0.01, ∗∗∗P < 0.001 versus control group (0 μmol/L). (E) The expressions of γH2AX, CHK2 and cyclin D1 in HCT-116 cells were measured by immunoblotting after treatment with WA. (F) The expressions of caspase3, cleaved caspase3, PARP and cleaved PARP in HCT-116 cells were measured by immunoblotting after treatment with WA.
3.8. Deficiency of PHGDH increases the oxidative stress of HCT-116 cells
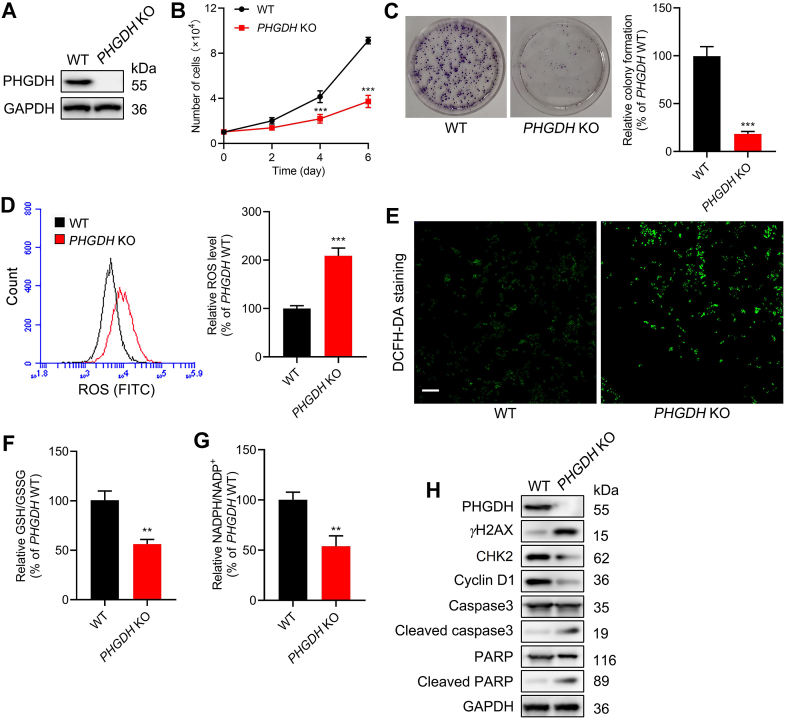
To further investigate the function of PHGDH in HCT-116 cells, the PHGDH gene knockout (KO) HCT-116 cells was generated by CRISPR/Cas9 system. The results of immunoblotting confirmed that PHGDH was effectively knockout in HCT-116 cells (Fig. 7A). The deletion of PHGDH notably inhibited the proliferation of HCT-116 cells (Fig. 7B and C). The ROS levels were also elevated in PHGDH KO HCT-116 cells (Fig. 7D and E), which were consistent with the results in the treatment of WA. The cellular ratio of GSH/GSSG and NADPH/NADP+ were decreased in PHGDH KO HCT-116 cells (Fig. 7F and G). Moreover, the expression of γH2AX, cleaved caspase3 and cleaved PARP were increased, while the expression of CHK2 and cyclin D1 were decreased in PHGDH KO HCT-116 cells (Fig. 7H and Supporting Information Fig. S9A). Collectively, these data indicate that the deficiency of PHGDH could induce the generation of ROS and inhibit the proliferation of HCT-116 cells.
Figure 7.
Effect of PHGDH knockout (KO) on the proliferation and oxidative stress of HCT-116 cells. (A) Immunoblotting analysis of the PHGDH in WT and PHGDH KO HCT-116 cells by CRISPR/Cas9 system. (B) Growth curve of WT and PHGDH KO HCT-116 cells. Data are presented as the mean ± SD, n = 6, ∗∗∗P < 0.001 versus PHGDH WT group. (C) Cell proliferative activity of PHGDH WT and PHGDH KO HCT-116 cells were evaluated by the colony formation assay. Data are presented as the mean ± SD, n = 3, ∗∗∗P < 0.001 versus PHGDH WT group. (D) The ROS levels of PHGDH WT and PHGDH KO HCT-116 cells were measured by flow cytometry. Data are presented as the mean ± SD, n = 3, ∗∗∗P < 0.001 versus PHGDH WT group. (E) The ROS levels of PHGDH WT and PHGDH KO HCT-116 cells were evaluated by fluorescence microscope (DCFH-DA staining). Scale bar = 50 μm. (F) GSH/GSSG ratio of PHGDH WT and PHGDH KO HCT-116 cells were measured by a glutathione assay kit. Data are presented as the mean ± SD, n = 3, ∗∗P < 0.01 versus PHGDH WT group. (G) NADPH/NADP+ ratio of PHGDH WT and PHGDH KO HCT-116 cells were performed by a NADPH assay kit. Data are presented as the mean ± SD, n = 3, ∗∗P < 0.01 versus PHGDH WT group. (H) The expressions of γH2AX, CHK2, cyclin D1, caspase3, cleaved caspase3, PARP and cleaved PARP in PHGDH WT and PHGDH KO HCT-116 cells were measured by immunoblotting.
3.9. WA-mediated generation of ROS and cytotoxicity is dependent on PHGDH
Since HCT-116 colon cells with overexpression of PHGDH (Fig. S1B), we further examined the cytotoxic selectivity of WA on the ZR-75-1 cells which had low expression of PHGDH17. Interestingly, WA also showed much lower cytotoxicity toward ZR-75-1 cells than HCT-116 cells (Fig. S9B). Furthermore, the immunoblotting analysis showed that the normal colonic epithelial NCM460 cells also had lower levels of PHGDH compared with HCT-116 cells (Fig. S1B), which was insensitive to WA in the aforementioned result (Fig. 1D). These data suggest that WA has lower cytotoxicity in the cells with low expression of PHGDH.
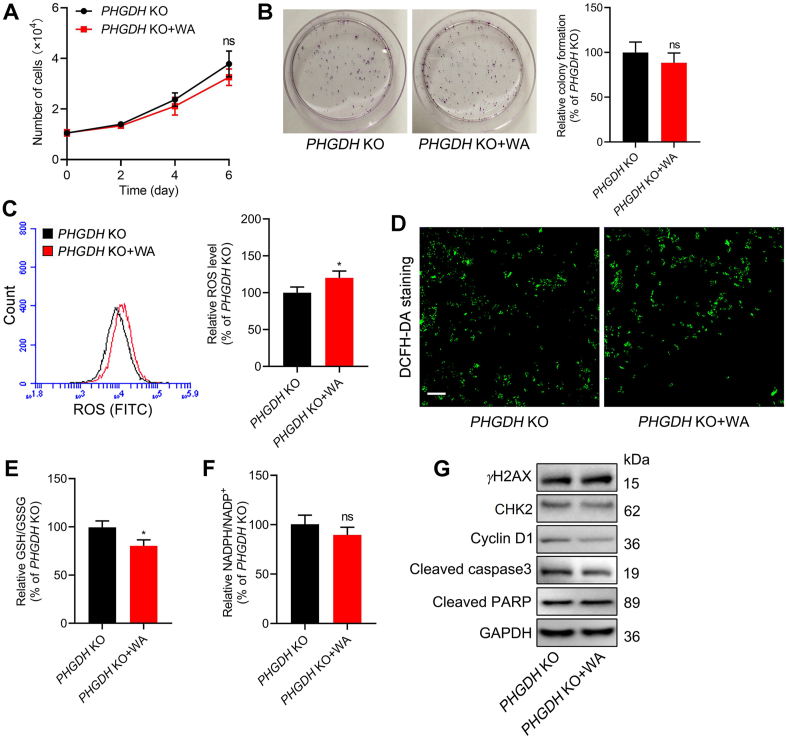
Notably, in comparison with the control group, PHGDH KO HCT-116 cells were insensitive to the treatment of WA in vitro (Fig. 8A and B). WA also had less effect on the ROS levels in PHGDH KO HCT-116 cells compared with that in PHGDH WT HCT-116 cells (Fig. 8C and D). Consistent with these, the cellular ratio of GSH/GSSG and NADPH/NADP+ also decreased much less than that in PHGDH WT HCT-116 cells (Fig. 8E and F). Moreover, WA had no obvious effect on the expression of γH2AX, cleaved caspase3, cleaved PARP, CHK2 and cyclin D1 in PHGDH KO HCT-116 cells (Fig. 8G and Fig. S9C), whereas WA remarkably inhibited the expression of CHK2 and cyclin D1, and elevated the expression of γH2AX, cleaved caspase3 and cleaved PARP in PHGDH WT HCT-116 cells (Fig. 6E and F). Taken together, these data indicate that PHGDH mainly contributes to the production of ROS and cytotoxicity mediated by WA in HCT-116 cells.
Figure 8.
Effect of WA on the proliferation and oxidative stress of PHGDH KO HCT-116 cells. (A) Growth curves of PHGDH KO HCT-116 cells after treatment with or without WA (2 μmol/L). Data are presented as the mean ± SD, n = 6, ns, no significance versus PHGDH KO group. (B) Cell proliferative activity of PHGDH KO HCT-116 cells after treatment with or without WA (2 μmol/L) by the colony formation assay. Data are presented as the mean ± SD, n = 3, ns, no significance versus PHGDH KO group. (C) The ROS levels in PHGDH KO HCT-116 cells after treatment with WA (2 μmol/L) were measured by flow cytometry. Data are presented as the mean ± SD, n = 3, ∗P < 0.05 versus PHGDH KO group. (D) The ROS levels in PHGDH KO HCT-116 cells after treatment with WA (2 μmol/L) were measured by fluorescence microscope (DCFH-DA staining). Scale bar = 50 μm. (E) GSH/GSSG ratio of PHGDH KO HCT-116 cells after treatment with WA (2 μmol/L) were measured by a Glutathione Assay Kit. Data are presented as the mean ± SD, n = 3, ∗P < 0.05 versus PHGDH KO group. (F) NADPH/NADP+ ratio of PHGDH KO HCT-116 cells after treatment with WA (2 μmol/L) were performed by a NADPH assay kit. Data are presented as the mean ± SD, n = 3, ns, no significance versus PHGDH KO group. (G) The expression of γH2AX, CHK2, cyclin D1, cleaved caspase3, and cleaved PARP in PHGDH KO HCT-116 cells were measured by immunoblotting after treatment with WA (2 μmol/L).
3.10. WA inhibits the proliferation of HCT-116 cells and colon tumorigenesis in vivo
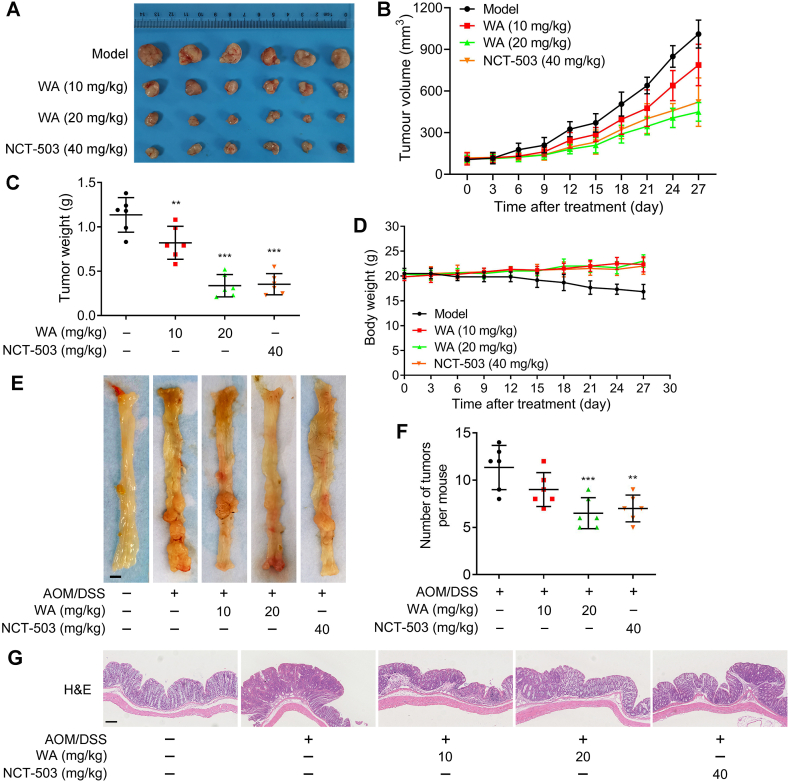
To evaluate the effect of WA on the proliferation of HCT-116 cells in vivo, we established a xenograft model of HCT-116 cells. Compared with the model group, WA considerably suppressed the growth of tumors, and the inhibitory effect of WA (20 mg/kg) was close to NCT-503 (40 mg/kg) which is a selective inhibitor of PHGDH (Fig. 9A–C). Meanwhile, the positive expression of Ki67 was reduced after the treatment of WA (Fig. S9D), suggesting that WA inhibited the proliferation of HCT-116 cells in vivo. As shown in Fig. 9D and Supporting Information Fig. S10, WA did not cause an obvious change in the mice body weight, and no obvious morphological changes were observed in the organs of the mice treated with WA, indicating WA had low toxicity to the mice.
Figure 9.
Effect of WA on the proliferation of colon cancer cells in vivo. (A) Tumor images of HCT-116 cells tumor-bearing mice after treated with WA (10 and 20 mg/kg) or NCT-503 (40 mg/kg). Changes of (B) tumor volume, (C) tumor weight, and (D) body weight are showed after treated with WA (10 and 20 mg/kg) or NCT-503 (40 mg/kg) in xenograft model. Data are presented as the mean ± SD, n = 6, ∗∗P < 0.01, ∗∗∗P < 0.001 versus model group. (E) Macroscopic images of colons from mice after treatment of WA (10 and 20 mg/kg) or NCT-503 (40 mg/kg) in the AOM/DSS-induced colon cancer. Scale bar = 2 mm. (F) The number of tumors in colon were counted. Data are presented as the mean ± SD, n = 6, ∗∗P < 0.01, ∗∗∗P < 0.001 versus AOM/DSS group. (G) Representative histological images of H&E-stained colon tumors in the AOM/DSS-induced colon cancer model. Scale bar = 50 μm.
We next generated an azoxymethane (AOM)-dextran sulfate sodium (DSS) mouse model for colitis-associated colonic tumorigenesis and evaluated the inhibitory effect of WA. The average number of tumors in the WA and NCT-503 groups were both less than that in the model group (Fig. 9E and F) and smaller tumors were observed in the WA and NCT-503-treated group (Supporting Information Fig. S11A). In the group with WA treatment, the mice showed weight loss after each DSS exposure but recovered more quickly than mice in the AOM/DSS group, suggesting a protective effect of WA on the mice (Supporting Information Fig. S11B). The results of hematoxylin and eosin (H&E) staining showed that there was a lower high-grade tumor proportion in the WA or NCT-503 treatment group compared with the AOM/DSS model group, indicating the incidence and proliferation of orthotopic colon tumors were also inhibited after treatment of WA in vivo (Fig. 9G).
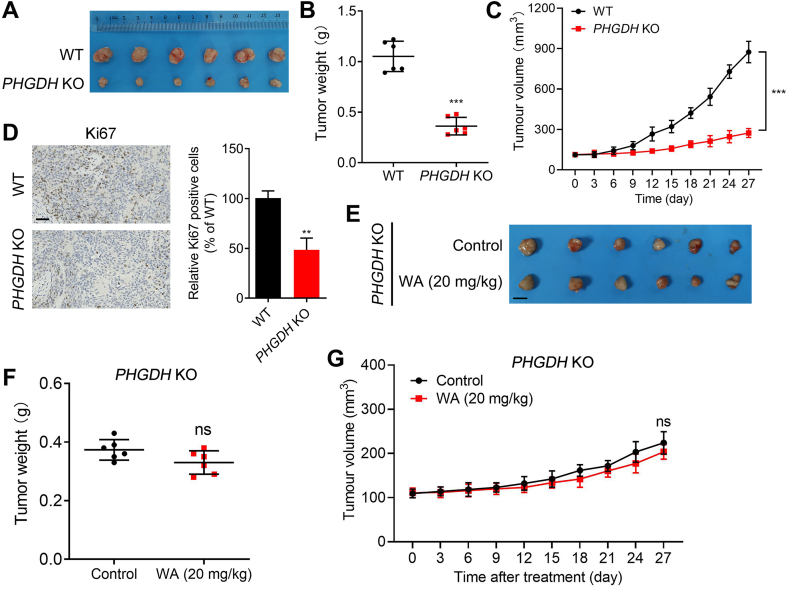
To further investigate the selectivity of WA on the inhibition of PHGDH, we established a xenograft model of PHGDH KO HCT-116 cells. As shown in Fig. 10A–C, the deficiency of PHGDH significantly inhibited the proliferation of HCT-116 cells in the xenograft model. Moreover, the positive expression of Ki67 was significantly reduced in the xenograft model of PHGDH KO HCT-116 cells (Fig. 10D). Notably, WA had no significant inhibitory effect on the cell proliferation in the xenograft model of PHGDH KO HCT-116 cells (Fig. 10E–G), further indicating the inhibition of WA on HCT-116 cells was dependent on PHGDH.
Figure 10.
Effect of WA on the proliferation of PHGDH KO HCT-116 cells in vivo. (A) Tumor images of PHGDH WT and PHGDH KO HCT-116 cells in xenograft model. (B) Tumor weight of PHGDH WT and PHGDH KO HCT-116 cells in xenograft model. Data are presented as the mean ± SD, n = 6, ∗∗∗P < 0.001 versus PHGDH WT group. (C) Changes of tumor volume were showed in xenograft model of PHGDH WT and PHGDH KO HCT-116 cells. Data are presented as the mean ± SD, n = 6, ∗∗∗P < 0.001 versus PHGDH WT group. (D) Histopathology of xenograft tumors stained with Ki67. Data are presented as the mean ± SD, n = 6, ∗∗P < 0.01 versus PHGDH WT group. Scale bar = 50 μm. (E) Tumor images, (F) tumor weight and (G) tumor volume of PHGDH KO HCT-116 cells tumor-bearing mice after treatment with WA (20 mg/kg). ns, no significance versus PHGDH KO group. Scale bar = 5 mm.
4. Discussion
The SSP is required for a number of biosynthetic and metabolic processes, which is crucial to cancer cell growth38. The hyperactivation of SSP has been reported in a variety of tumors, which is pivotal to sustain the proliferation of tumor cells7. PHGDH as the first rate-limiting enzyme in the SSP3, is also overexpressed in multiple tumors10. The specific inhibitors of PHGDH suppressed SSP in tumor cells and were selectively toxic to tumors with highly activated SSP24. In consequence, PHGDH is identified as a fascinating metabolic target in cancer that has hyperactive SSP.
Here, we first report that WA, a natural small molecule, is a novel covalent inhibitor of PHGDH. Currently, there have been a few reports of small molecule PHGDH covalent inhibitors. CBR-5884 was reported as a covalent inhibitor of PHGDH, whereas there was no direct evidence to support the covalent binding of PHGDH and CBR-588411. Although disulfiram was reported as a covalent inhibitor of PHGDH19, the PHGDH may not be the specific target of disulfiram due to the extensive activities and targets of disulfiram. In this study, through the chemical proteomics, we discovered that WA could directly bind to PHGDH and inhibit the activity of PHGDH.
In the previous study, we identified SERCA2 as a potential target of WA in MDA-MB-231 cells39. However, the WA-conjugated sepharose beads were used as an affinity reagent in the previous study, which further incubated with cell lysis rather than live cells and might easily produce nonspecific binding. In the present study, we applied an alkyne-tagged WA-based probe to identify the cellular targets of WA in live cells. Meanwhile, PHGDH expression was lower in MDA-MB-231 cells and higher in HCT-116 cells20. Based on these differences, we conclude that PHGDH is a novel target of WA in HCT-116 cells.
The specific PHGDH inhibitors, such as NCT-503, were selectively toxic toward cell lines with overexpression of PHGDH17. WA also had more efficient cytotoxicity toward cell lines with overexpression of PHGDH, indicating that PHGDH is a selective target of WA. Due to the dependency of SSP on PHGDH, the inhibition of PHGDH by WA also effectively restrained the SSP in HCT-116 cells. GSH, as an important intracellular antioxidant, is a major downstream product of the SSP6,10. The inhibition of PHGDH could significantly elevate the intracellular ROS levels and suppress the intracellular GSH levels15,16,40,41. Our findings reveal that WA dramatically elevated the intracellular ROS levels and reduced the intracellular ratio of GSH/GSSG and NADPH/NADP+. Meanwhile, deficiency of PHGDH in HCT-116 cells also induced the generation of ROS and decreased the ratio of GSH/GSSG and NADPH/NADP+. However, WA had no obvious effect on the intracellular ROS and the ratio of GSH/GSSG in PHGDH KO HCT-116 cells, indicating that WA-mediated production of ROS was dependent on PHGDH.
Our study further substantiated that WA is covalently bound to PHGDH via the α,β-unsaturated ketone moiety. The α,β-unsaturated ketone moiety is considered as a Michael acceptor that covalently binds to thiol of cysteine36. Previous studies had identified the Cys234 and Cys116 as the active and allosteric sites of PHGDH17,19. Unlike the previous study, we demonstrated that WA was covalently bound to the Cys295 residue of PHGDH and inactivated the activity of PHGDH by LC–MS/MS analysis and point mutation. Since the Cys295 residue is located in the SBD of PHGDH, WA maybe occupy the SBD of PHGDH and blocked the capacity of the substrate binding. Further MD simulation revealed that WA exerted an allosteric regulation effect on PHGDH upon binding to the Cys295 residue, suggesting that Cys295 is a novel allosteric site of PHGDH. Therefore, the identification of Cys295 by WA is valuable for the development of novel allosteric inhibitor of PHGDH.
5. Conclusions
Collectively, our findings reveal that WA is a novel PHGDH covalent inhibitor, which could serve as a promising lead compound in the development of new therapeutic agents for PHGDH-dependent cancers and also provides a novel chemical tool to investigate the functions of PHGDH and SSP.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81872983 and 81903861), the Natural Science Foundation of Jiangsu Province (BK20181329, China), and the Program for Changjiang Scholars and Innovative Research Team in University (IRT_15R63, China).
Author contributions
Lingyi Kong and Jianguang Luo initiated and supervised the research. Chen Chen designed and performed the experiments, interpreted the data, and drafted the manuscript. Tianyu Zhu designed the chemical synthesis strategy and interpreted the compound data. Xiaoqin Liu and Dongrong Zhu performed partial cellular assays. Yi Zhang and Sifang Wu extracted and purified the research reagents. Chao Han and Hao Zhang were involved in data analysis. All authors read and approved the final manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.06.008.
Contributor Information
Jianguang Luo, Email: luojg@cpu.edu.cn.
Lingyi Kong, Email: cpu_lykong@126.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhu J., Thompson C.B. Metabolic regulation of cell growth and proliferation. Nat Rev Mol Cell Biol. 2019;20:436–450. doi: 10.1038/s41580-019-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Counihan J.L., Grossman E.A., Nomura D.K. Cancer Metabolism: current understanding and therapies. Chem Rev. 2018;118:6893–6923. doi: 10.1021/acs.chemrev.7b00775. [DOI] [PubMed] [Google Scholar]
- 3.Mullen A.R., DeBerardinis R.J. Genetically-defined metabolic reprogramming in cancer. Trends Endocrinol Metab. 2012;23:552–559. doi: 10.1016/j.tem.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amelio I., Cutruzzola F., Antonov A., Agostini M., Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39:191–198. doi: 10.1016/j.tibs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang M., Vousden K.H. Serine and one-carbon metabolism in cancer. Nat Rev Cancer. 2016;16:650–662. doi: 10.1038/nrc.2016.81. [DOI] [PubMed] [Google Scholar]
- 6.Newman A.C., Maddocks O.D.K. Serine and functional metabolites in cancer. Trends Cell Biol. 2017;27:645–657. doi: 10.1016/j.tcb.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Mattaini K.R., Sullivan M.R., Vander Heiden M.G. The importance of serine metabolism in cancer. J Cell Biol. 2016;214:249–257. doi: 10.1083/jcb.201604085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Locasale J.W. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grant G.A. d-3-Phosphoglycerate dehydrogenase. Front Mol Biosci. 2018;5:110. doi: 10.3389/fmolb.2018.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li A.M., Ye J. The PHGDH enigma: do cancer cells only need serine or also a redox modulator? Cancer Lett. 2020;476:97–105. doi: 10.1016/j.canlet.2020.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao X., Fu J., Du J., Xu W. The role of d-3-phosphoglycerate dehydrogenase in cancer. Int J Biol Sci. 2020;16:1495–1506. doi: 10.7150/ijbs.41051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Locasale J.W., Grassian A.R., Melman T., Lyssiotis C.A., Mattaini K.R., Bass A.J., et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Possemato R., Marks K.M., Shaul Y.D., Pacold M.E., Kim D., Birsoy K., et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia X.Q., Zhang S., Zhu H.J., Wang W., Zhu J.H., Wang X.D., et al. Increased expression of PHGDH and prognostic significance in colorectal cancer. Transl Oncol. 2016;9:191–196. doi: 10.1016/j.tranon.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engel A.L., Lorenz N.I., Klann K., Munch C., Depner C., Steinbach J.P., et al. Serine-dependent redox homeostasis regulates glioblastoma cell survival. Br J Cancer. 2020;122:1391–1398. doi: 10.1038/s41416-020-0794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samanta D., Park Y., Andrabi S.A., Shelton L.M., Gilkes D.M., Semenza G.L. PHGDH expression is required for mitochondrial redox homeostasis, breast cancer stem cell maintenance, and lung metastasis. Cancer Res. 2016;76:4430–4442. doi: 10.1158/0008-5472.CAN-16-0530. [DOI] [PubMed] [Google Scholar]
- 17.Pacold M.E., Brimacombe K.R., Chan S.H., Rohde J.M., Lewis C.A., Swier L.J., et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol. 2016;12:452–458. doi: 10.1038/nchembio.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravez S., Spillier Q., Marteau R., Feron O., Frederick R. Challenges and opportunities in the development of serine synthetic pathway inhibitors for cancer therapy. J Med Chem. 2017;60:1227–1237. doi: 10.1021/acs.jmedchem.6b01167. [DOI] [PubMed] [Google Scholar]
- 19.Spillier Q., Vertommen D., Ravez S., Marteau R., Themans Q., Corbet C., et al. Anti-alcohol abuse drug disulfiram inhibits human PHGDH via disruption of its active tetrameric form through a specific cysteine oxidation. Sci Rep. 2019;9:4737. doi: 10.1038/s41598-019-41187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid M.A., Allen A.E., Liu S., Liberti M.V., Liu P., Liu X., et al. Serine synthesis through PHGDH coordinates nucleotide levels by maintaining central carbon metabolism. Nat Commun. 2018;9:5442. doi: 10.1038/s41467-018-07868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravez S., Corbet C., Spillier Q., Dutu A., Robin A.D., Mullarky E., et al. alpha-Ketothioamide derivatives: a promising tool to interrogate phosphoglycerate dehydrogenase (PHGDH) J Med Chem. 2017;60:1591–1597. doi: 10.1021/acs.jmedchem.6b01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu D., Guo H., Chang Y., Ni Y., Li L., Zhang Z.M., et al. Cell- and tissue-based proteome profiling and dual imaging of apoptosis markers with probes derived from venetoclax and idasanutlin. Angew Chem Int Ed Engl. 2018;57:9284–9289. doi: 10.1002/anie.201802003. [DOI] [PubMed] [Google Scholar]
- 23.Liao L.X., Song X.M., Wang L.C., Lv H.N., Chen J.F., Liu D., et al. Highly selective inhibition of IMPDH2 provides the basis of antineuroinflammation therapy. Proc Natl Acad Sci U S A. 2017;114:E5986–E5994. doi: 10.1073/pnas.1706778114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q., Liberti M.V., Liu P., Deng X., Liu Y., Locasale J.W., et al. Rational design of selective allosteric inhibitors of PHGDH and serine synthesis with anti-tumor activity. Cell Chem Biol. 2017;24:55–65. doi: 10.1016/j.chembiol.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma T., Zhang Y., Zhang C., Luo J.G., Kong L.Y. Downregulation of TIGAR sensitizes the antitumor effect of physapubenolide through increasing intracellular ROS levels to trigger apoptosis and autophagosome formation in human breast carcinoma cells. Biochem Pharmacol. 2017;143:90–106. doi: 10.1016/j.bcp.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Qi S., Guo L., Yan S., Lee R.J., Yu S., Chen S. Hypocrellin A-based photodynamic action induces apoptosis in A549 cells through ROS-mediated mitochondrial signaling pathway. Acta Pharm Sin B. 2019;9:279–293. doi: 10.1016/j.apsb.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bottcher T., Pitscheider M., Sieber S.A. Natural products and their biological targets: proteomic and metabolomic labeling strategies. Angew Chem Int Ed Engl. 2010;49:2680–2698. doi: 10.1002/anie.200905352. [DOI] [PubMed] [Google Scholar]
- 28.Ziegler S., Pries V., Hedberg C., Waldmann H. Target identification for small bioactive molecules: finding the needle in the haystack. Angew Chem Int Ed Engl. 2013;52:2744–2792. doi: 10.1002/anie.201208749. [DOI] [PubMed] [Google Scholar]
- 29.Lonsdale R., Ward R.A. Structure-based design of targeted covalent inhibitors. Chem Soc Rev. 2018;47:3816–3830. doi: 10.1039/c7cs00220c. [DOI] [PubMed] [Google Scholar]
- 30.Wright M.H., Sieber S.A. Chemical proteomics approaches for identifying the cellular targets of natural products. Nat Prod Rep. 2016;33:681–708. doi: 10.1039/c6np00001k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Wang Z., Ma X., Yang S., Hu X., Tao J., et al. Glycyrrhetinic acid binds to the conserved P-loop region and interferes with the interaction of RAS-effector proteins. Acta Pharm Sin B. 2019;9:294–303. doi: 10.1016/j.apsb.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kambe T., Correia B.E., Niphakis M.J., Cravatt B.F. Mapping the protein interaction landscape for fully functionalized small-molecule probes in human cells. J Am Chem Soc. 2014;136:10777–10782. doi: 10.1021/ja505517t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan S., Jang S.Y., Wang D., Liew S.S., Li Z., Lee J.S., et al. A suite of “minimalist” photo-crosslinkers for live-cell imaging and chemical proteomics: case study with brd4 inhibitors. Angew Chem Int Ed Engl. 2017;56:11816–11821. doi: 10.1002/anie.201706076. [DOI] [PubMed] [Google Scholar]
- 34.Chang J., Kim Y., Kwon H.J. Advances in identification and validation of protein targets of natural products without chemical modification. Nat Prod Rep. 2016;33:719–730. doi: 10.1039/c5np00107b. [DOI] [PubMed] [Google Scholar]
- 35.Gersch M., Kreuzer J., Sieber S.A. Electrophilic natural products and their biological targets. Nat Prod Rep. 2012;29:659–682. doi: 10.1039/c2np20012k. [DOI] [PubMed] [Google Scholar]
- 36.Maurais A.J., Weerapana E. Reactive-cysteine profiling for drug discovery. Curr Opin Chem Biol. 2019;50:29–36. doi: 10.1016/j.cbpa.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman A.C., Maddocks O.D.K. One-carbon metabolism in cancer. Br J Cancer. 2017;116:1499–1504. doi: 10.1038/bjc.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maddocks O.D.K., Berkers C.R., Mason S.M., Zheng L., Blyth K., Gottlieb E., et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2012;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu T., Chen C., Wang S., Zhang Y., Zhu D., Li L., et al. Cellular target identification of withangulatin A using fluorescent analogues and subsequent chemical proteomics. Chem Commun (Camb) 2019;55:8231–8234. doi: 10.1039/c9cc03653a. [DOI] [PubMed] [Google Scholar]
- 40.Wu X., Xia J., Zhang J., Zhu Y., Wu Y., Guo J., et al. Phosphoglycerate dehydrogenase promotes proliferation and bortezomib resistance through increasing reduced glutathione synthesis in multiple myeloma. Br J Haematol. 2020;190:52–66. doi: 10.1111/bjh.16503. [DOI] [PubMed] [Google Scholar]
- 41.Vandekeere S., Dubois C., Kalucka J., Sullivan M.R., Garcia-Caballero M., Goveia J., et al. Serine synthesis via PHGDH is essential for heme production in endothelial cells. Cell Metab. 2018;28:573–587. doi: 10.1016/j.cmet.2018.06.009. e13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.