Abstract
Metabolic homeostasis requires dynamic catabolic and anabolic processes. Autophagy, an intracellular lysosomal degradative pathway, can rewire cellular metabolism linking catabolic to anabolic processes and thus sustain homeostasis. This is especially relevant in the liver, a key metabolic organ that governs body energy metabolism. Autophagy's role in hepatic energy regulation has just begun to emerge and autophagy seems to have a much broader impact than what has been appreciated in the field. Though classically known for selective or bulk degradation of cellular components or energy-dense macromolecules, emerging evidence indicates autophagy selectively regulates various signaling proteins to directly impact the expression levels of metabolic enzymes or their upstream regulators. Hence, we review three specific mechanisms by which autophagy can regulate metabolism: A) nutrient regeneration, B) quality control of organelles, and C) signaling protein regulation. The plasticity of the autophagic function is unraveling a new therapeutic approach. Thus, we will also discuss the potential translation of promising preclinical data on autophagy modulation into therapeutic strategies that can be used in the clinic to treat common metabolic disorders.
KEY WORDS: Autophagy, Liver metabolism, Signaling proteins, Lysosome, Nutrient regeneration, Quality control, Farnesoid X receptor, Cryptochrome 1
Abbreviations: AIM, Atf8 interacting motif; ATGL, adipose triglyceride lipase; ATL3, Atlastin GTPase 3; ATM, ATM serine/threonine kinase; BA, bile acid; BCL2L13, BCL2 like 13; BNIP3, BCL2 interacting protein 3; BNIP3L, BCL2 interacting protein 3 like; CAR, constitutive androstane receptor; CCPG1, cell cycle progression 1; CLN3, lysosomal/endosomal transmembrane protein; CMA, chaperonin mediated autophagy; CREB, cAMP response element binding protein; CRY1, cryptochrome 1; CYP27A1, sterol 27-hydroxylase; CYP7A1, cholesterol 7α-hydroxylase; DFCP1, double FYVE-containing protein 1; FAM134B, family with sequence similarity 134, member B; FFA, free fatty acid; FOXO1, Forkhead box O1; FUNDC1, FUN14 domain containing 1; FXR, farnesoid X receptor; GABARAPL1, GABA type A receptor associated protein like 1; GIM, GABARAP-interacting motif; LAAT-1, lysosomal amino acid transporter 1 homologue; LALP70, lysosomal apyrase-like protein of 70 kDa; LAMP1, lysosomal-associated membrane protein-1; LAMP2, lysosomal-associated membrane protein-2; LD, lipid droplet; LIMP1, lysosomal integral membrane protein-1; LIMP3, lysosomal integral membrane protein-3; LIR, LC3 interacting region; LXRa, liver X receptor a; LYAAT-1, lysosomal amino acid transporter 1; MCOLN1, mucolipin 1; MFSD1, major facilitator superfamily domain containing 1; mTORC1, mammalian target of rapamycin complex 1; NAFLD, non-alcoholic fatty liver disease; NBR1, BRCA1 gene 1 protein; NCoR1, nuclear receptor co-repressor 1; NDP52, calcium-binding and coiled-coil domain-containing protein 2; NPC-1, Niemann-Pick disease, type C1; OPTN, optineurin; PEX5, peroxisomal biogenesis factor 5; PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; PINK1, phosphatase and tensin homolog (PTEN)-induced kinase 1; PKA, protein kinase A; PKB, protein kinase B; PLIN2, perilipin 2; PLIN3, perilipin 3; PP2A, protein phosphatase 2a; PPARα, peroxisomal proliferator-activated receptor-alpha; PQLC2, PQ-loop protein; PXR, pregnane X receptor; RETREG1, reticulophagy regulator 1; ROS, reactive oxygen species; RTN3, reticulon 3; RTNL3, a long isoform of RTN3; S1PR2, sphingosine-1-phosphate receptor 2; S6K, P70-S6 kinase; S6RP, S6 ribosomal protein; SCARB2, scavenger receptor class B member 2; SEC62, SEC62 homolog, preprotein translocation factor; SIRT1, sirtuin 1; SLC36A1, solute carrier family 36 member 1; SLC38A7, solute carrier family 38 member 7; SLC38A9, sodium-coupled neutral amino acid transporter 9; SNAT7, sodium-coupled neutral amino acid transporter 7; SPIN, spindling; SQSTM1, sequestosome 1; STBD1, starch-binding domain-containing protein 1; TBK1, serine/threonine-protein kinase; TEX264, testis expressed 264, ER-phagy receptor; TFEB/TFE3, transcription factor EB; TGR5, takeda G protein receptor 5; TRAC-1, thyroid-hormone-and retinoic acid-receptor associated co-repressor 1; TRPML1, transient receptor potential mucolipin 1; ULK1, Unc-51 like autophagy activating kinase 1; UPR, unfolded protein response; V-ATPase, vacuolar-ATPase; VDR, vitamin D3 receptor; VLDL, very-low-density lipoprotein; WIPI1, WD repeat domain phosphoinositide-interacting protein 1
Graphical abstract
Three major modes of regulation of hepatic metabolism by autophagy. Autophagy can regenerate nutrients, quality control cellular organelles, and regulate levels of signaling proteins related to hepatic metabolism.
1. Introduction
Autophagy—loosely translated “self-eating”—is the highly conserved lysosome-mediated intracellular degradative pathway which plays a pivotal role to maintain hepatic metabolic homeostasis. Autophagy comes in three main forms: macroautophagy, chaperone-mediated autophagy (CMA), and microautophagy, which are differentiated by the mechanism of cargo sequestration. In general, macroautophagy hereinafter referred to as autophagy can be selective or non-selective, where selective autophagy targets damaged or extraneous organelles whereas non-selective autophagy are responsible for the bulk recycling of cytoplasmic components under cellular stress conditions. The detailed autophagic signaling pathway and cellular events responsible for its execution and regulation are well established and out of the scope of this review. In this review, we discuss how autophagy particularly contributes to hepatic metabolism. We will discuss three different modalities by which autophagy directly contribute to hepatic metabolism: 1) how autophagy degrades the nutrient macromolecules (proteins, carbohydrates, lipids, and nucleic acids) to maintain intracellular metabolites pools; 2) how autophagy maintains quality control of metabolic powerhouse organelles; and 3) how autophagy selectively modulates various crucial signaling proteins to directly regulate the expression levels of metabolic enzymes. We also discuss current therapeutic strategies, potential targets, their limitations, and challenges related to autophagy modulation for the treatment of common liver diseases. We begin with the current knowledge about autophagic degradation and nutrient recycling.
2. Autophagic transport, lysosomal degradation, and nutrient recycling
Fundamentally, autophagy is involved in the sequestration and transport of cytosolic components into the lysosome for degradation and nutrient recycling. In this section, we discuss autophagic degradation, generation of molecular metabolites, and post-degradative events and their contribution to hepatic metabolism, which has not been well characterized and discussed in the field of autophagy and liver metabolism.
2.1. Metabolic substrates and products of autophagic degradation
Non-selective and selective autophagic degradation of cytosolic macromolecules generally maintain intracellular metabolite pools to support anabolic pathways during starvation or cellular stress. Autophagic digestion of energy-dense substrates like glycogen, proteins, lipids, and nucleic acids in lysosomes generates various catabolites such as glucose, amino acids, fatty acids, and nucleosides that are liberated into the cytoplasm for reutilization (Fig. 1A). The detailed process of selective degradation of nutrient components such as glycogen, lipid droplets, and protein aggregates will be discussed later in Sections 3.3, 3.4, 3.5. Though selective autophagy can occur by different mechanisms, it generally involves a receptor that either binds the substrate cargo or is an integral part of the cargo and interacts with a scaffold protein to link the cargo to complexes involved in autophagy1.
Figure 1.
Autolysosomal degradation, transport, and nutrient recycling. (A) Various cellular nutrient dense macromolecules and lysosomal hydrolases responsible for generation of small biomolecules. (B) Lysosomal transporters for efflux of cellular nutrients from lysosomal lumen into cytosol for reuse by cells. Glycocalyx coats the inner leaflet of the lysosomal membrane. The activity of most lysosomal nutrient transporters varies according to membrane voltage or intra-lysosomal protein levels generated by vacuolar-ATPase (V-ATPases).
2.2. Lysosomal degradative enzymes, nutrient processing in hepatic liver metabolism
Lysosomes are dynamic, membrane-bound organelles ubiquitously found in hepatocytes. They are the degradative and nutrient-sensing centers for autophagy induction and execution. Lysosomes not only hydrolytically degrade the macromolecular components received from autophagosomes, but also pinocytosis or endocytosed extracellular macromolecules.
Lysosomal hydrolytic degradation is mediated by the concerted action of approximately 60 different, mostly soluble, acid hydrolases such as proteases, lipases, glycosidases, sulphatases, phosphatases, and nucleases. The functions of most of these hydrolases require an acidic lumen with a pH of around 4.5, which is established and maintained by an ATP-driven proton pump, the vacuolar-ATPase(V-ATPase) proton pump located on the perimeter limited membrane2,3. To protect the perimeter membrane and its resident membrane proteins from degradation, the inner leaflet of the lysosomal membrane is coated with a polysaccharide layer called the glycocalyx4 (Fig. 1B). These hydrolases degrade multiple types of cytoplasmic components simultaneously and release the resultant molecular building blocks, such as amino acids, glucose, nucleotides, and fatty acids into cytosol either for further degradation to provide energy or reutilization by biosynthetic pathways to produce complex molecules, depending upon cellular needs.
2.3. Efflux of lysosomal degradative biomolecules via lysosomal membrane protein
Lysosomal membrane proteins are integral components of the lysosomal membrane that maintain stability and integrity of the lysosomal limiting membrane. Major lysosomal membrane proteins include lysosome-associated membrane glycoprotein 1 (LAMP1), lysosome-associated membrane glycoprotein 2 (LAMP2), lysosomal integral membrane protein-1 (LIMP1), lysosomal integral membrane protein-3 (LIMP3). Other minor integral components of lysosomal membranes include V-ATPase, Niemann-Pick disease, type C (NPC)-1, cystinosin, sialin, lysosomal/endosomal transmembrane protein (CLN3), lysosomal apyrase-like protein of 70 kDa (LALP70), etc.5. Besides the hydrolytic enzymes that reside within the lysosomal lumen, these integral lysosomal proteins are believed to protect the membrane from degradation by lysosomal hydrolases. Likely, the heavy glycosylation of these integral proteins contributes to this protective function. Indeed, histological analysis showed a coat of glycoconjugates on the inner surface of the lysosomal membrane4.
Besides these various lysosomal membrane proteins, lysosomes also contain specific nutrient transporters or permeases for the export of low-molecular-weight basic metabolites from the lysosomal lumen to the cytosol (Fig. 1B). New details on how intra-lysosomal catabolites are exported to cytosol for reutilization are slowly emerging. Amino acids generated from lysosomal protein degradation efflux via amino acid transporters such as sodium-coupled neutral amino acid transporter 9 (SLC38A9)6. SLC38A9 mediates the transport of many essential amino acids out of lysosomes, including leucine, in an arginine-regulated fashion. Lysosomes have other amino acids transporters such as lysosomal amino acid transporter 1 (LYAAT-1) (also known as SLC36A1), lysosomal amino acid transporter 1 homolog (LAAT-1; also known as PQLC2), and sodium-coupled neutral amino acid transporter 7 (SNAT7, also known as SLC38A7) to efflux the luminal amino acids.
Similarly, sugar channels including spindlin (SPIN) and ion channels including mucolipin 1 (MCOLN1, also known as TRPML1) maintain stores of sugars and ions, such as Cu+, Fe+, and Ca2+, within the lysosomal lumen7.
Lipid, particularly cholesterol, egress occurs through NPC1, NPC2, and LIMP2/SCARB28. Lipids can also egress from the lysosomal membrane to other organellar membranes directly via membrane contact sites9. Little is known about the transport of other lipids such as FFA. Lysosomes also express polytopic transmembrane/transporter proteins such as major facilitator superfamily domain containing 1 (MFSD1) protein that belongs to the major facilitator superfamily of transporters10. MFSD1 mediates secondary active or passive transport across the lysosomal membrane.
No specific metabolite tracing of these effluxed catabolites has been studied yet. Hence, the details of these catabolites’ fates are poorly understood and have been speculative. However, it is widely accepted in the field that these metabolites released by autolysosomal degradation can be directly utilized to yield energy or for new macromolecule synthesis (Fig. 1). Nutrients released from lysosomal degradation such as amino acids, glucose or cholesterol, etc., can initiate the lysosomal nutrient-sensing signaling pathway such as mammalian target of rapamycin complex 1 (mTORC1)-transcription factor EB (TFEB/TFE3) signaling that can feedback inhibit the autophagy process11, 12, 13. The details about lysosomal nutrient sensing and the signaling pathways involved are discussed in more detail in the recent review14,15.
Lysosome-derived nutrient catabolites can also be transported out of lysosomes extracellularly through vesicular membrane trafficking (secretory lysosomes). The extracellular lysosomal degradative products are released either into the bile or systemic circulation through the apical region. Notably, in the liver, these simpler biomolecules can be converted into glucose and ketone bodies for distribution elsewhere in the body. On the other hand, defective transport of these small molecules across the lysosomal membrane results in the accumulation of undigested lipids or glycogen inside the lysosome and toxicity, leading to the development of various liver pathologies16,17.
We next discuss autophagy's role in 1) nutrient regeneration by degradation of cellular macromolecules, 2) quality control of metabolically central organelles, and 3) regulation of signaling molecules to support hepatic metabolism.
3. Autophagy regulates energetic homeostasis by nutrient regeneration
Hepatic autophagy is an essential cellular process responsible not only for the degradation of cytoplasmic constituents but also for the maintenance of metabolic and energy homeostasis. With the considerable strides in the understanding of molecular mechanisms of autophagy, there is growing interest to understand the mechanisms of autophagy's contributions to metabolic and energetic homeostasis.
3.1. Autophagy and normal hepatic metabolism
Autophagy was initially thought to only be induced in response to certain stressors such as starvation, but it has since been demonstrated that autophagy exhibits constitutive activity in the healthy liver18. Studies have demonstrated that dysregulation of hepatic autophagy disrupts cellular homeostasis. For example, in a murine autophagy-related gene 7 (Atg7), knockout model, autophagy impairment has been shown to induce cellular abnormalities including mitochondria deformation, membrane structural alteration, and ubiquitin-positive aggregate formation19. This suggests that autophagy serves a homeostatic role by controlling the level of intracellular organelles and protein aggregates.
Autophagy in the healthy liver serves three main functions which are both inextricably linked to hepatic metabolism: support of energy balance, quality control, and regulation of signaling proteins. Autophagy also sustains hepatic energetic needs by catabolism of glycogen, lipids, and proteins. The extent to which autophagy degrades intracellular components depends on the fasting–feeding cycle, which affects plasma levels of hormones such as insulin and glucagon, as well as levels of amino acids20,21. Additionally, ammonia derived from glutaminolysis supports basal autophagy independently of mTORC1 and Unc-51 like autophagy activating kinase 1 (ULK1)22,23.
3.2. Hepatic autophagy mobilizes nutrients during starvation
Autophagy is required to sustain circulating nutrients during fasting, which has been demonstrated to be critical for the survival of adult mice. Hepatic autophagy induces glycogenolysis, lipolysis, and protein catabolism in that order to meet the body's energetic needs during severe nutrient deprivation20. The autophagosome membrane engulfs glycogen, lipid droplets, or protein, which are then degraded in a lysosome for nutrient recycling and sustaining the critical metabolic pathways.
The liver serves essential metabolic functions for the body which are strictly regulated by metabolic hormones such as insulin and glucagon. Glucose is converted to pyruvate by glycolysis in the cytoplasm, and pyruvate is then oxidized through the citric acid cycle and oxidative phosphorylation in the mitochondria to provide ATP and sustain cellular energy needs in the liver. In the fed state when carbohydrates are abundant, hepatocytes also convert glucose into fatty acids through de novo lipogenesis which is then incorporated into complex lipids and stored in lipid droplets and membrane structures or released into circulation as very low-density lipoprotein (VLDL)24. During the fasted state, the liver utilizes glycogenolysis and gluconeogenesis to mobilize and secrete glucose to support the body's energetic needs24. In response to prolonged fasting, endogenous glucose production is primarily performed via hepatic gluconeogenesis24. In the fasted state, lipolysis is promoted in adipocytes, resulting in the release of non-esterified fatty acids which the liver uses to generate ketone bodies by mitochondrial β-oxidation and ketogenesis to sustain extrahepatic energy needs24. Hepatocytes are highly capable of catabolizing proteins to release their amino acids which can be used to provide energy after disposal of nitrogenous wastes, which can be accomplished via the liver urea cycle25. The carbon skeletons of glucogenic amino acids can be incorporated into the citric acid cycle and diverted into gluconeogenesis, while ketogenic amino acids can be used as substrates for ketogenesis; these amino acid sources include the liver as well as other tissues such as skeletal muscle and intestinal tissue25. Insulin induces glycolysis and lipogenesis while suppressing gluconeogenesis and glucagon exerts the opposite effects on hepatic metabolism. Additionally, activation of the sympathetic nervous system stimulates hepatic gluconeogenesis whereas the parasympathetic nervous system suppresses this process24.
In the following section, we review autophagy-mediated selective degradation of nutrient components such as glycogen, lipid droplets, and protein aggregates, as well as note some relevant regulators that have been identified (Fig. 2).
Figure 2.
Autophagy regulates macromolecule degradation and maintains intracellular metabolite pools to sustain metabolism. Selective degradation of glycogen (A) by glycophagy, protein (B) and lipid droplets (LD) (C) generates glucose, amino acids (AA), and lipid metabolites (such as free fatty acids [FFA], glycerol and free cholesterol) respectively. Negative regulator molecules or pharmacological agents are indicated by red lines.
3.3. Glycophagy and hepatic glycogen metabolism for glucose homeostasis
Hepatic autophagy has a major role in blood glucose homeostasis. Glycogen breakdown has long been known to occur in the liver to mobilize nutrients under fasted conditions by the classic metabolic process of glycogenolysis. Glycogen can also be selectively degraded into glucose in autophagosomes to sustain energetic needs by “glycophagy”20 (Fig. 2A). Starch-binding domain-containing protein 1 (STBD1) is highly expressed in tissues that store glycogen such as the liver and are mainly responsible for glycogen transport to lysosomes in the liver26. STBD1 contains a C-terminal CBM20 glycan-binding domain that can bind glycogen and an ATG8 interacting motif (AIM, also referred to as an LC3 interacting region [LIR]), through which it can bind GABA type A receptor associated protein like 1 (GABARAPL1), a member of the Atg8 family26, 27, 28. STBD1 is thought to act as an adaptor protein for anchoring glycogen to lysosomes during glycophagy and has been shown to colocalize with GABARAPL1, glycogen, and late endosomal and lysosomal marker LAMP127,29.
The cyclic AMP (cAMP)/protein kinase A (PKA) signaling pathway is an established activator of both cytosolic glycogenolysis and glycophagy, which generally act in parallel. The binding of glucagon to the glucagon receptor on hepatocytes activates adenyl cyclase which increases the intracellular concentration of the secondary messenger cAMP, thus activating PKA which phosphorylates glycogen phosphorylase and glycogen synthase to promote cytosolic glycogenolysis and inhibit glycogenesis, respectively20. Although glycogen phosphorylase and cytosolic debranching enzymes are mainly responsible for the breakdown of hepatic glycogen to glucose in adults and children, the degradation of glycogen to glucose in animals during the neonatal period is primarily facilitated by the lysosomal enzyme α-1,4-glucosidase (also called acid glucosidase)30,31. Catabolism of glycogen is known to be essential for survival in neonatal animals a few hours after birth when the maternal nutrient supply is cut off, causing hypoglycemia and glucagon release; in studies on newborn rat hepatocytes, glucagon and cAMP increase the size and number of autophagosomes containing glycogen granules and the activity of α-1,4-glucosidase glycogenolysis20,32, 33, 34, 35. Administration of cAMP signaling inhibitors such as propranolol and parenteral glucose incur the opposite effects in newborn rat hepatocytes, inhibiting an increase in the volume of autophagosomes and the activity of α-1,4-glucosidase, further substantiating the role of cAMP/PKA signaling in glycophagy36,37. The class I-PI3-kinase (PI3K)–AKT/protein kinase B (PKB)-mTOR signaling pathway, through which insulin indirectly activates mTOR and suppresses autophagy, is also known to regulate glycophagy29. Furthermore, activation of mTOR reduces glycophagy by inhibiting protein phosphatase 2a (PP2A), which functions downstream of mTOR as a common target of glucagon, insulin, and cAMP, and promotes the synthesis of α-1,4-glucosidase when active29,38. Lysosomal calcium levels also regulate autophagy and glycophagy, with the administration of agents such as phorbol myristate acetate, which increases lysosomal calcium, increasing the total volume of autophagosomes and α-1,4-glucosidase activity and decreasing the fractional volume of undigested glycogen in autophagosomes and the activity of acid mannose 6-phosphatase in newborn rat hepatocytes39. Ionophore A23187, which reduces lysosomal calcium, accordingly elicited the opposite influence on enzyme activity in newborn rat hepatocytes, further illustrating the effects of lysosomal calcium levels on glycophagy39. Glycophagy and autophagy of proteins (discussed later in this section) fuel nutrient mobilization during early starvation, but there is a gradual shift from these autophagy substrates to degradation of lipid droplets (LD) after 8 h of starvation40. This sequence of selective autophagic substrates degradation during starvation somewhat aligns with the progression of conventional metabolic degradative processes in the liver in response to prolonged nutrient deprivation: glycogenolysis and gluconeogenesis at first, and eventually mitochondrial β oxidation and ketogenesis, as well as gluconeogenesis24.
3.4. Autophagy in amino acid and protein metabolism
The liver is the principal source of serum proteins and breaks down internal proteins to supply the body with amino acids for energy in response to prolonged fasting41 (Fig. 2B). Basal autophagic function degrades ~1.5% of total hepatic protein per hour under nutrient-rich conditions but in response to starvation this rate increases to ~4.5%42. After 48 h of fasting, increased autophagy can account for the catabolism of ~40% of the total liver protein in wild-type mice43. In wild-type mouse livers, a surge of amino acid levels in liver tissue and blood was observed as a result of autophagy after 24 h of starvation and blood glucose levels were maintained within the normal range after this surge, whereas these levels continued to decrease in liver-specific autophagy-deficient mice, which did not exhibit an amino acid surge or decrease in liver protein44. Furthermore, when serine (a glucogenic amino acid) was administered to wild-type and liver-specific autophagy-deficient mice that had been starved for 24 h, both groups showed increased blood glucose concentrations, indicating that the observed difference in blood glucose maintenance is not due to differences in ability to use glucogenic amino acids for gluconeogenesis44. Out of 18 amino acids that were analyzed with respect to the amino acids released after 24 h of starvation in wild-type mice, 11 showed definite surge responses: valine, leucine, isoleucine, serine, threonine, methionine, asparagine, phenylalanine, tyrosine, lysine, and arginine (all of which are glucogenic except leucine and lysine)44. Though alanine, which is also a major glucogenic amino acid, only exhibited a slight increase, this result could potentially be explained by more efficient conversion to pyruvic acid compared to serine44. During 24 h of fasting, amino acids released by liver autophagy were also excreted directly into circulation and transported to skeletal muscle, in addition to the portion of released glucogenic amino acids that were converted to glucose in the liver by gluconeogenesis and secreted into circulation44. The maintenance of blood glucose levels during fasting by starvation-induced autophagy is a notable liver-specific metabolic contribution to energy homeostasis. Autophagy-derived amino acids are also used for the synthesis of proteins and a 2008 study demonstrated that in early mice embryos amino acids liberated by autophagy of maternal proteins are utilized to produce essential preimplantation embryonic proteins45.
Liver autophagy of proteins can be controlled by hormones that act on other metabolic processes in the liver, such as insulin and glucagon, and plasma amino acid levels. Experiments in cultured hepatocytes and ex vivo rat livers have identified the capability of amino acid concentrations alone to regulate autophagy, with maximal suppression achieved at twice the normal concentration and the amino acids leucine, phenylalanine/tyrosine, glutamine, proline, methionine, tryptophan, and histidine acting as major inhibitors42,43,46,47. However, animal studies have suggested that insulin and glucagon may be more important in regulating autophagy since plasma amino acid levels do not vary enough to exert this observed regulatory effect in vivo44. Multiple studies have established that insulin reduction is sufficient to induce liver autophagy. Rats administered streptozotocin, which depresses serum insulin, exhibit a 70% increase in degradation of endogenous proteins by autophagy48. Insulin activates class I-PI3-kinase and PKB (also known as AKT) signaling, eventually causing activation of mTOR by phosphorylation to suppress autophagy49,50. In mice starved for 24 h, insulin fell to its lowest level while glucagon was maintained at a high, stable level; AKT, P70-S6 kinase (S6K), and S6 ribosomal protein (RPS6), which are phosphorylated in an insulin-dependent manner, were dephosphorylated, suggesting inactivation of the mTOR/S6 kinase pathway by decreased plasma insulin-mediated induction of liver autophagy whereas they were phosphorylated or activated in the livers of fed mice44. In an additional experiment, mice were administered glucose after both 18 and 21 h of starvation, which substantially suppressed liver autophagy and the corresponding amino acid surge that has been observed during starvation. Glucagon remained constant under these experimental conditions, but insulin transiently increased after the second glucose administration, surpassing normal levels of fed mice, lending support to the major regulatory role of insulin in liver autophagy44. Glucagon introduction has been shown to not only induce autophagosome formation but also to upregulate acidic phosphatase and cathepsin D in lysosomes and increase lysosome fragility in rat livers51. Although the consensus has been that insulin-mediated activation of the class I PI3K–AKT–mTOR axis is the main regulator for inhibition of autophagosome formation, a 2013 study indicated that though insulin administration affected this pathway in muscle tissue, it had a weak effect on the liver and amino acids had a larger role in regulating this pathway in the liver52. Follow-up research has demonstrated that plasma amino acid levels can regulate liver autophagy independently from the action of hormones20.
Besides bulk degradation of protein aggregates by hepatic autophagy and their contribution to hepatic metabolism, CMA also degrades metabolic enzymes involved in lipid, carbohydrate, and amino acid metabolism. Studies on livers unable to perform CMA have shown increased levels of glycolytic enzymes including glyceraldehyde-3-phosphate dehydrogenase, pyruvate kinase, aldolase A, and malate dehydrogenase 1 (enolase 1), causing higher rates of glycolysis40. During nutrient deprivation, CMA degrades glycolytic enzymes to reduce hepatic glucose use so that it can be mobilized for use by other organs40. In addition to autophagy, another main degradative process for proteins is the ubiquitin–proteasome system, which is considered the principal proteolytic pathway in eukaryotic cells53. The proteasome generally degrades proteins with shorter half-lives, while autophagy primarily degrades long-lived proteins53. These processes sometimes degrade the same proteins, such as protein aggregates which can become less recognizable substrates resistant to proteasomal degradation and more easily degraded by autophagy53. With respect to energetic metabolism, both proteosome-mediated protein degradation and autophagic degradation can help maintain amino acid pools during nutrient deprivation, but hepatic autophagy primarily supports energetic homeostasis during chronic starvation54.
3.5. Lipophagy facilitates hepatic lipid metabolism
A major role of the liver is processing and storing fat. Hepatic lipid homeostasis is supported by coordinated regulation of lipid uptake, de novo synthesis, storage, and catabolism55. Hepatic cytosolic lipolysis, a type of ‘neutral’ lipolysis, is the conventionally known process of hepatic LD catabolism to release FAs by lipases and is still being characterized in comparison to canonical adipose tissue lipolysis56,57. Lipophagy is an alternative method for hepatic LD breakdown and refers to the catabolism of LDs via autophagy, using ‘acid’ lipolysis in lysosomes56. It is considered a central regulator of liver lipid metabolism and presumably to contribute to lipid turnover as significantly as conventional lipase-driven lipolysis57 (Fig. 2C). In this process, an autophagosome forms and pinches off part of a LD then fuses with a lysosome to supply the liver with free fatty acids that can be catabolized by β-oxidation20. Singh et al.58 first substantiated the claim that autophagy is critical for hepatic lipid homeostasis from experiments in both cultured hepatocytes in vitro and mouse liver in vivo. The group demonstrated that during nutrient deprivation, LDs associated with autophagic pathway components such as LC3 and that inhibition of autophagy heightened the hepatic triglyceride and LD content58. Additionally, CMA targets and degrades the LD-associated proteins perilipin 2 (PLIN2) and perilipin 3 (PLIN3); this process is enhanced during starvation and concerted with increased cytosolic adipose triglyceride lipase (ATGL) levels and association of macroautophagy proteins on LDs, suggesting that these processes work cooperatively59. Moreover, blockage of this CMA-mediated degradation of PLIN2 and PLIN3 was shown to reduce the association of macrolipophagy proteins and ATGL with lipid droplets, leading to decreased lipid oxidation and increased LDs in both cultured hepatocytes in vitro and mouse livers in vivo, indicating this CMA-mediated process removing the perilipin coat that surrounds LDs is needed for either conventional lipolysis or macrolipophagy to occur57,59.
In recent years, studies have demonstrated the interplay between lipophagy and conventional lipolysis. In 2016, Martinez-Lopez et al.60 found that cold exposure in the hypothalamus of mice complementarily activated lipophagy and cytosolic lipases in the liver and brown adipose tissue, with ATGL requiring interaction with the autophagosome marker LC3 to catalyze LD lipolysis effectively. Furthermore, the 2017 work of Sathyanarayan et al.61 shows that hepatic ATGL signaling via sirtuin 1 (SIRT1) is both necessary and sufficient to induce autophagy and lipophagy, thus majorly controlling LD catabolism and subsequent fatty acid oxidation in the liver. Schott et al.'s recent findings62 have identified a synergistic interaction between conventional lipolysis and lipophagy in hepatocytes as tandem pathways for LD breakdown, in which lipolysis targets larger-sized LDs (too large for direct lipophagic degradation) generating LDs smaller in size which can be catabolized by lipophagy. Bile acids, which are the end products of hepatic lipid metabolism (cholesterol catabolism) inhibit autophagy through farnesoid X receptor (FXR) sensing and by altering Rab7 association with autophagosomes to decrease autophagosome-lysosome fusion (see Section 5.1 below). Although the mechanisms by which lipophagy are modulated have not yet been fully elucidated, small Rab GTPase proteins, which belong to the Ras superfamily and are involved in membrane trafficking processes, are thought to directly regulate lipophagy; Rab7 (as mentioned previously), Rab10, Rab18, and Rab32 have been proposed to have major roles in lipophagy based on their identification in LD proteomic screens57. In addition to nutrient deprivation, lipophagy is also activated during the opposing condition: to prevent hepatic lipotoxicity when high levels of dietary lipids are delivered to the liver40. Additionally, an alternate method of autophagic LD degradation in hepatocytes has been newly discovered by Schulze et al.63 and termed “direct lysosome-based autophagy”. In this process, stable contacts between LDs and lysosomes in hepatocytes can allow the transfer of both lipids and proteins from LDs directly into the lysosome under nutrient-limited conditions, without the presence of an autophagosomal intermediate. This was shown to occur even when core macroautophagy (ATG5) or CMA (LAMP2A) components were knocked down by siRNA, indicating this as an additional mechanism for lysosome-mediated LD turnover which may have the therapeutic potential63.
4. Autophagy-mediated quality control supports metabolism
The role of autophagy in hepatic quality control is mediated by catabolism of intracellular substituents which are in excess, aging, or damaged. The products of this degradation process subsequently are diverted into alternate metabolic pathways to be recycled or to generate cellular energy. Catabolism of organelles and other intracellular components such as specific proteins and invading pathogens by selective autophagy maintains cellular homeostasis and protects the cells from malfunctioning substituents51.
4.1. Mitochondrial quality control via mitophagy maintains metabolic homeostasis
Various metabolic pathways such as free fatty acid (FFA) oxidation, the citric acid cycle, and oxidative phosphorylation occur in mitochondria. Hepatocytes contain numerous mitochondria, which essentially regulate the flux of various cellular metabolites to accommodate energetic demand, to sustain hepatic metabolic activities. Thus, maintaining healthy mitochondria is crucial for metabolic homeostasis. Autophagy clears unhealthy mitochondria through mitophagy to accommodate energetic demand and support liver metabolic pathways and function (Fig. 3A).
Figure 3.
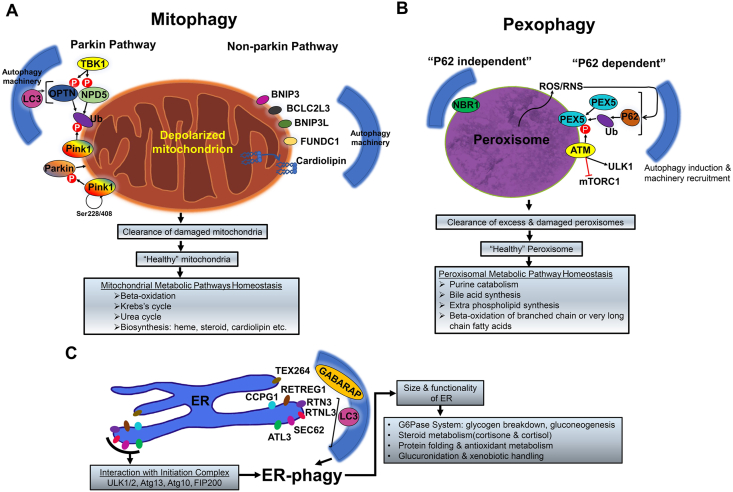
Autophagy maintains cellular organelle quality control to impact hepatic metabolism. (A) Two major pathways of mitochondrial quality control occur, via parkin or non-parking pathway. (B) Peroxisome mediated increased ROS or RNS activate P62 mediated pexophagy. NBR1 mediated P62 independent pexophagy can also occur for peroxisomal quality control. (C) ER-quality control maintenance by ER-phagy is mediated by various adaptor proteins-RETREG1, RTN3, RTNL3, SEC62, CCPG1, TEX264, and ATL3.
Mitophagy refers to the selective degradation of mitochondria by autophagy. Quality control and support of energetic homeostasis are inextricably linked in the process of mitophagy, which not only regulates the population size of the organelle but also selectively removes depolarized and damaged mitochondria, and thus limiting free radical generation and preventing wasteful consumption of ATP by mitochondrial ATP synthase40.
A well-characterized division of mitophagy is parkin-mediated mitophagy which degrades depolarized mitochondria. Healthy mitochondria normally cleave and degrade phosphatase and tensin homolog (PTEN)-induced kinase 1 (PINK1), but in mitochondria which are depolarized or contain misfolded protein aggregates, this protein becomes stabilized on the outer mitochondrial membrane and autophosphorylates its Ser228 and Ser408 residues, recruiting parkin (a cytosolic E3 ubiquitin ligase)64. PINK1 phosphorylates parkin to activate its E3 ligase activity and phosphorylates ubiquitin attached to mitochondrial proteins, further facilitating the labeling of the damaged mitochondria since parkin exhibits a high affinity for phosphorylated ubiquitin20,64. This phospho-ubiquitin signature generated by PINK1 and increased by parkin, which generates more chains of ubiquitin on the mitochondria, recruits the adaptor proteins optineurin (OPTN) and calcium-binding and coiled-coil domain-containing protein 2 (NDP52) to induce mitophagy65. Serine/threonine-protein kinase TBK1 then phosphorylates OPTN and NDP52 which heightens their binding affinity to LC3, a core autophagy protein20. OPTN and NDP52 are also involved in the recruitment and assembly of other core autophagy proteins such as ULK1, WD repeat domain phosphoinositide-interacting protein 1 (WIPI1), and double FYVE-containing protein 1 (DFCP1), which mediate autophagosome maturation around the damaged mitochondria20,51,65.
Alternatively, mitophagy can also occur via a non-parkin-mediated pathway (Fig. 3A). Various mitochondrial adaptor proteins are also known as mitophagy receptors such as BCL2 interacting protein 3 (BNIP3), BCL2 interacting protein 3 like (BNIP3L), BCL2 like 13 (BCL2L13), and FUN14 domain containing 1 (FUNDC1) can recruit autophagic machinery to damaged mitochondria for mitophagy66,67. Also, the translocation of specific lipids such as cardiolipin from the inner mitochondrial membrane to the outer mitochondrial membrane of damaged mitochondria can interact with autophagosomes via lipidated LC3 for selective degradation68.
Taken together, the role of autophagy in selective degradation of damaged mitochondria and control of the mitochondrial population size supports cellular quality control, maintains metabolic homeostasis, and bolsters the liver's resiliency to injury and disease. Pharmacological targeting of mitophagy may ameliorate liver injury in hepatic pathologies which involve mitochondrial impairment and/or oxidative stress.
4.2. Pexophagy manages peroxisomal catabolism and metabolic pathway recurring in peroxisome
The selective autophagy of peroxisomes, termed pexophagy, is an important pathway for the removal of peroxisomes and has important implications for homeostasis of hepatic metabolism (Fig. 3B). Peroxisomes are single-membrane-bound organelles that contain enzymes involved in a variety of metabolic reactions including purine catabolism, bile acid synthesis, ether phospholipid synthesis, and β-oxidation of branched or very long fatty acids which cannot be catabolized by mitochondria.
Although the mechanisms of pexophagy have not been fully elucidated, it has been established that reactive oxygen species (ROS) augment the interaction between peroxisomal targeting signal I receptor (PEX5) and extranuclear serine-protein kinase Ataxia Telangiectasia mutated protein kinase (ATM)20,69. This complex translocates onto peroxisomes and ATM signaling activates ULK1 and inhibits mTORC1 to induce autophagy in response to ROS or reactive nitrogen species (RNS) presence69,70. ATM kinase not only suppresses the autophagy inhibitor mTORC1 but also induces pexophagy by causing PEX5 phosphorylation and then ubiquitylation which induces P62 to bind and target the peroxisomes for degradation69. ATM kinase's role in inactivating mTORC1 via ATM-AMPK-tuberous sclerosis (TSC) signaling in addition to activating ULK1 suggests that ATM can modulate pexophagy through both ATM-AMPK–TSC signaling and P62-mediated autophagy20,69. Additionally, the next to BRCA1 gene 1 protein (NBR1) has been identified as a specific adaptor protein for pexophagy which can mediate pexophagy when it is in excess without requiring P62, but which increases the efficiency of pexophagy when bound by P6271.
Pexophagy not only maintains the number of peroxisomes in the cell as a quality control process but also responds to the energetic state of the cell. Long-term restriction of protein in rats causes peroxisome loss and disrupts metabolic function in the liver, and a mouse model has identified increased PEX2 and pexophagy levels during severe malnutrition, which may partially elucidate the mechanism behind this observed phenotype72. Though understanding of the mechanisms of pexophagy and its contribution to hepatic metabolism necessitates further investigation, we now understand that inhibition of autophagy by pharmacological or genetic targeting rescues peroxisome number and function73. These findings, taken in conjunction with the liver's major role in fat metabolism and other catabolic processes, suggest pexophagy significantly contributes to the support of not only quality control, but also metabolic and energetic homeostasis.
4.3. ER-phagy supports hepatic metabolism
ER-phagy (reticulophagy) is the selective autophagic degradation of the endoplasmic reticulum (ER) and also contributes to metabolic homeostasis in the liver (Fig. 3C). ER-phagy is mediated by direct interaction of multiple ER-phagy adaptor molecules such as reticulophagy regulator 1 (RETREG1, also known as FAM134B), reticulon 3 (RTN3), RTNL3 (a long isoform of RTN3), SEC62 homolog, preprotein translocation factor (SEC62) and cell cycle progression 1 (CCPG1), testis expressed 264, ER-phagy receptor (TEX264), and atlastin GTPase 3 (ATL3). These ER-phagy adaptor molecules are ER-membrane resident proteins and contain at least one LC3 or GABARAP-interacting region (LIR) or Atg8 interacting motif, that can bind to autophagosomal LC3/GABARAP family proteins. These adaptor proteins can also interact with most-upstream autophagy initiation complex components such as ULK1/2, Atg13, etc.
ER-phagy preserves the size and functionality of the ER. However, the pathophysiological relevance of ER-phagy is under investigation. One of the liver's main functions is drug metabolism, which often involves the proliferation of smooth ER where catabolism of the administered drug takes place, in addition to upregulation of cytochrome P-450 (Cyp450) genes involved in the catabolism of the given drug74. After discontinuance of the drug, the ER is reduced to its original size within several days. Multiple studies have confirmed that selective autophagy removes the extra ER membranes, though the mechanism for this process has not been fully elucidated74, 75, 76. Additionally, selective autophagy was also found to degrade the typical ER membrane proteins phenobarbital (PB)-inducible CYP450 and NADPH-cytochrome P-450 reductase (FP2) after cessation of phenobarbital treatment, further supporting the homeostatic relevance of autophagy in the reestablishment of normal ER structure and function after drug treatment77.
The ER of hepatocytes is also subject to continuous stress due to the extensive production of secretory proteins. In addition to activation of the unfolded protein response (UPR) which helps alleviate ER proteotoxicity, proteotoxic stress in the ER has been shown to trigger autophagy which may help degrade misfolded proteins and selective portions of the ER membrane-mediated to some extent by transcription factors such as c-Jun, but also by factors involved in the UPR such as eukaryotic translation initiation factor 2 alpha (eIF2alpha)18,78, 79, 80. The liver degrades proteins via autophagy at tremendous rates which also contributes to quality control and proteostasis, as outlined in more detail in the earlier section in the context of support of energetic homeostasis.
5. Autophagy-mediated regulation of signaling proteins impact metabolism
Conventionally, autophagy is well known to contribute to hepatic metabolism by the generation of the metabolite pool by simple degradation of macromolecules such as LD, proteins, or glycogen. Emerging evidence indicates that autophagy could also regulate hepatic metabolism in a previously uncharacterized way. We discuss this concept that autophagic selective modulation of signaling molecules directly impacts the hepatic metabolism below.
5.1. Autophagy regulates FXR signaling, and bile acid metabolism
Bile acid (BA) metabolism is another important function of the liver, relevant to both hepatic autophagy and metabolic homeostasis. BAs are synthesized in the liver (exclusively in hepatocytes) from cholesterol, either by the classic bile acid synthesis pathway (initiated by the rate-limiting enzyme cholesterol 7α-hydroxylase, CYP7A1) or the alternative bile acid synthesis pathway (initiated by mitochondrial sterol 27-hydroxylase, CYP27A1)81. Because BAs are derived from cholesterol, BA synthesis and secretion is the primary route of cholesterol excretion81,82. Hence, modulation of BA level indirectly impacts cholesterol metabolism.
BAs are non-energetic biomolecules but rather, detergent molecules, which aid in intestinal nutrient digestion and absorption of fats, steroids, and lipid-soluble vitamins81. BAs also serve as endogenous signaling molecules and regulate hepatic metabolism. BA signaling activates both nuclear receptors and G protein-coupled receptors. These nuclear receptors are FXR, pregnane X receptor (PXR), vitamin D3 receptor (VDR), and constitutive androstane receptor (CAR), while the G protein-coupled receptors are Takeda G protein receptor 5 [TGR5, also known as G protein bile acid receptor-1 (GPBAR-1)] and sphingosine-1-phosphate receptor 2 (S1PR2)81. Since BA signaling in its entirety is far beyond the scope of this review, we will further discuss the only FXR in consideration of its relevance to hepatic autophagy and metabolism.
Under normal physiology, BA homeostasis is mainly achieved through bile-acid mediated feedback inhibition of intrahepatic BA synthesis. Newly synthesized BAs are secreted by hepatocytes into bile for subsequent release into the small intestine. The majority of BAs are reabsorbed into enterocytes and transported back to the liver via the portal circulation. Intrahepatic BAs activate FXR to inhibit its biosynthesis and increase extrahepatic BA secretion.
Autophagy function has recently been shown to be necessary to maintain bile acid (BA) homeostasis83 (Fig. 4A). Deficiency of hepatic autophagy as seen in autophagy-related gene 7 (Atg7) or autophagy-related gene 5 (Atg5) knocked-out livers showed impaired FXR function, intrahepatic cholestasis with increased levels of serum BA, a higher ratio of tauro-muricholic acid/taurocholic acid, increased hepatic total bile acid load, and alteration of intestinal bile acid compositions83. BA level alteration due to autophagy impairment also significantly changes the composition of gut microbiota84. These phenotypic observations suggest basal autophagy is necessary for efficient BA handling via sustenance of basal expression and activity of FXR. FXR expression is transcriptionally repressed in these genetic models, suggesting autophagy could contribute to BA metabolism via transcriptionally regulating signaling proteins such as FXR to hepatic metabolism (Fig. 4A). Notably, this event occurs independently of macromolecule recycling. Accordingly, hepatic autophagy does more than just recycle macromolecules and cellular organelles.
Figure 4.
Autophagy regulates signal proteins' level and their activity to directly contribute to hepatic metabolism. (A) Autophagy transcriptionally modulates FXR levels and activity to impact bile acid (BA) metabolism. (B) Autophagy selectively degrades CRY1, a negative regulator of glucogenesis. (C) Selective degradation of corepressor NCoR1 impact hepatic lipid metabolism.
BA metabolism is not only critical for hepatic metabolic homeostasis in the direct manner briefly discussed above, but also because of its influence on autophagy. BAs can regulate autophagy via three major processes. A) Transcriptional inhibition: BA transcriptionally inhibits hepatic autophagy gene expression via an FXR-mediated mechanism. Post-prandial reabsorption of BA attenuates autophagy transcriptionally via the activation of FXR. FXR activation suppresses the expression of various autophagy-related proteins by antagonizing the function of their transcription factors such as cAMP response element-binding protein (CREB) and peroxisomal proliferator-activated receptor-alpha (PPARa)85,86. B) Autophagy flux impairment: at a cellular level, elevated BAs can also directly repress hepatic autophagy via inhibition of autophagosome to lysosome fusion. The flux impairment depends on the BA-mediated reduction of Rab GTPases such as Rab7, which associates with autophagosomes to facilitate autophagosome-lysosome fusion. FXR-dependent induction of Rubicon has also been found to be responsible for the impairment of autophagosome-lysosome maturation87,88. Interestingly, the inhibition of Rubicon releases the BA-mediated autophagy inhibition indicating that Rubicon could be a potential therapeutic target for autophagy inhibited at the flux level. In quite a contrast, accelerated BA synthesis via Cyp7A1 induction attenuates AKT/mTOR signaling leading to autophagy activation89. C) Hepatic cholesterol accumulation: autophagy is sensitive to changes in hepatic cholesterol metabolism and is impaired by hepatic cholesterol accumulation82. Decreased cholesterol can activate autophagy by a phosphatidylinositol 3-kinase-dependent mechanism90 and further studies have demonstrated various cholesterol-lowering agents decrease AKT–mTOR signaling and increase autophagy in hepatocytes82,89,91,92. On contrary, cholesterol accumulation can impair autophagy via lysosomal dysfunction. Excessive lysosomal cholesterol accumulation impairs autolysosome clearance and may also induce autophagy to compensate for this impaired autophagic flux82,89,93,94.
In sum, BA homeostasis is crucial to protect tissues from cholesterol and BA toxicity and maintain hepatic metabolic homeostasis. Dysregulation of BA amount or BA pool composition poses dramatic metabolic and pathologic implications. Thus, pharmacological strategies targeting bile acid metabolism and hepatic autophagy pose as promising therapeutic solutions for diseases associated with impaired hepatic metabolism.
5.2. Autophagy regulates glucose metabolism by selective turnover of clock protein cryptochrome 1 (CRY1) transcriptional repressor level
It is well known that autophagy is necessary to maintain the levels of glucose in blood and tissues and hence prevent systemic hypoglycemia in neonates and adults95,96. Autophagy contributes to glucose metabolism by glycogen breakdown (see above Section 3.3). Recently, studies have shown autophagy can directly regulate systemic glucose level by modulating the CRY1 signaling protein (Fig. 4B). Autophagic digestion selectively targets proteins controlling the circadian clock, most notably CRY1. The autophagic degradation of CRY1 is in agreement with previous observations where AMPK, a nutrient sensor and a key inducer of autophagy promotes CRY1 degradation97. CRY1 is a core clock repressor that maintains clock periodicity and robustly suppresses hepatic gluconeogenesis by regulating CREB/cAMP and glucocorticoid receptor signaling, as well as by reducing levels of FOXO1, a transcription factor that drives gluconeogenesis98, 99, 100. Hence, autophagic degradation of CRY1 releases its repression in gluconeogenesis and increases blood glucose levels. Physiologically, autophagy drives glucose production in the liver by timely degradation of CRY1 in the postprandial situation.
CRY1 has several light chains 3 (LC3)-interacting region (LIR) motifs to interact with LC3 for its selective degradation by autophagy. Hence, CRY1 is accumulated to cause lowered blood glucose levels in mice genetically deficient in autophagy. Interestingly, high-fat diet feeding, which is well known to block autophagic flux, decreases CRY1 protein levels in an autophagy-dependent manner, and restoring hepatic CRY1 levels reverses obesity-associated hyperglycemia.
5.3. Autophagy regulates lipid metabolism by regulating nuclear receptor co-repressor 1 (NCoR1) degradation
It is well established that autophagy plays a central role in lipid metabolism by selectively degrading lipid droplets via lipophagy. Selective degradation of LD releases FFA and glycerol which are then transported to mitochondria for their β-oxidation and hence to fuel metabolic processes. FFA can also contribute to membrane biosynthesis. Also, autophagy, or some element of the autophagic machinery, participates in LD biogenesis, particularly in hepatocytes101.
Apart from this, autophagy also regulates the synthesis of FFAs and triglycerides at the transcriptional level by fine-tuning levels of NCoR1 (Fig. 4C). NCoR1 is a transcriptional coregulatory protein which is also known as thyroid hormone and retinoic acid-receptor associated co-repressor 1 (TRAC-1) as it binds thyroid hormone receptor (TR) and retinoic acid receptor (RAR) and inhibits their respective genes. Hence, NCoR1 can interact with several nuclear receptors. Moreover, NCoR1 can recruit histone deacetylases to DNA promoter regions and thus downregulate gene expression.
NCoR1 inhibits transactivation of peroxisome proliferator-activated receptor (PPARα) and liver X receptor an (LXRa), both of which play gene regulatory role for β-oxidation and lipogenesis respectively102.
The transcriptional corepressor role of NCoR1 is modulated by autophagy. In autophagy-activated conditions such as starvation, both nuclear and cytosolic NCoR1 is markedly decreased due to autophagic selective degradation. This reduction is largely inhibited by genetic knockdown of Atg7 or by lysosomal protease inhibitors E64d and pepstatin treatment. Mechanistically, NCoR1 translocates to the cytoplasm where it directly interacts with GABARAP but not with LC3B for selective degradation by autophagic machinery. The N-terminal region of NCoR1 contains a GABARAP-interacting motif (GIM) that aids in its interaction with GABARAP for selective degradation.
Selective degradation of NCoR1 by autophagy is indispensable for the transactivation of various nuclear receptors, particularly PPARα and LXRa in response to starvation. Indeed, suppression of liver autophagy is accompanied by repression of multiple downstream target genes of PPARα and LXRa (our unpublished data). Since PPARα increases, FFA oxidation, and LXRa promotes hepatic de novo lipogenesis, the regulation of NCoR1, and hence the modulation of these two nuclear receptors may be a tertiary autophagic means to modulate lipid anabolism and catabolism.
6. Recent advances in translating autophagy research into therapeutics for metabolic liver disease
Autophagy plays an important role not only in normal liver physiology but also in the pathogenesis of many metabolic liver diseases such as alcoholic liver disease (ALD) and nonalcoholic fatty liver disease (NAFLD). Generally, autophagy balances the metabolism and senses stress in the liver. Autophagy mediated turnover of macromolecules, and selective degradation of organelles in the liver maintains hepatic homeostasis. In the context of ALD or NAFLD, this homeostasis is impaired and results in the accumulation of hepatic lipids (steatosis) or cytotoxic proteins (Mallory-Denk bodies) that could further stimulate hepatic injury, recruitment of inflammatory cells resulting in alcoholic steatohepatitis, nonalcoholic steatohepatitis (NASH). Enhancement of autophagy by pharmacological approaches has been already proven to alleviate liver steatosis and injury in alcoholic and non-alcoholic fatty liver in preclinical models103,104. Hence, therapeutic targeting of autophagy could be beneficial for treating metabolic liver diseases.
Autophagy could be strategically modulated at four different levels of the process: 1) Transcription factors or upstream signaling pathway regulating autophagy induction, e.g., TFEB, AMPK, or mTORC1 complex. 2) Autophagy initiation, phagophore, and autophagosome formation, e.g., VPS34 or Beclin1 or Atg4. 3) Autophagic flux level, i.e., autophagosome and lysosomal fusion, e.g., lysosomal hydrolases. 4) Targeting autophagy targeted signaling proteins (see Section 5 above). However, because of the broad range of cellular functions regulated by autophagy in physiological and pathological conditions, targeting autophagy remains considerably challenging. Moreover, the drugs targeting the individual components of the autophagic signaling pathway such as VPS34 or the autophagic flux can cause autophagy-independent effects. Yet, with increased specific understanding of the role of autophagy in liver metabolisms such as regulation of signaling proteins, targeting autophagy regulated substrates such as CRY1, NCoR1 or FXR could be a novel strategy to alter autophagic impact in liver metabolism and metabolic disease pathogenesis. In a preclinical model of NAFLD where rodents are fed a high-fat diet, accelerated autophagic degradation of CRY1 was observed. Interestingly, restoring the CRY1 level prevented obesity-associated dysregulation of gluconeogenesis. Hence, drug designs targeting the LIR motif and preventing autophagic CRY1 degradation should help stabilize CRY1 levels and lower blood glucose levels in obese or NAFLD patients. Nevertheless, there could be a limitation of these compounds as the signaling proteins have a non-autophagy specific function, and hence targeting these molecules could affect processes that are distinct from autophagy.
To date, several autophagy inducers and inhibitors have been identified and successfully implemented in disease models, though most are in preclinical stages. So far, the lysosomotropic autophagy inhibitors chloroquine and its derivative hydroxychloroquine and the rapamycin or rapalogs which stimulate autophagy are the most widely used in clinical trials aiming at modulating autophagy (https://clinicaltrials.gov).
Autophagy's crucial role in support of quality control, energetic homeostasis, and regulation of signaling molecules in hepatic metabolism makes it an attractive therapeutic target for metabolic liver diseases. The beneficial and negative effects of autophagy in various liver diseases and cell types complicate this goal, but research on this topic in the past decade has yielded many encouraging findings. One target for pharmacological therapy of various metabolic liver diseases, which is regulated by the autophagy process is FXR. Agonism of FXR by GW4064 was shown to significantly correct cholestasis and liver injury caused by deficient autophagy in a murine model83, and several other studies indicate FXR as a therapeutic target for hepatic cholestasis, with many FXR agonists undergoing preclinical and clinical investigation105. Protecting effects of FXR agonists have been demonstrated in several experiments and trials on cholestasis, cirrhosis, and NAFLD81. Conversely, for conditions exhibiting harmful effects of FXR activation, such as obstructive cholestatic liver disorders, experiments indicate FXR antagonism is beneficial105,106. Aside from the FXR modulation, other autophagy-modulating compounds have shown therapeutic promise for the treatment of metabolic liver diseases. Table 1 contains a comprehensive list of potential pharmacological treatments to target autophagy and combat metabolic liver disease pathogenesis.
Table 1.
Potential pharmacological treatments to target autophagy in metabolic liver diseases.
| Compound | For treatment of | Chemical properties | Function | Effect on autophagy | Ref. |
|---|---|---|---|---|---|
| Carbamazepine |
|
Tricyclic compound, anticonvulsant | mTOR independent autophagy inducer acting by reducing inositol levels | Induce | 103,107, 108, 109 |
| Curcumin |
|
Natural phyto-polylphenol pigment | Activates PPARα, and regulates upstream signaling pathways of AMPK and mTOR | Induce | 110,111 |
| Ezetimibe |
|
Beta-lactam | NPC1L1 inhibitor and potent NRF2 activator | Induce | 107,112,113 |
| Fibrates |
|
Amphipathic carboxylic acids | Ligands for the nuclear receptor PPARα | Induce | 114 |
| Fluphenazine |
|
Phenothiazine, dopamine receptor antagonist | mTORC1 inhibition | Induce | 107,115 |
| Metformin |
|
Biguanide | Indirectly inhibits mTOR, induces SIRT1-mediated autophagy | Induce | 116,117 |
| Norursodeoxycholic acid (norUDCA) |
|
Side chain shortened UDCA derivative | Appears to induce autophagy via activation of AMPK via mTOR/ULK1 | Induce | 114,118,119 |
| Pimozide |
|
Diphenylbutyl-piperidine derivative | mTOR-independent autophagy enhancer | Induce | 107 |
| Rapamycin (sirolimus) |
|
Organic heterotricyclic compound | Binds to mTOR and activates autophagy | Induce | 108,114 |
| Resveratrol |
|
Natural phenol | Sirtuin-1-dependent induction of autophagy | Induce | 40,115,116,120 |
| Suberoylanilide hydroxamic acid (vorinostat) |
|
Histone deacetylase inhibitor | Autophagy upregulation via AKT/mTOR signaling | Induce | 107 |
| Tat-beclin-1 peptide |
|
Peptide | Binds negative regulator of autophagy, GAPR-1 | Induce | 107,121,122 |
| Trehalose |
|
Disaccharide | mTOR-independent autophagy enhancer | Induce | 51,115,123 |
| Ursodeoxycholic acid (UDCA) |
|
Hydrophilic bile acid | FXR antagonist, active autophagy induction | Induce | 107,114,118,124,125 |
| Torin 1 |
|
Organofluorine, pyridoquinoline | mTORC1 and mTORC2 inhibition, increases liver TFEB levels | Induce | 126, 127, 128 |
| PP242 |
|
Pyrazolo-pyrimidine | mTORC1 and mTORC2 inhibition | Induce | 128,129 |
| BH3 mimetics (ABT-737) |
|
Peptides and small molecules | Stimulates Beclin 1-dependent activation of the pro-autophagic lipid kinase VPS34 | Induce | 130 |
| Lithium and L-690330 |
|
Alkali metal and phosphonic acid (respectively) | Inositol monophosphatase inhibition, lowering inositol and myo-inositol-1,4,5-triphosphate (IP3) levels | Induce | 109,131 |
| Sodium valproate |
|
Valproic acid | Lowers inositol and myo-inositol-1,4,5-triphosphate (IP3) levels | Induce | 109 |
| INT-767 |
|
Semisynthetic bile acid derivative | Dual FXR and TGR5 agonist | Presumed suppression | 81,124 |
| Theonellasterol |
|
4-Methylene-24-ethylsteroid | Selective FXR antagonist | Presumed suppression | 132 |
| Cilophexor |
|
Non-steroidal FXR agonist | FXR agonist | Suppress | 105,133 |
| GW4064 |
|
Synthetic isoxazole | FXR agonist | Suppress | 83,124 |
| Hydroxychloroquine |
|
Chloroquine derivative | Regulates lipid metabolism via peroxisome proliferator-activated receptor gamma pathway, blocks autophagosome-lysosome fusion | Suppress | 116,134 |
| Obeticholic acid (OCA) |
|
Semi-synthetic bile acid analog | FXR agonist | Suppress | 114,118 |
| Tropifexor |
|
Non-bile acid FXR agonist | FXR agonist | Suppress | 105,135 |
7. Concluding remarks, future directions, and open questions
It is now well established that autophagy function is quintessential for hepatic normal functioning and its impairment contributes directly to liver disease pathogenesis. With a better understanding of the mechanisms underlying autophagy induction and execution, great interest is emerging to understand the pathophysiological role of autophagy in liver function and therapeutically target autophagy in metabolic disease conditions. Novel concepts about how autophagy contributes to hepatic metabolism are unraveling. Besides conventional macromolecular recycling and organellar quality control, autophagy also selectively modulates various signaling proteins. The selective targeting of these signaling proteins that lie in the crux of regulating metabolic pathways is of special interest and could be the basis for future autophagy-based therapeutics. An open question that remains to be tested is whether specific LIR motifs responsible for autophagic degradation of the signaling protein could be developed or not. Further in-depth studies will be required to identify more downstream selective autophagy target proteins that act as signaling proteins to impact liver metabolic homeostasis.
Acknowledgments
This work was in part supported by the Tulane University School of Medicine Endowment Fund (BK, USA), American Society for Investigative Pathology (ASIP/SROPP) (KB & SB, USA), and BeHEARD Rare Disease challenge 2020 (BK, USA). We apologize to those whose work was not cited in this review due to the scope and the space limitation.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Author contributions
Katherine Byrnes: Investigation, Writing–original draft; Sophia Blessinger, Niani Bailey, Russell Scaife, and Gang Liu: Writing–review & editing; Bilon Khambu: Conceptualization, Writing–original draft.
Conflicts of interest
The authors declare no conflict of interest.
References
- 1.Gatica D., Lahiri V., Klionsky D.J. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20:233–242. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao J., Benlekbir S., Rubinstein J.L. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature. 2015;521:241–245. doi: 10.1038/nature14365. [DOI] [PubMed] [Google Scholar]
- 3.Mellman I. Organelles observed: lysosomes. Science. 1989;244:853–854. doi: 10.1126/science.244.4906.853. [DOI] [PubMed] [Google Scholar]
- 4.Neiss W.F. A coat of glycoconjugates on the inner surface of the lysosomal membrane in the rat kidney. Histochemistry. 1984;80:603–608. [PubMed] [Google Scholar]
- 5.Eskelinen E.L., Tanaka Y., Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–145. doi: 10.1016/s0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 6.Wyant G.A., Abu-Remaileh M., Wolfson R.L., Chen W.W., Freinkman E., Danai L.V., et al. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell. 2017;171:642–654. doi: 10.1016/j.cell.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrence R.E., Zoncu R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat Cell Biol. 2019;21:133–142. doi: 10.1038/s41556-018-0244-7. [DOI] [PubMed] [Google Scholar]
- 8.Meng Y., Heybrock S., Neculai D., Saftig P. Cholesterol handling in lysosomes and beyond. Trends Cell Biol. 2020;30:452–466. doi: 10.1016/j.tcb.2020.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Thelen A.M., Zoncu R. Emerging roles for the lysosome in lipid metabolism. Trends Cell Biol. 2017;27:833–850. doi: 10.1016/j.tcb.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massa Lopez D., Thelen M., Stahl F., Thiel C., Linhorst A., Sylvester M., et al. The lysosomal transporter MFSD1 is essential for liver homeostasis and critically depends on its accessory subunit GLMP. Elife. 2019;8:1–27. doi: 10.7554/eLife.50025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martina J.A., Chen Y., Gucek M., Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martina J.A., Diab H.I., Lishu L., Jeong A.L., Patange S., Raben N., et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal. 2014;7 doi: 10.1126/scisignal.2004754. ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roczniak-Ferguson A., Petit C.S., Froehlich F., Qian S., Ky J., Angarola B., et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savini M., Zhao Q., Wang M.C. Lysosomes: signaling hubs for metabolic sensing and longevity. Trends Cell Biol. 2019;29:876–887. doi: 10.1016/j.tcb.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin H.R., Zoncu R. The lysosome at the intersection of cellular growth and destruction. Dev Cell. 2020;54:226–238. doi: 10.1016/j.devcel.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Ploeg A.T., Reuser A.J. Pompe's disease. Lancet. 2008;372:1342–1353. doi: 10.1016/S0140-6736(08)61555-X. [DOI] [PubMed] [Google Scholar]
- 17.Ruivo R., Anne C., Sagné C., Gasnier B. Molecular and cellular basis of lysosomal transmembrane protein dysfunction. Biochim Biophys Acta. 2009;1793:636–649. doi: 10.1016/j.bbamcr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Schneider J.L., Cuervo A.M. Liver autophagy: much more than just taking out the trash. Nat Rev Gastroenterol Hepatol. 2014;11:187–200. doi: 10.1038/nrgastro.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., et al. Impairment of starvation-induced and constitutive autophagy in Atg 7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueno T., Komatsu M. Autophagy in the liver: functions in health and disease. Nat Rev Gastroenterol Hepatol. 2017;14:170–184. doi: 10.1038/nrgastro.2016.185. [DOI] [PubMed] [Google Scholar]
- 21.Pfeifer U. Inverted diurnal rhythm of cellular autophagy in liver cells of rats fed a single daily meal. Virchows Arch B. 1972;10:1–3. doi: 10.1007/BF02899710. [DOI] [PubMed] [Google Scholar]
- 22.Cheong H., Lindsten T., Wu J., Lu C., Thompson C.B. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci U S A. 2011;108:11121–11126. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eng C.H., Yu K., Lucas J., White E., Abraham R.T. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal. 2010;3:ra31. doi: 10.1126/scisignal.2000911. [DOI] [PubMed] [Google Scholar]
- 24.Rui L. Energy metabolism in the liver. Comp Physiol. 2014;4:177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trefts E., Gannon M., Wasserman D.H. The liver. Curr Biol. 2017;27:R1147–R1151. doi: 10.1016/j.cub.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun T., Yi H., Yang C., Kishnani P.S., Sun B. Starch binding domain-containing protein 1 plays a dominant role in glycogen transport to lysosomes in liver. J Biol Chem. 2016;291:16479–16484. doi: 10.1074/jbc.C116.741397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang S., Heller B., Tagliabracci V.S., Zhai L., Irimia J.M., DePaoli-Roach A.A., et al. Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J Biol Chem. 2010;285:34960–34971. doi: 10.1074/jbc.M110.150839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang S., Wells C.D., Roach P.J. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: identification of the Atg 8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem Biophys Res Commun. 2011;413:420–425. doi: 10.1016/j.bbrc.2011.08.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao H., Tang M., Liu M., Chen L. Glycophagy: an emerging target in pathology. Clin Chim Acta. 2018;484:298–303. doi: 10.1016/j.cca.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 30.Khambu B., Yan S., Huda N., Liu G., Yin X.M. Homeostatic role of autophagy in hepatocytes. Semin Liver Dis. 2018;38:308–319. doi: 10.1055/s-0038-1669939. [DOI] [PubMed] [Google Scholar]
- 31.Schworer C.M., Cox J.R., Mortimore G.E. Alteration of lysosomal density by sequestered glycogen during deprivation-induced autophagy in rat liver. Biochem Biophys Res Commun. 1979;87:163–170. doi: 10.1016/0006-291x(79)91661-9. [DOI] [PubMed] [Google Scholar]
- 32.Kotoulas A.O., Kotoulas O.B., Kalamidas S. An electron microscopic and biochemical study of the effects of cyclic 3′,5′-AMP, ergotamine or propranolol on the lysosomes of newborn rat hepatocytes. Histol Histopathol. 1991;6:421–426. [PubMed] [Google Scholar]
- 33.Kalamidas S.A., Kotoulas O.B., Kotoulas A.O., Maintas D.B. The breakdown of glycogen in the lysosomes of newborn rat hepatocytes: the effects of glucose, cyclic 3′,5′-AMP and caffeine. Histol Histopathol. 1994;9:691–698. [PubMed] [Google Scholar]
- 34.Kondomerkos D.J., Kalamidas S.A., Kotoulas O.B., Hann A.C. Glycogen autophagy in the liver and heart of newborn rats. The effects of glucagon, adrenalin or rapamycin. Histol Histopathol. 2005;20:689–696. doi: 10.14670/HH-20.689. [DOI] [PubMed] [Google Scholar]
- 35.Devos P., Hers H.G. Random, presumably hydrolytic, and lysosomal glycogenolysis in the livers of rats treated with phlorizin and of newborn rats. Biochem J. 1980;192:177–181. doi: 10.1042/bj1920177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalamidas S.A., Kondomerkos D.J. The administration of nonmetabolizable glucose analogues fails to suppress the development of glycogen autophagy in newborn rat hepatocytes. Microsc Res. 2010;73:1009–1014. doi: 10.1002/jemt.20825. [DOI] [PubMed] [Google Scholar]
- 37.Kotoulas O.B., Kalamidas S.A., Miles P., Hann A.C. An electron microscopic and biochemical study of the effects of propranolol on the glycogen autophagy in newborn rat hepatocytes. Histol Histopathol. 2003;18:811–818. doi: 10.14670/HH-18.811. [DOI] [PubMed] [Google Scholar]
- 38.Kalamidas S.A., Kondomerkos D.J., Kotoulas O.B., Hann A.C. Electron microscopic and biochemical study of the effects of rapamycin on glycogen autophagy in the newborn rat liver. Microsc Res. 2004;63:215–219. doi: 10.1002/jemt.20032. [DOI] [PubMed] [Google Scholar]
- 39.Kalamidas S.A., Kotoulas O.B., Hann A.C. Studies on glycogen autophagy: effects of phorbol myristate acetate, ionophore A23187, or phentolamine. Microsc Res. 2002;57:507–511. doi: 10.1002/jemt.10104. [DOI] [PubMed] [Google Scholar]
- 40.Madrigal-Matute J., Cuervo A.M. Regulation of liver metabolism by autophagy. J Gastroenterol. 2016;150:328–339. doi: 10.1053/j.gastro.2015.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komatsu M. Liver autophagy: physiology and pathology. J Biochem. 2012;152:5–15. doi: 10.1093/jb/mvs059. [DOI] [PubMed] [Google Scholar]
- 42.Schworer C.M., Shiffer K.A., Mortimore G.E. Quantitative relationship between autophagy and proteolysis during graded amino acid deprivation in perfused rat liver. J Biol Chem. 1981;256:7652–7658. [PubMed] [Google Scholar]
- 43.Mortimore G.E., Hutson N.J., Surmacz C.A. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc Natl Acad Sci U S A. 1983;80:2179–2183. doi: 10.1073/pnas.80.8.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ezaki J., Matsumoto N., Takeda-Ezaki M., Komatsu M., Takahashi K., Hiraoka Y., et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy. 2011;7:727–736. doi: 10.4161/auto.7.7.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsukamoto S., Kuma A., Murakami M., Kishi C., Yamamoto A., Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 46.Mortimore G.E., Pösö A.R. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu Rev Nutr. 1987;7:539–564. doi: 10.1146/annurev.nu.07.070187.002543. [DOI] [PubMed] [Google Scholar]
- 47.Seglen P.O., Gordon P.B., Poli A. Amino acid inhibition of the autophagic/lysosomal pathway of protein degradation in isolated rat hepatocytes. Biochim Biophys Acta. 1980;630:103–118. doi: 10.1016/0304-4165(80)90141-5. [DOI] [PubMed] [Google Scholar]
- 48.Lenk S.E., Bhat D., Blakeney W., Dunn W.A. Effects of streptozotocin-induced diabetes on rough endoplasmic reticulum and lysosomes of rat liver. Am J Physiol Endocrinol Metab. 1992;263:E856–E862. doi: 10.1152/ajpendo.1992.263.5.E856. [DOI] [PubMed] [Google Scholar]
- 49.Scott P.H., Brunn G.J., Kohn A.D., Roth R.A., Lawrence J.C., Jr. Evidence of insulin-stimulated phosphorylation and activation of the mammalian target of rapamycin mediated by a protein kinase B signaling pathway. Proc Natl Acad Sci U S A. 1998;95:7772–7777. doi: 10.1073/pnas.95.13.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raught B., Gingras A.C., Sonenberg N. The target of rapamycin (TOR) proteins. Proc Natl Acad Sci U S A. 2001;98:7037–7044. doi: 10.1073/pnas.121145898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ke P.Y. Diverse functions of autophagy in liver physiology and liver diseases. Int J Mol Sci. 2019;20:1–102. doi: 10.3390/ijms20020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naito T., Kuma A., Mizushima N. Differential contribution of insulin and amino acids to the mTORC1-autophagy pathway in the liver and muscle. J Biol Chem. 2013;288:21074–21081. doi: 10.1074/jbc.M113.456228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donohue T.M., Jr., Thomes P.G. Ethanol-induced oxidant stress modulates hepatic autophagy and proteasome activity. Redox Biol. 2014;3:29–39. doi: 10.1016/j.redox.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lilienbaum A. Relationship between the proteasomal system and autophagy. Int J Biochem Mol Biol. 2013;4:1–26. [PMC free article] [PubMed] [Google Scholar]
- 55.He A., Dean J.M., Lu D., Chen Y., Lodhi I.J. Hepatic peroxisomal β-oxidation suppresses lipophagy via RPTOR acetylation and MTOR activation. Autophagy. 2020;16:1727–1728. doi: 10.1080/15548627.2020.1797288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zechner R., Madeo F., Kratky D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat Rev Mol Cell Biol. 2017;18:671–684. doi: 10.1038/nrm.2017.76. [DOI] [PubMed] [Google Scholar]
- 57.Schulze R.J., Drižytė K., Casey C.A., McNiven M.A. Hepatic lipophagy: new insights into autophagic catabolism of lipid droplets in the liver. Hepatol Commun. 2017;1:359–369. doi: 10.1002/hep4.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaushik S., Cuervo A.M. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17:759–770. doi: 10.1038/ncb3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinez-Lopez N., Garcia-Macia M., Sahu S., Athonvarangkul D., Liebling E., Merlo P., et al. Autophagy in the CNS and periphery coordinate lipophagy and lipolysis in the brown adipose tissue and liver. Cell Metabol. 2016;23:113–127. doi: 10.1016/j.cmet.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sathyanarayan A., Mashek M.T., Mashek D.G. ATGL Promotes autophagy/lipophagy via sirt1 to control hepatic lipid droplet catabolism. Cell Rep. 2017;19:1–9. doi: 10.1016/j.celrep.2017.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schott M.B., Weller S.G., Schulze R.J., Krueger E.W., Drizyte-Miller K., Casey C.A., et al. Lipid droplet size directs lipolysis and lipophagy catabolism in hepatocytes. J Cell Biol. 2019;218:3320–3335. doi: 10.1083/jcb.201803153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schulze R.J., Krueger E.W., Weller S.G., Johnson K.M., Casey C.A., Schott M.B., et al. Direct lysosome-based autophagy of lipid droplets in hepatocytes. Proc Natl Acad Sci U S A. 2020;117:32443. doi: 10.1073/pnas.2011442117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams J.A., Ding W.X. Targeting Pink1–Parkin-mediated mitophagy for treating liver injury. Pharmacol Res. 2015;102:264–269. doi: 10.1016/j.phrs.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Novak I., Kirkin V., McEwan D.G., Zhang J., Wild P., Rozenknop A., et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu L., Feng D., Chen G., Chen M., Zheng Q., Song P., et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 68.Kagan V.E., Jiang J., Huang Z., Tyurina Y.Y., Desbourdes C., Cottet-Rousselle C., et al. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell Death Differ. 2016;23:1140–1151. doi: 10.1038/cdd.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang J., Tripathi D.N., Jing J., Alexander A., Kim J., Powell R.T., et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol. 2015;17:1259–1269. doi: 10.1038/ncb3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tripathi D.N., Chowdhury R., Trudel L.J., Tee A.R., Slack R.S., Walker C.L., et al. Reactive nitrogen species regulate autophagy through ATM–AMPK–TSC2-mediated suppression of mTORC1. Proc Natl Acad Sci U S A. 2013;110:E2950–E2957. doi: 10.1073/pnas.1307736110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deosaran E., Larsen K.B., Hua R., Sargent G., Wang Y., Kim S., et al. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci. 2013;126:939–952. doi: 10.1242/jcs.114819. [DOI] [PubMed] [Google Scholar]
- 72.Sargent G., van Zutphen T., Shatseva T., Zhang L., Di Giovanni V., Bandsma R., et al. PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J Cell Biol. 2016;214:677–690. doi: 10.1083/jcb.201511034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Law K.B., Bronte-Tinkew D., Di Pietro E., Snowden A., Jones R.O., Moser A., et al. The peroxisomal AAA ATPase complex prevents pexophagy and development of peroxisome biogenesis disorders. Autophagy. 2017;13:868–884. doi: 10.1080/15548627.2017.1291470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yin X.M., Ding W.X., Gao W. Autophagy in the liver. J Hepatol (Baltimore) 2008;47:1773–1785. doi: 10.1002/hep.22146. [DOI] [PubMed] [Google Scholar]
- 75.Feldman D., Swarm R.L., Becker J. Elimination of excess smooth endoplasmic reticulum after phenobarbital administration. J Histochem Cytochem. 1980;28:997–1006. doi: 10.1177/28.9.7410819. [DOI] [PubMed] [Google Scholar]
- 76.Bolender R.P., Weibel E.R. A morphometric study of the removal of phenobarbital-induced membranes from hepatocytes after cessation of treatment. J Cell Biol. 1973;56:746–761. doi: 10.1083/jcb.56.3.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Masaki R., Yamamoto A., Tashiro Y. Cytochrome P-450 and NADPH-cytochrome P-450 reductase are degraded in the autolysosomes in rat liver. J Cell Biol. 1987;104:1207–1215. doi: 10.1083/jcb.104.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kouroku Y., Fujita E., Tanida I., Ueno T., Isoai A., Kumagai H., et al. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 79.Fuest M., Willim K., MacNelly S., Fellner N., Resch G.P., Blum H.E., et al. The transcription factor c-Jun protects against sustained hepatic endoplasmic reticulum stress thereby promoting hepatocyte survival. J Hepatol. 2012;55:408–418. doi: 10.1002/hep.24699. [DOI] [PubMed] [Google Scholar]
- 80.Yorimitsu T., Nair U., Yang Z., Klionsky D.J. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chiang J.Y.L., Ferrell J.M. Bile Acid metabolism in liver pathobiology. Gene Expr. 2018;18:71–87. doi: 10.3727/105221618X15156018385515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y., Ding W.X., Li T. Cholesterol and bile acid-mediated regulation of autophagy in fatty liver diseases and atherosclerosis. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863:726–733. doi: 10.1016/j.bbalip.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khambu B., Li T., Yan S., Yu C., Chen X., Goheen M., et al. Hepatic autophagy deficiency compromises farnesoid X receptor functionality and causes cholestatic injury. Hepatology. 2019;69:2196–2213. doi: 10.1002/hep.30407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yan S., Khambu B., Chen X., Dong Z., Guo G., Yin X.M. Hepatic autophagy deficiency remodels gut microbiota for adaptive protection via FGF15-FGFR4 signaling. Cell Mol Gastroenterol Hepatol. 2020;11:973–997. doi: 10.1016/j.jcmgh.2020.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seok S., Fu T., Choi S.E., Li Y., Zhu R., Kumar S., et al. Transcriptional regulation of autophagy by an FXR–CREB axis. Nature. 2014;516:108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee J.M., Wagner M., Xiao R., Kim K.H., Feng D., Lazar M.A., et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Manley S., Ni H.M., Kong B., Apte U., Guo G., Ding W.X. Suppression of autophagic flux by bile acids in hepatocytes. Toxicol Sci. 2014;137:478–490. doi: 10.1093/toxsci/kft246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Panzitt K., Jungwirth E., Krones E., Lee J.M., Pollheimer M., Thallinger G.G., et al. FXR-dependent Rubicon induction impairs autophagy in models of human cholestasis. J Hepatol. 2020;72:1122–1131. doi: 10.1016/j.jhep.2020.01.014. [DOI] [PubMed] [Google Scholar]
- 89.Wang Y., Ding Y., Li J., Chavan H., Matye D., Ni H.M., et al. Targeting the enterohepatic bile acid signaling induces hepatic autophagy via a CYP7A1–AKT–mTOR axis in mice. Cell Mol Gastroenterol Hepatol. 2017;3:245–260. doi: 10.1016/j.jcmgh.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheng J., Ohsaki Y., Tauchi-Sato K., Fujita A., Fujimoto T. Cholesterol depletion induces autophagy. Biochem Biophys Res Commun. 2006;351:246–252. doi: 10.1016/j.bbrc.2006.10.042. [DOI] [PubMed] [Google Scholar]
- 91.Yamamura T., Ohsaki Y., Suzuki M., Shinohara Y., Tatematsu T., Cheng J., et al. Inhibition of Niemann-Pick-type C1-like 1 by ezetimibe activates autophagy in human hepatocytes and reduces mutant α1-antitrypsin Z deposition. J Hepatol. 2014;59:1591–1599. doi: 10.1002/hep.26930. [DOI] [PubMed] [Google Scholar]
- 92.Wang H.J., Park J.Y., Kwon O., Choe E.Y., Kim C.H., Hur K.Y., et al. Chronic HMGCR/HMG-CoA reductase inhibitor treatment contributes to dysglycemia by upregulating hepatic gluconeogenesis through autophagy induction. Autophagy. 2015;11:2089–2101. doi: 10.1080/15548627.2015.1091139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fucho R., Martínez L., Baulies A., Torres S., Tarrats N., Fernandez A., et al. ASMase regulates autophagy and lysosomal membrane permeabilization and its inhibition prevents early stage non-alcoholic steatohepatitis. J Hepatol. 2014;61:1126–1134. doi: 10.1016/j.jhep.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marí M., Caballero F., Colell A., Morales A., Caballeria J., Fernandez A., et al. Mitochondrial free cholesterol loading sensitizes to TNF- and Fas-mediated steatohepatitis. Cell Metabol. 2006;4:185–198. doi: 10.1016/j.cmet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 95.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 96.Karsli-Uzunbas G., Guo J.Y., Price S., Teng X., Laddha S.V., Khor S., et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Canc Discov. 2014;4:914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lamia K.A., Sachdeva U.M., DiTacchio L., Williams E.C., Alvarez J.G., Egan D.F., et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Toledo M., Batista-Gonzalez A., Merheb E., Aoun M.L., Tarabra E., Feng D., et al. Autophagy regulates the liver clock and glucose metabolism by degrading CRY1. Cell Metabol. 2018;28:268–281. doi: 10.1016/j.cmet.2018.05.023. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lamia K.A., Papp S.J., Yu R.T., Barish G.D., Uhlenhaut N.H., Jonker J.W., et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang E.E., Liu Y., Dentin R., Pongsawakul P.Y., Liu A.C., Hirota T., et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shibata M., Yoshimura K., Furuya N., Koike M., Ueno T., Komatsu M., et al. The MAP1–LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun. 2009;382:419–423. doi: 10.1016/j.bbrc.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 102.Mottis A., Mouchiroud L., Auwerx J. Emerging roles of the corepressors NCoR 1 and SMRT in homeostasis. Genes Dev. 2013;27:819–835. doi: 10.1101/gad.214023.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lin C.W., Zhang H., Li M., Xiong X., Chen X., Chen X., et al. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol. 2013;58:993–999. doi: 10.1016/j.jhep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ding W.X., Li M., Chen X., Ni H.M., Lin C.W., Gao W., et al. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. J Gastroenterol. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stofan M., Guo G.L. Bile acids and FXR: novel targets for liver diseases. Front Med. 2020;7:1–12. doi: 10.3389/fmed.2020.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stedman C., Liddle C., Coulter S., Sonoda J., Alvarez J.G., Evans R.M., et al. Benefit of farnesoid X receptor inhibition in obstructive cholestasis. Proc Natl Acad Sci U S A. 2006;103:11323–11328. doi: 10.1073/pnas.0604772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Patel D., Teckman J.H. Alpha-1-antitrypsin deficiency liver disease. Clin Liver Dis. 2018;22:643–655. doi: 10.1016/j.cld.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 108.Khambu B., Wang L., Zhang H., Yin X.M. The activation and function of autophagy in alcoholic liver disease. Curr Mol Pharmacol. 2017;10:165–171. doi: 10.2174/1874467208666150817112654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sarkar S., Floto R.A., Berger Z., Imarisio S., Cordenier A., Pasco M., et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kong D., Zhang Z., Chen L., Huang W., Zhang F., Wang L., et al. Curcumin blunts epithelial-mesenchymal transition of hepatocytes to alleviate hepatic fibrosis through regulating oxidative stress and autophagy. Redox Biol. 2020;36:101600. doi: 10.1016/j.redox.2020.101600. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 111.Lee D.E., Lee S.J., Kim S.J., Lee H.S., Kwon O.S. Curcumin ameliorates nonalcoholic fatty liver disease through inhibition of O-GlcNAcylation. Nutrients. 2019;11:2702. doi: 10.3390/nu11112702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chang E., Kim L., Park S.E., Rhee E.J., Lee W.Y., Oh K.W., et al. Ezetimibe improves hepatic steatosis in relation to autophagy in obese and diabetic rats. World J Gastroenterol. 2015;21:7754–7763. doi: 10.3748/wjg.v21.i25.7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee D.H., Han D.H., Nam K.T., Park J.S., Kim S.H., Lee M., et al. Ezetimibe, an NPC1L1 inhibitor, is a potent Nrf 2 activator that protects mice from diet-induced nonalcoholic steatohepatitis. Free Radic Biol Med. 2016;99:520–532. doi: 10.1016/j.freeradbiomed.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 114.Panzitt K., Fickert P., Wagner M. Regulation of autophagy by bile acids and in cholestasis—CholestoPHAGY or CholeSTOPagy. Biochim Biophys Acta Mol Basis Dis. 2021;1867:166017. doi: 10.1016/j.bbadis.2020.166017. [DOI] [PubMed] [Google Scholar]
- 115.Panda P.K., Fahrner A., Vats S., Seranova E., Sharma V., Chipara M., et al. Chemical screening approaches enabling drug discovery of autophagy modulators for biomedical applications in human diseases. Front Cell Dev Bio. 2019;7:1–19. doi: 10.3389/fcell.2019.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Weiskirchen R., Tacke F. Relevance of autophagy in parenchymal and non-parenchymal liver cells for health and disease. Cells. 2019;8:1–13. doi: 10.3390/cells8010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li Y., Liu L., Wang B., Wang J., Chen D. Metformin in non-alcoholic fatty liver disease: a systematic review and meta-analysis. Biomed Rep. 2013;1:57–64. doi: 10.3892/br.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chiang J.Y.L. Bile acid metabolism and signaling. Comprehensive physiology. 2013;3:1191–1212. doi: 10.1002/cphy.c120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tang Y., Blomenkamp K.S., Fickert P., Trauner M., Teckman J.H. NorUDCA promotes degradation of α1-antitrypsin mutant Z protein by inducing autophagy through AMPK/ULK1 pathway. PLoS One. 2018;13 doi: 10.1371/journal.pone.0200897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Faghihzadeh F., Hekmatdoost A., Adibi P. Resveratrol and liver: a systematic review. J Res Med Sci. 2015;20:797–810. doi: 10.4103/1735-1995.168405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Soria L.R., Perocheau D.P., De Sabbata G., De Angelis A., Bruno G., Polishchuk E., et al. Beclin-1-mediated activation of autophagy improves proximal and distal urea cycle disorders. bioRxiv. 2020 doi: 10.15252/emmm.202013158. https://www.biorxiv.org/content/10.1101/2020.07.22.216382v1 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shoji-Kawata S., Sumpter R., Leveno M., Campbell G.R., Zou Z., Kinch L., et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sarkar S., Davies J.E., Huang Z., Tunnacliffe A., Rubinsztein D.C. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 124.Han C.Y. Update on FXR biology: promising therapeutic target? Int J Mol Sci. 2018;19:2069. doi: 10.3390/ijms19072069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu W.B., Chen Y.Y., Zhu B., Peng X.M., Zhang S.W., Zhou M.L. Excessive bile acid activated NF-kappa B and promoted the development of alcoholic steatohepatitis in farnesoid X receptor deficient mice. Biochimie. 2015;115:86–92. doi: 10.1016/j.biochi.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 126.Chao X., Wang S., Zhao K., Li Y., Williams J.A., Li T., et al. Impaired TFEB-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. J Gastroenterol. 2018;155:865–879. doi: 10.1053/j.gastro.2018.05.027. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thoreen C.C., Kang S.A., Chang J.W., Liu Q., Zhang J., Gao Y., et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yan S., Huda N., Khambu B., Yin X.M. Relevance of autophagy to fatty liver diseases and potential therapeutic applications. Amino Acids. 2017;49:1965–1979. doi: 10.1007/s00726-017-2429-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Feldman M.E., Apsel B., Uotila A., Loewith R., Knight Z.A., Ruggero D., et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Malik S.A., Orhon I., Morselli E., Criollo A., Shen S., Mariño G., et al. BH3 mimetics activate multiple pro-autophagic pathways. Oncogene. 2011;30:3918–3929. doi: 10.1038/onc.2011.104. [DOI] [PubMed] [Google Scholar]
- 131.Narayanan P., Mistry P.K. Update on alpha-1 antitrypsin deficiency in liver disease. Clin Liver Dis. 2020;15:228–235. doi: 10.1002/cld.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Renga B., Mencarelli A., D'Amore C., Cipriani S., D'Auria M.V., Sepe V., et al. Discovery that theonellasterol a marine sponge sterol is a highly selective FXR antagonist that protects against liver injury in cholestasis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Patel K., Harrison S.A., Elkhashab M., Trotter J.F., Herring R., Rojter S.E., et al. Cilofexor, a nonsteroidal FXR agonist, in patients with noncirrhotic NASH: a phase 2 randomized controlled trial. J Hepatol. 2020;72:58–71. doi: 10.1002/hep.31205. [DOI] [PubMed] [Google Scholar]
- 134.Qiao X., Zhou Z.C., Niu R., Su Y.T., Sun Y., Liu H.L., et al. Hydroxychloroquine improves obesity-associated insulin resistance and hepatic steatosis by regulating lipid metabolism. Front Pharmacol. 2019;10:855. doi: 10.3389/fphar.2019.00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tully D.C., Rucker P.V., Chianelli D., Williams J., Vidal A., Alper P.B., et al. Discovery of tropifexor (LJN452), a highly potent non-bile acid fxr agonist for the treatment of cholestatic liver diseases and nonalcoholic steatohepatitis (NASH) J Med Chem. 2017;60:9960–9973. doi: 10.1021/acs.jmedchem.7b00907. [DOI] [PubMed] [Google Scholar]