Abstract
The relationship between chronic psychological stress and tumorigenesis has been well defined in epidemiological studies; however, the underlying mechanism remains underexplored. In this study, we discovered that impaired macrophage phagocytosis contributed to the psychological stress-evoked tumor susceptibility, and the stress hormone glucocorticoid (GC) was identified as a principal detrimental factor. Mechanistically, GC disturbed the balance of the “eat me” signal receptor (low-density lipoprotein receptor-related protein-1, LRP1) and the “don't eat me” signal receptor (signal regulatory protein alpha, SIRPα). Further analysis revealed that GC led to a direct, glucocorticoid receptor (GR)-dependent trans-repression of LRP1 expression, and the repressed LRP1, in turn, resulted in the elevated gene level of SIRPα by down-regulating miRNA-4695-3p. These data collectively demonstrate that stress induces the imbalance of the LRP1/SIRPα axis and entails the disturbance of tumor cell clearance by macrophages. Our findings provide the mechanistic insight into psychological stress-evoked tumor susceptibility and indicate that the balance of LRP1/SIRPα axis may serve as a potential therapeutic strategy for tumor treatment.
KEY WORDS: Psychological stress, Tumorigenesis, Macrophages, Phagocytosis, LRP1, SIRPα, Therapeutic target
Graphical abstract
Stress-induced glucocorticoid inhibits the expression of “eat me” signal receptor LRP1 by GR-dependent trans-repression, which in turn raises the protein level of “don't eat me” signal receptor SIRPα in an miRNA-4695-3p-dependent manner. The imbalance of LRP1 and SIRPα disrupted by glucocorticoid thus suppresses the phagocytosis of tumor cells by macrophages.
1. Introduction
The relationship between chronic psychological stress and tumorigenesis has been intensively investigated in epidemiological researches1, 2, 3, 4. Fundamental studies further revealed that chronic stress-induced hormones promote tumor metastasis via the activation of their receptors5, and increase stem-like properties in tumor cells via enhancing the lactate dehydrogenase A dependent metabolic rewriting6. However, research so far has mainly focused on the detrimental influence of stress on tumor cells themselves, ignoring the role of stress on immune cells in the tumor microenvironment. Recently, increasing attention has been paid to tumor immunosurveillance in the prevention and treatment of tumors7,8. In particular, macrophages, the most infiltrating immune cells in tumor tissues, can recognize and phagocytose tumor cells through non-specific immunity9, followed by T cell-mediated specific immunity, which involves the degradation of tumor cells and the expression of major histocompatibility complex classes I and II on the cell surface10,11. It is worth to note that a large number of macrophages are required to phagocyte apoptotic tumor cells timely during radiotherapy or chemotherapy, which is critical in promoting the therapeutic effect12. These lines of evidence indicate that macrophage-based phagocytosis is a critical mediator of tumor immunosurveillance. Therefore, disturbance to macrophage function is expected to affect tumor growth and development. Earlier studies from our laboratory and other groups have suggested that psychological stress deteriorated the phagocytosis capacity of macrophages in mice13, 14, 15, raising the possibility that chronic stress could facilitate tumor growth by inhibiting macrophage function. However, whether stress-augmented tumor growth is associated with the dysfunction of macrophage-induced phagocytosis has not been rigorously tested.
Tremendous progress has been made in recent years in identifying a constellation of “eat me” and “don't eat me” signals expressed on tumor cells and their corresponding receptors on macrophages16,17. As a dominant “eat me” signal, calreticulin is highly expressed on the surface of drug-treated or apoptotic tumor cells and binds to its receptor, low-density lipoprotein receptor-related protein-1 (LRP1) on the surface of macrophages, thus promoting the reorganization of actin and phagocytosis of tumor cells18, 19, 20. On the contrary, CD47 and its receptor signal regulatory alpha (SIRPα) act as a pivotal “don't eat me” signal in counteracting the “eat me” signal-elicited macrophages' phagocytosis16,21,22. Interestingly, a previous study revealed that blocking the interaction of calreticulin with LRP1 prevented anti-CD47 antibody-mediated phagocytosis, highlighting the balance between pro- and anti-phagocytic signals in immune evasion of tumor20. This intriguing observation serves as a starting point for us to speculate whether there lies a certain axis between “eat me” and “don't eat me” signal receptors on macrophages, through which they are counterbalanced by each other.
In this study, we discovered that impaired macrophage phagocytosis contributed to the psychological stress-evoked tumor susceptibility, and the stress hormone glucocorticoid (GC) was identified as a principle detrimental factor. In deciphering GC-restrained phagocytosis of tumor cells by macrophages, a disturbed balance of the LRP1–SIRPα axis was revealed to be the main culprit. GC led to a direct, glucocorticoid receptor (GR)-dependent trans-repression of LRP1 expression, and the repressed LRP1 in turn resulted in the elevated protein level of SIRPα by down-regulating miRNA-4695-3p. Our findings, from the perspective of macrophage phagocytosis, uncover that “eat me” and “don't eat me” signal receptors expressed on macrophages are limited by each other. These results shed significant insights into the mechanism underlying chronic stress-evoked tumorigenesis and provide potential innovative strategies for preventing and treating tumors.
2. Materials and methods
2.1. Animals and treatment
Female BALB/c wild-type (WT) mice (6–8 weeks old) and nude mice (4–5 weeks old) were purchased from Guangdong Medical Laboratory Animal Center and Laboratory Animal Center of Southern Medical University. The mice were maintained a 12 h light/dark cycle at 22 ± 2 °C and adaptive feed for a week before the experiment. The animal experiment protocol has been reviewed and approved by Laboratory Animal Ethic Committee of Jinan University, Guangzhou, China.
WT mice were randomly divided into five groups: control, stress, corticosterone (CORT), “mifepristone (Ru486, an antagonist of GR) + stress”, “CORT + Ru486”. CORT and Ru486 were purchased from Sigma–Aldrich (MO, USA) and dissolved in polyethylene glycol 400. As shown in Fig. 1A, animals were daily received 2-h restraint stress, CORT (2 mg/kg/day, s.c.) or Ru486 (25 mg/kg/day, s.c.) treatment for 28 days. On the 7th day, mice were subcutaneously injected with 4T1 cells (1 × 106 cells in 200 μL). Mice in stress group were restraint in a ventilated plastic centrifuge tubes of 50 mL, maintained horizontally in their home cages during the restraint sessions and released into the same cage during the free sessions.
Figure 1.
Stress hormone GC-promoted tumor growth is link to macrophage function. (A) Experiment procedures for establishing restraint stress-induced tumor susceptibility in WT mice. Animals were divided into five groups (n = 5), including control, stress, CORT, “Stress + Ru486” and “CORT + Ru486”. They were received daily 2 h restraint stress, CORT (2 mg/kg, s.c.) or GR antagonist Ru486 (25 mg/kg, s.c.) treatment for 28 days, 4T1 cells were subcutaneously inoculated to mice on the 7th day of stress. (B) Tumor growth of all mice was monitored every 3 days for 21 days. (C) Tumor weight was recorded after mice sacrificed on the 22nd day. (D) Plasma CORT level was detected by HPLC–UV method. (E) Schematic protocol in T lymphocyte-deficient nude mice. Details are described in the Materials and methods section. (F) Tumor growth, (G) tumor weight and (H) photographs of excised MDA-MB-231 solid tumor from nude mice (n = 5). Scale bar = 1 cm. (I) Schematic protocol in macrophages-depleted mice. Details are described in the Materials and methods section. (J) Tumor growth, (K) tumor weight and (L) photographs of excised 4T1 solid tumors (n = 6). Scale bar = 1 cm. GC, glucocorticoid; CORT, corticosterone (the main type of GC in rodents); Ru486, mifepristone; VBL, vinblastine; CL, clodronate liposomes. The values are represented as mean ± SD. ns, not significant. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
In order to exclude the involvement of T cells in stress-evoked tumor growth, T lymphocyte-deficient nude mice were utilized. They were randomly divided into four groups, including control, CORT, vinblastine (VBL) and “VBL + CORT”. Mice in CORT and “CORT + VBL” groups were subcutaneously injected with 2 mg/kg CORT for 28 days. MDA-MB-231 cells (2 × 106 cells in 200 μL) were subcutaneously injected to each mouse after 7 days of CORT treatment. VBL (0.5 mg/kg) were intratumorally injected to mice every 3 days on the 7th day after tumor cells injection (Fig. 1E).
Subsequently, macrophages-depleted mice were established by clodronate liposomes (CL, Liposoma BV, Amsterdam, The Netherlands) to explore the potential role of macrophages. As seen from Fig. 2I, mice were received 28 days of CORT (2 mg/kg/day, s.c., daily). On the 7th day, they were subcutaneously injected with 4T1 cells (1 × 106 cells in 200 μL). Seven days before 4T1 cells inoculation, macrophages in mice were depleted by a 200-μL injection (i.p.) of CL. This depletion was conducted every 6 days.
Figure 2.
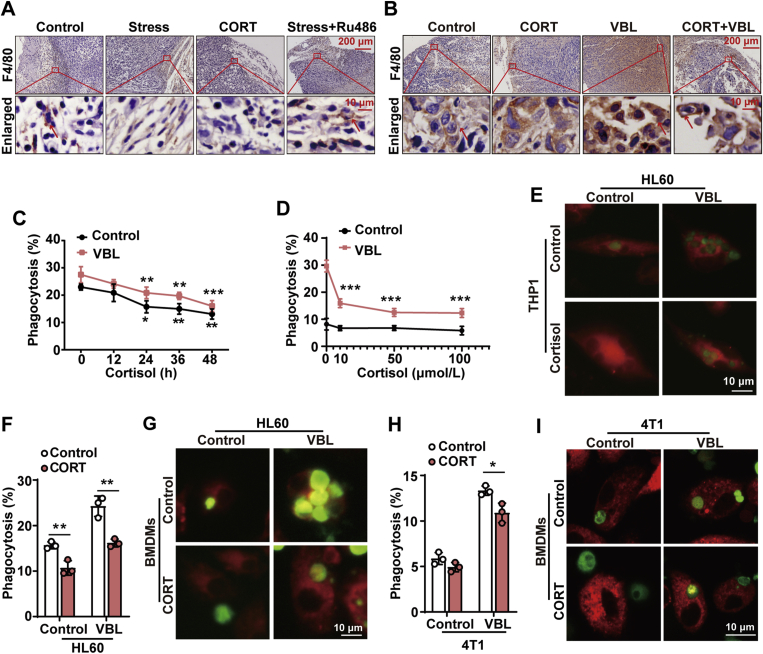
Diminished macrophages-mediated phagocytosis of tumor cells contributes to stress-provoked tumor growth. (A) and (B) Immunohistochemistry analysis of macrophages stained with F4/80 in tumor tissue section from WT mice and nude mice. Tumor cell phagocytosis by macrophages was observed and indicated by red arrow. (C)–(E) THP1 macrophages were co-incubated with CMFDA-labeled HL60 tumor cells (pretreated with or without VBL) for 2 h, and then macrophages phagocytosis was assessed by flow cytometry. (C) Macrophages were treated with or without cortisol (50 μmol/L) for different time periods (12, 24, 36, and 48 h). (D) THP1 macrophages were treated with or without different concentrations of cortisol (10, 50, and 100 μmol/L) for 24 h. (E) Representative phagocytosis of HL60 tumor cells by THP1 macrophages was recorded by confocal microscopy. Scale bar = 10 μm. (F)–(I) BMDMs were treated with CORT (100 μmol/L) for 24 h and co-incubated with HL60 cells (F, G) or 4T1 cells (H, I) for 2 h. Phagocytosis was assessed by flow cytometry, and representative images were recorded by confocal microscopy. Scale bar = 10 μm. The values are represented as mean ± SD (n = 3). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. WT, wildtype; CORT, corticosterone; Ru486, mifepristone; VBL, vinblastine; cortisol, the main type of GC in human being. BMDMs, bone marrow-derived macrophages.
The body weight of each mouse was measured every day. Tumor size was measured every 3 days using calipers, and tumor volume was calculated using Eq. (1):
| (1) |
Mice were necropsied on the 21st day after tumor injection. Tumor tissues were collected, weighted, photographed.
2.2. Cell culture and treatment
4T1 cells and HL60 cells were cultured in RPMI medium 1640 (Thermo Fisher Scientific, MA, USA), and MDA-MB-231 cells were cultured in DMEM (Thermo Fisher Scientific), containing 10% fetal bovine serum (PAN biotech, Baglia, Germany) in an incubator containing 5% CO2 at 37 °C. Human peripheral blood mononuclear cell line (THP1 cells) was maintained at 1640-RPMI medium supplemented with 10% fetal bovine serum, 2 mmol/L l-glutamine and 50 μmol/L β-mercaptoethanol. THP1 cells were differentiated into macrophages using 200 nmol/L phorbol 12-myristate 13-acetate (Sigma–Aldrich) for 72 h.
2.3. Histomorphology and immunohistochemistry
After 4% paraformaldehyde-fixation, tumor samples were embedded in paraffin, sliced in a thickness of 4 μm. For the immunohistochemistry on studying macrophages phagocytosis in tumor tissue, sections of tumor tissue were deparaffinized using xylene, hydrated with a graded series of alcohol, and boiled in citrate solution (pH = 6) for 8 min for antigen retrieval. Anti-F4/80 antibody (#ab6640, Abcam, MA, USA) was staining at 4 °C overnight. Staining was visualized using DAB Detection Kit (Gene Tech, Shanghai, China) according to the manufacturer's instructions. The images of tissues were characterized by the M8 Microscope and Scanner (PreciPoint, Freising, Germany).
2.4. Immunofluorescence staining
For the cell immunofluorescence staining, cells were fixed in 4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton-X100 for 5 min, and then incubated with glucocorticoid receptor (D6H2L) XP rabbit mAb (#12041, Cell Signaling Technology, MA, USA) at 4 °C overnight. Then, cells were incubated with goat anti-rabbit IgG (H + L) cross-adsorbed secondary antibody, Alexa Fluor 488 (A-11008, Thermo Fisher Scientific) for 1 h. Last, the cell nuclei were stained using DAPI staining solution (Beyotine, Shanghai, China). Immunofluorescence images of cells were recorded using Laser Scanning Confocal Microscopy (LSM800, Zeiss, Oberkohen, Germany).
For the tissue immunofluorescence staining, tumor tissue was fixed on freeing microtome using embedding medium, sliced in a thickness of 6 μm. The sections of tumor tissue were fixed with 4% paraformaldehyde for 15 min, and then incubated with recombinant anti-LRP1 antibody (#ab92544, Abcam) and anti-F4/80 antibody (#ab6640, Abcam) at 4 °C overnight. Then, cells were incubated with donkey anti-rat IgG (H + L) highly cross-adsorbed secondary antibody, Alexa Fluor 594 (A-21209, Thermo Fisher Scientific) for 1 h. Last, the section was stained using DAPI for 10 min and mounted using anti-fluorescence quenching agent. The immunofluorescence images were recorded using LX51 Inverted Microscope (Olympus, Tokyo, Japan) equipped with DP70 Digital Camera System (Olympus, Tokyo, Japan).
2.5. Detection of CORT level in the plasma
Blood was collected from mice and centrifuged at 2500×g for 10 min to obtain plasma. Cortisone (Sigma–Aldrich) was added to plasma as internal standard. The steroids were extracted by ethylacetate, washed with 0.1 mol/L NaOH solution and double distilled water, dried with nitrogen. The samples were reconstituted with liquid phase (acetonitrile:water = 45:55, v/v). CORT level in the plasma of mice was quantitatively analyzed by high performance liquid chromatography–ultraviolet, equipped with C18 reversed phase column (5 μm, 4.6 mm I.D. × 250 mm). The flow rate was 0.8 mL/min, and the detection wavelength was 254 nm.
2.6. Western bolt analysis
Total proteins from cells or tumor tissue were isolated using whole cell lysis buffer (Beyotine, Shanghai, China) and quantified by Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Briefly, the equal amounts proteins of each group were separated on sodium dodecyl sulfate-polyacrylamide gel and transferred into PVDF membrane (Merk Millipore, MA, USA). The membranes were blocked with non-fat milk for 2 h and incubated with recombinant anti-LRP1 antibody and recombinant anti-SIRP alpha antibody (#ab92544 and ab191419, Abcam) overnight at 4 °C, and then incubated with HRP AffiniPure goat anti-rabbit IgG (H + L) (FD0128, Fude Biological Technology, Hangzhou, China) at room temperature for 2 h. Immunoreactive proteins were detected using FDbio-Pico ECL Kit (FD8000, Fude Biological Technology, Hangzhou, China) and Tanon 5200 Chemiluminescent Imaging System (Tanon, Shanghai, China).
2.7. Quantitative polymerase chain reaction (qPCR) analysis
Total RNA from cells and tumor tissues were isolated by TRIzol reagent (15596026, Thermo Fisher Scientific) and converted into cDNA using TranScript One-Step gDNA Removal and cDNA Synthesis Supermix Kit (AT311-02, Transgen Biotech, Beijing, China) according to the manufacturer's instructions. qPCR was performed using TransStart Top Green qPCR SuperMix Kit (AQ131-01, Transgen Biotech, Beijing, China) and CFX Connect Real-Time PCR Detection System (Bio-Rad Laboratories, CA, USA). The specific primers (Genray Biotech, Shanghai, China) used in qPCR are described in the Supporting Information Table S1. Relative mRNA expression of each gene was normalized with housekeeping gene through the delta cycle threshold method.
2.8. Isolation and differentiation of mouse bone marrow-derived macrophages (BMDMs)
The isolation and differentiation of BMDMs was performed according to the reported method23. Mice (6–8 weeks old, C57BL/6) were anesthetized and immersed in 75% alcohol for disinfection. Two femurs of each mouse were obtained in clean beach and put into a Petri dish containing sterile and cold phosphate buffer solution (PBS). Removal of muscles on femurs, the bone marrow cavity of femurs was flushed using cold PBS and passing through Falcon Cell Strainer (352350, BD Biosciences, NJ, USA) to obtain single cell suspensions. Cells were centrifuged at 200×g for 5 min, suspended in complete medium and counted. After adjusting the concentration, cells were plated in a six-well plate, cultured in DMEM/F12-complete containing recombinant murine M-CSF (100 ng/mL, Peprotech, NJ, USA) for 7 days, marked with F4/80 and identified by flow cytometry.
2.9. Assessment of phagocytosis capacity of tumor cells by macrophages
The assessment of phagocytosis was performed as previously described24,25. Tumor cells (HL60 cells) were labeled with 1 μmol/L CellTracker Green CMFDA Dye (C7025, Thermo Fisher Scientific), and treated in the presence or in the absence of VBL (an apoptosis inducer) for 18 h. Tumor cells were collected, washed with PBS to remove excess dye, and co-cultured with macrophages (THP1 macrophages or BMDMs) (macrophages: tumor cells = 1:10) in serum-free medium for 2 h at 37 °C in a CO2 incubator. Next, after removing the unbound/unengulfed tumor cells, macrophages were labeled with PE anti-human CD11b antibody or PE anti-mouse F4/80 antibody (301,306 and 123,110, BioLegend, CA, USA) and analyzed by flow cytometry (FACSAria II, BD Biosciences, NJ, USA). Data analysis was performed using FlowJo software (v.10.0.8). F4/80+CMFDA+ or CD11b+CMFDA+ double-positive ratio indicates the phagocytosis percentage of tumor cells by macrophages.
For imaging the capacity of macrophage phagocytosis, tumor cells and macrophages were incubated with CMFDA (1 μmol/L) and CellTracker Red CMPTX dyes (1 μmol/L, C34552, Thermo Fisher Scientific) for 30 min at 37 °C, respectively. Cells were washed with PBS to remove excess dye, and co-cultured at a ratio of 1:10 (macrophages:tumor cells) in serum-free medium for 2 h. The unbound/unengulfed targets were removed and macrophages were fixed with 4% paraformaldehyde before imaging. The phagocytosis of tumor cells by macrophages was recorded by Laser Scanning Confocal Microscopy (LSM800, Zeiss, Oberkohen, Germany).
2.10. Luciferase reporter assay
HEK293 cells in a 48-well plate were transfected with LRP1-promoter luciferase reporter plasmids (200 ng), pRL-TK vector (an internal control with renilla luciferase gene, 50 ng), and GR plasmids (200 ng). The transfection assays were performed using DNA transfection reagent (Neofect Biotech) according to the manufacturer's protocols. On the next day, the DMEM was changed with or without cortisol (100 μmol/L). The luciferase activities were determined by the Dual-Luciferase Reporter Assay System and GloMax 20/20 luminometer (Promega, WI, USA). The relative luciferase activity values were normalized to control group.
2.11. miRNA expression profiling
Total RNA from THP1 macrophages were isolated by TRIzol Reagent (Thermo Fisher Scientific) and precipitated in equivalent isopropanol (v/v) with 1 μL glycogen overnight at −20 °C. mRNA library was constructed using MGIEasy Small RNA Library Prep Kit (MGI, Shenzhen, China) following the manufacturer's instructions. Libraries were sequenced on an BGISEQ 500 sequencer for paired-end RNA-seq with a read length of 100 bp. Adapters were trimmed from the reads, and the reads shorter than 17 nt were discarded. The reads were mapped to the human mRNA reference database using FANSe3 algorithm26 on Chi-Cloud NGS Analysis Platform (Chi-Biotech, Shenzhen, China). The significant changed miRNAs were screened using edgeR method, and the screening threshold for significant gene was set as following: log (fold change) > 1 and P value < 0.05. The significantly changed miRNAs were verified by qPCR in THP1 macrophages.
2.12. Transfection of siRNA, plasmids, miRNA mimic and inhibitor
The small interfering RNA (siRNA) targeting LRP1 (GenePharma, Shanghai, China) were transfected to THP1 macrophages or HEK293 cells, and a non-targeting siRNA was used as negative control. The transfection was performed using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer's instructions. Membrane-containing LRP1 minireceptors (mLRP1, mLRP2, mLRP3, and mLRP4) plasmids were constructed according to previous research27, 28, 29, transfected to HEK293 cells treated with or without cortisol (50 μmol/L) using DNA transfection reagent. miRNA-4695-3p mimic (25 nmol/L), miRNA-4695-3p inhibitor (100 nmol/L) and scrambled oligonucleotides (negative control, NC) (Ribo Bio, Guangzhou, China) were transfected to THP1-differentiated macrophages. Cells were harvested 24 h after transfection for qPCR and Western blot analysis.
2.13. Statistical analysis
Data were represented by mean ± standard deviation (SD) and graphed using GraphPad Prism 6. P values were determined using unpaired two-tailed t-tests or one-way analysis of variance (ANOVA) followed by a post hoc analysis with Turkey's multiple comparison test. Overall-free survival and relapse-free survival were generated using Kaplan–Meier (KM) plotter database (2017 version)30. Statistical significance was set at P < 0.05.
3. Results
3.1. Diminished macrophage-mediated phagocytosis contributes to stress-provoked tumor growth
In this study, we firstly established 4T1 breast cancers in wild type (WT) mice to explore the role of psychological stress in gynecological oncology. Mice received daily 2-h restraint stress for 28 days and were inoculated with 4T1 breast cancer cells on the 7th day of stress (Fig. 1A). In consistent with previous reports31, 32, 33, 34, chronic restraint stress dramatically promoted tumorigenesis as shown by significantly increased tumor volume and tumor weight (Fig. 1B and C). In the meantime, the level of corticosterone (CORT), a typical stress GC hormone in rodents, was significantly elevated in the plasma of stressed mice (Fig. 1D). However, stress-induced tumor growth was significantly relieved by the GC receptor antagonist mifepristone (Ru486, 25 mg/kg, s.c). To confirm that stress induced GC contributes to stress-provoked tumor growth, mice were administered with CORT (2 mg/kg, s.c.) daily for 28 days and inoculated with 4T1 cells (Fig. 1A). Results show that CORT treatment significantly elevated tumor weight and tumor volume in mice, in a pattern similar to restraint stress (Fig. 1B–D). Collectively, these findings demonstrate that stress-enhanced tumor growth is linked to the production of GC.
To investigate the exact role of GC, we took advantage of T lymphocyte-deficient nude mice (Fig. 1E) and macrophage-depleted mice by CL (Fig. 1I and Supporting Information Fig. S1) in this study. Unexpectedly, results show that tumor growth was also evoked by GC treatment in nude mice (Fig. 1F–H), similar to the results from WT mice. By sharp contrast, the detrimental effect of GC on tumor growth was abolished in macrophage-depleted mice (Fig. 1J–L). Based on these strong results, we therefore infer that deterioration of macrophage-based phagocytosis might be critical for stress-induced tumor growth. To test this hypothesis, we assessed the impact of stress/GC on macrophages-mediated phagocytosis of tumor cells in vivo and in vitro. Results from in vivo experiments demonstrated that both restraint stress and GC treatment obviously inhibited the phagocytosis of tumor cells by macrophages in tumor tissues of both WT mice (Fig. 2A) and T lymphocyte-deficient nude mice (Fig. 2B). Our previous studies have found that stress in mice led to a significant reduction of macrophages13. In order to explore the effect of GC on phagocytic function of macrophages in vitro, GC with no significant effect on the cell viability and cell number of macrophages were applied in this study (Supporting Information Fig. S2). In vitro results revealed that cortisol, a typical type of GC in human beings when experiencing stress, decreased the phagocytosis of HL60 cells by THP1 macrophages in a time- and dose-dependent manner (Fig. 2C and D). Confocal imaging further confirmed this phenomenon (Fig. 2E). In accordance with the inhibitory effect of cortisol on THP1 macrophages, CORT (100 μmol/L, 24 h) also inhibited mouse bone marrow-derived macrophages (BMDM)-mediated phagocytosis of HL60 cells (Fig. 2F and G) and 4T1 cells (Fig. 2H and I). These results highly suggest an essential role of GC-inhibited macrophage phagocytosis in stress-augmented tumor growth.
3.2. Stress hormone GC disrupts the balance of “eat me” and “don't eat me” signal receptors on macrophages
Subsequently, we sought to characterize the pattern by which stress/GC affect the phagocytic capacity of macrophages. Since efficient engulfment of tumor cells by macrophages is exquisitely regulated by a constellation of “eat me” and “don't eat me” signals and their corresponding receptors expressed on macrophages, we firstly determined the effect of stress on “eat me” signal receptors expressed on THP1 macrophages. Result found that stress hormone cortisol remarkably inhibited the expression of “eat me” signal receptors on macrophages (Fig. 3A). Especially, cortisol treatment obviously decreased the protein and mRNA levels of LRP1 in a dose- and time-dependent manner (Fig. 3B–E). A consistent result was also found in HL60 cell-co-cultured macrophages (Fig. 3F and G). Further, we employed a public database, KM plotter, to analyze the correlation among LRP1 mRNA expressions, relapse-free survival, and overall survival in human breast cancer patients. Results revealed that LRP1 mRNA level is positively correlated with the relapse-free survival of breast cancer patients and this correlation extended to overall survival (Fig. 3H and I). Data from in vivo experiments confirm that both stress and CORT treatment remarkably decreased LRP1 mRNA and protein levels in breast cancer tissues of WT mice (Fig. 3J and K) and T lymphocyte-deficient nude mice (Supporting Information Fig. S3A and S3B). Immunofluorescence images further demonstrated that LRP1 protein expressed on macrophages was decreased by stress/CORT treatment (Fig. 2L and Fig. S3C).
Figure 3.
Stress hormone GC disturbs the balance of “eat me” and “don't eat me” signal receptors in macrophages. (A) Expressions of “eat me” signal receptors in cortisol-treated-THP1 macrophages. (B)–(G) Effects of GC on the mRNA and protein levels of LRP1 in THP1 macrophages, respectively. (B, D) Macrophages were treated with different concentrations of cortisol (10, 50, and 100 μmol/L) for 24 h. (C, E) Macrophages were treated with cortisol (50 μmol/L) for different time periods (12, 24, and 36 h). (F, G) THP1 macrophages were treated with cortisol (50 μmol/L) for 24 h, followed by co-culture with HL60 cell for 2 h. (H) Relapse-free survival and (I) overall survival of LRP1 mRNA levels were generated using KM plotter (2017 version). (J, K) LRP1 protein and mRNA expressions in tumor tissues of WT mice. (L) LRP1 protein expression in macrophages was monitored by F4/80 and LRP1 double immunostaining in tumor tissues. SIRPα protein expression in THP1 macrophages treated with cortisol for different (M) concentrations or (N) time periods. (O) Schematic model depicting the imbalance of LRP1 and SIRPα in macrophages caused by stress hormone GC. The values are represented as mean ± SD (n = 3). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. GC, glucocorticoid; CORT, corticosterone; Ru486, mifepristone; LRP1, low density lipoprotein receptor-related protein 1; CD14, cluster of differentiation 14; CD36, cluster of differentiation 36; TREM2, triggering receptor expressed on myeloid cells 2; GAS6, growth arrest-specific 6; TIM4, T-cell membrane protein 4; DD1α, death domain1α; MERTK, mer receptor tyrosine kinase; MFGE8, milk fat globule EGF and factor V/VIII domain containing; SIRPα, signal-regulatory protein alpha; F4/80, the marker of macrophage.
Contrary to the reduced expression of “eat me” signal receptor LRP1, an interesting finding caught our attention is that the protein level of SIRPα, an important “don't eat me” signal receptor, was obviously elevated by stress/CORT treatment in breast cancer tissues of WT mice (Fig. 3J) and T lymphocyte-deficient nude mice (Fig. S3A and S3B). Data from THP1 macrophages further confirmed that cortisol treatment obviously increased the protein expression of SIRPα in a time- and dose-dependent manner (Fig. 3M and N). Nevertheless, no significant alteration of its mRNA level was noticed (Fig. S3F, S3G and S3I). Similar results were observed in tumor cell-macrophage co-culture systems (Fig. S3H), Results from KM plotter also revealed that SIRPA mRNA level has no correlation with the relapse-free survival and the overall survival of breast cancer patients (Fig. S3D and S3E). Collectively, these results show that stress hormone GC induced opposite effects on the expression levels of LRP1 and SIRPα, indicating an imbalance of “eat me” and “don't eat me” receptors on macrophages (Fig. 3O).
3.3. GC leads to a direct, GR-dependent trans-repression of LRP1 expression
The above results strongly indicate that GC downregulated the mRNA expression of LRP1. The biological action of GC is mainly mediated by its receptor (GR)35, which can be translocated into the nucleus and binds to GR element (GRE) to regulate the transcription of targeted genes35. Thus, we asked whether the effect of GC on LRP1 mRNA expression was associated with GR-dependent gene transcription. To prove this hypothesis, we firstly examined the location of GR in THP1 macrophages treated with cortisol. Confocal images indicated that GR was translocated to the nucleus (Fig. 4A). In addition, we uncovered that both GR antagonist Ru486 treatment and GR knockdown by siRNA completely abrogated cortisol-induced LRP1 elevation in THP1 macrophages (Fig. 4B–D). These data support our notion that the effect of GC on LRP1 mRNA level is GR-dependent. Furthermore, protein synthesis inhibitor cycloheximide (CHX, 10 μg/mL) and transcription inhibitor actinomycin D (ActD, 100 ng/mL) were utilized to explore whether this action was related to the repression of LRP1 transcription. As shown in Fig. 4E, cortisol still inhibited the gene expression of LRP1 in THP1 macrophages pretreated with CHX. To exclude the possibility that GC modulates LRP1 expression on post-transcriptional level, the decay of LRP1 mRNA in the presence or absence of GC after the inhibition of transcription by ActD was analyzed. As expected, GC had no significant impact on the half-life of LRP1 mRNA (Fig. 4F). These data indicate that GC-induced decrease of LRP1 expression is associated with GR-mediated transcription.
Figure 4.
GC leads to direct, GR-dependent trans-repression of LRP1 expression. (A) Cortisol (50 μmol/L, 6 h)-treated THP1 macrophages were stained with GR antibody and recorded by confocal microscopy. (B) LRP1 protein expression was detected in THP1 macrophages treated with cortisol (50 μmol/L, 24 h) in presence or absence of Ru486 (5 μmol/L). (C) The sequences of GR siRNA for THP1 macrophages were verified by Western blot. (D) LRP1 mRNA level in GR siRNA transfected THP1 macrophages. (E) LRP1 mRNA level was analyzed in THP1 macrophages treated with cortisol (50 μmol/L) in presence or absence of protein synthesis inhibitor (CHX, 10 μg/mL, 6 h). (F) The decay of LRP1 mRNA were analyzed in THP1 macrophages treated with cortisol (50 μmol/L) for 2 h before RNA synthesis inhibition (ActD, 100 ng/mL). (G) Schematic map of putative nGREs within the 2000-bp region of human LRP1 promoter. Luciferase activities of LRP1 were determined in (H) cortisol (100 μmol/L) or (I) GR plasmids (200 ng) treated HEK293 cells. (J) Schematic of GC/GR-dependent trans-repression of LRP1 expression. The values are represented as mean ± SD (n = 3). ∗P < 0.05, ∗∗P < 0.01. GR, glucocorticoid receptor; LRP1, low density lipoprotein receptor-related protein 1; NC, negative control; CHX, cycloheximide; ActD, actinomycin D; nGRE, negative glucocorticoid receptor element.
Genome-wide analyses revealed that GC can evoke the repression of target genes through a negative GRE (CTCC(n)0–2GGAGA)36. In our study, in silico promoter analysis revealed that three possible nGREs were located at a 2 kb region upstream of the transcription start site of LRP1 (Fig. 4G). To assess whether GR can directly transcriptionally regulate LRP1, the promoter region (~2 kb) of LRP1 was cloned into a luciferase reporter (pGL3 basic). Result from luciferase activity assay shows that both cortisol and GR overexpression repressed the transcription of LRP1 (Fig. 4H and I). Taken together, these findings demonstrate that GC leaded to a direct, GR-dependent trans-repression of LRP1 expression (Fig. 4J).
3.4. LRP1 counterbalances SIRPα protein expression
Above results indicate a solid relation between LRP1 and SIRPα (Fig. 3). In order to clearly delineate the pattern, gain- and loss-of-function experiments were performed in THP1 macrophages and HEK293 cells. The sequence of LRP1 siRNA (siLRP1) was verified by Western blot (Fig. 5A). As shown in Fig. 5B and C, depletion of LRP1 in THP1 macrophages increased SIRPα protein levels without significant change on its mRNA level. LRP1 is composed of a 515-kD chain possessing four extracellular ligand binding domains and an 85-kD membrane anchored chain. LRP1 minireceptors (mLRP2 and mLRP4) are deemed to fold and traffic similarly to endogenous full-length LRP1 and binds many of its physiological ligands27. As we expected, transfection of mLRP2 and mLRP4 decreased the expression of SIRPα protein (Fig. 5D). In turn, the elevation of SIRPα protein level induced by cortisol treatment (Fig. 5E) or siLRP1 transfection (Fig. 5F) was blocked by both mLRP2 and mLRP4 transfection. These data confirm that LRP1 counterbalances SIRPα protein expression, forming a delicate balance of the LRP1–SIRPα axis.
Figure 5.
LRP1 counterbalances SIRPα protein expression. (A) The sequence of GR siRNA for THP1 macrophages was verified by Western blot. (B) and (C) The mRNA and protein levels of LRP1 and SIRPα in siLRP1-transfected THP1 macrophages were detected by qPCR and Western blot, respectively. (D) LRP1 and SIRPα protein expressions in HEK293 cells transfected with mLRP1, mLRP2, mLRP3, mLRP4 plasmid for 24 h were detected. (E) Cortisol (100 μmol/L) or (F) siLRP1-treated HEK293 cells were transfected with mLRP2 or mLRP4 plasmids. Protein expressions of LRP1 and SIRPα were detected by Western blot. The values are represented as mean ± SD (n = 3). ∗∗∗P < 0.001. LRP1, low density lipoprotein receptor-related protein 1; GR, glucocorticoid receptor; SIRPα, signal-regulatory protein alpha.
3.5. miRNA-4695-3p plays a key role in GC-disturbed balance of the LRP1-SIRPα axis
MicroRNAs (miRNAs) have emerged as key regulators of biological process37,38 and LRP1 has been reported to induce a considerable change of miRNAs39. Importantly, the expression of SIRPα protein can be regulated by miRNAs through binding to the 3′-untranslational regions of SIRPA mRNA and blocking its translation40,41. An intriguing finding in our study is that GC increased SIRPα protein expression with no significant effect on its mRNA level (Fig. 3J and K), prompting us to test whether miRNAs contribute to GC-disturbed balance of the LRP1-SIRPα axis. In order to profile miRNA changes, cortisol-treated (50 μmol/L, 24 h) or siLRP1-transfected THP1 macrophages were collected to conduct miRNA transcriptomic analysis using ANSe3 algorithm on Chi-Cloud NGS Analysis Platform. As shown in the volcano plot (Fig. 6A), we screened out that a total of 38 miRNAs were significantly altered, among which 31 miRNAs were downregulated in cortisol-treated macrophages. In the meantime, a total of 101 miRNAs were remarkably changed in siLRP1 treatment, with 43 miRNAs in downregulation. Cortisol or siLRP1-induced downregulated miRNAs were displayed in the heat maps (Fig. 6B and C). It is noteworthy that both cortisol and siLRP1 simultaneously repressed the expression of miRNA-4695-3p (Fig. 6D). This result was further confirmed by qPCR analysis in THP1 macrophages and HEK293 cells (Fig. 6E and F). A similar alteration of miRNA-4695-3p was also discovered in the tumor tissues of WT mice (Fig. 6G) and nude mice (Fig. 6H) subjected to restraint stress/GC treatment. To investigate whether miRNA-4695-3p could regulate the expression of SIRPα, miRNA-4695-3p inhibitor and mimic were used to treat THP1 macrophages and their expressions were verified (Supporting Information Fig. S4). We found that the expression of SIRPα protein was inhibited by miRNA 4695-3p mimic but improved by the miRNA-4695-3p inhibitor (Fig. 6I), accompanied with no changes on SIRPA mRNA expression (Fig. 6J). Notably, cortisol or siLRP1-caused elevation in SIRPα protein was blocked by the miRNA-4695-3p mimic (Fig. 6K). These results identify miRNA-4695-3p as a critical determinant of the LRP1-SIRPα axis disruption caused by GC (Fig. 6L).
Figure 6.
miRNA-4695-3p plays a key role in GC-disturbed balance of the LRP1–SIRPα axis. (A) Volcano map of differentially expressed genes in cortisol (50 μmol/L, 24 h) vs. control, and siLRP1 vs. NC. (B) and (C) Significant downregulated miRNAs in THP1 macrophages treated with cortisol or siLRP1 are summarized and displayed in the heat map. (D) Venn diagram of significant down-regulated miRNAs between cortisol treatment and siLRP1 transfection. (E) miRNA-4695-3p level in cortisol-treated THP1 macrophages (50 μmol/L, 24 h) and HEK293 cells (100 μmol/L, 24 h) were verified by qPCR. (F) miRNA-4695-3p level in siLRP1-transfected THP1 macrophages and HEK293 cells were verified by qPCR. (G) and (H) miRNA-4695-3p level in tumor tissue of WT mice and nude mice. (I) SIRPα protein and (J) mRNA expressions in THP1 macrophages treated with miRNA-4695-3p mimic (25 nmol/L) or inhibitor (100 nmol/L) were analyzed by Western blot and qPCR. (K) Cortisol (50 μmol/L) or siLRP1 treated THP1 macrophages were transfected with miRNA-4695-3p mimic (25 nmol/L) for 24 h, and SIRPα protein expression was detected by Western blot. (L) Schematic picture illustrating the key role of miRNA-4695-3p in GC-disturbed balance of the LRP1–SIRPα axis. The values are represented as mean ± SD (n = 3). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. LRP1, low density lipoprotein receptor-related protein 1; NC, negative control; WT, wildtype; signal-regulatory protein alpha; GC, glucocorticoid.
4. Discussion
Chronic psychological stress as a susceptible factor in tumorigenesis has been well established by mounting evidence in recent years1, 2, 3, 4. However, mechanistic understanding of the underlying causes is lacking. In this research, we characterized stress hormone GC as the major culprit in stress-promoted cancer susceptibility, similar to our previous discovery regarding stress-induced virus infection42. By taking advantage of T lymphocyte-deficient nude mice and macrophage-depleted mice, we further revealed the importance of macrophages in stress-provoked tumorigenesis. In fact, the role of macrophage phagocytosis as a critical mediator of tumor immunosurveillance, especially during chemotherapy, has been well clarified43, 44, 45, 46. Through a collection of experiments in vivo and in vitro, we confirmed that stress/GC depresses the phagocytosis of macrophages to eliminate tumor cells. Our results, for the first time, innovatively provide a key link between macrophage's phagocytosis and stress-evoked tumor progression.
The exquisite balance between “eat me” and “don't eat me” signals and their corresponding receptors serves as a crucial factor in deciding the phagocytosis capacity of tumor cells by macrophages16,20,47. Our data identify that the LRP1–SIRPα axis is disrupted by stress/GC, resulting in an imbalanced engulfment signal. This finding well supports for our perspective that stress increases disease susceptibility by disturbing host inner Yin–Yang balance48. An emerging question is how GC decreases LRP1 expression at gene level in macrophages. Our data show that GC-caused LRP1 downregulation is in a direct GR-dependent trans-repression manner. Three possibly nGREs but not GRE were found at the 2 kb region upstream of transcription start site of LRP1 through in silico promoter analysis. However, it still remains unknown which specific sequence of nGRE is bound by GR in the transcription start site of LRP1. The truncated and binding site mutant of LRP1 promoter may help us better understand the exact binding site of GC trans-repressed LRP1.
Another pivotal question we have solved is how stress/GC elevates SIRPα protein levels without affecting its mRNA? An earlier study proposed that LRP1-activated Notch signaling induced SIRPα protein downregulation49, 50, 51, raising the possibility that the LRP1–SIRPα axis might be disrupted, and this disturbance might be relevant in our experimental settings. We thus resorted to gain- and loss-of-function experiments in HEK293 and THP1-derived macrophages. Our data reveal that GC-induced elevation in SIRPα protein expression is resultant from LRP1 changes. The firsthand evidence establishes the role of “eat me” signals in counteracting “don't eat me” signals, demonstrating their subtle relationship.
A previous report has pointed out that LRP1 can affect the expression of miRNAs39, while the expression of SIRPα protein can be regulated by miRNAs40,41. In our study, no significant change on SIRPA mRNA expression was noticed in GC-treated or siLRP1-transfected macrophages, raising the possible role of miRNAs in the imbalance of the LRP1-SIRPα axis. miRNA transcriptomic and gain-/loss-of-function experiments underpinned our hypothesis and pointed out that miRNA-4695-3p is likely the bridging factor. Nevertheless, the binding site between miRNA-4695-3p and SIRPA mRNA and how LRP1 affects this miRNA remains unclear, which is worth being addressed in future studies.
5. Conclusions
Our study illustrates an essential role of macrophage-dependent phagocytosis in stress-evoked tumor growth. Mechanistically, GC, as a crucial regulator in response to stress status, inhibits LRP1 expression by GR-dependent transcription. GC-downregulated LRP1 in turn raises SIRPα protein level in a miRNA-4695-3p dependent manner. The imbalance of “eat me” and “don't eat me” signal receptors thus suppress the phagocytosis of tumor cells by macrophages. Our findings, from the perspective of macrophage-mediated tumor cell phagocytosis, take deep insights into the mechanism for stress-induced tumorigenesis and provide the potential of harnessing phagocytosis-related immunotherapy in preventing and treating tumors.
Acknowledgments
This work was supported, in part, by National Natural Science Foundation of China (grant numbers 81673709, 82004231, U1801284 and 81873209), Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (grant number 2017BT01Y036, China) and GDUPS (2019, China), Guangdong Science and Technology Foundation for Distinguished Young Scholars (grant number 2017A030306004, China), Science and Technology Program of Guangzhou (grant numbers 201903010062 and 202102010116, China), Fellowship of China Postdoctoral Science Foundation (grant number 2020M683204, China), Guangdong Basic and Applied Basic Research Fund (grant number 2020A1515110388, China), Fundamental Research Funds for the Central Universities (grant number 21620448, China). The authors (RRH and YFL) also gratefully acknowledge the support of K. C. Wong Education Foundation.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.06.002.
Contributor Information
Jiaxu Chen, Email: chenjx@bucm.edu.cn.
Yi-Fang Li, Email: liyifang706@jnu.edu.cn.
Rong-Rong He, Email: rongronghe@jnu.edu.cn.
Author contributions
Rong-Rong He conceived and designed the experiments. Yanping Wu performed the experiments and prepared the manuscript. Xiang Luo, Qingqing Zhou, Haibiao Gong and Huaying Gao assisted the experiments and data analysis. Rong-Rong He and Yi-Fang Li supervised the study. Rong-Rong He and Yi-Fang Li revised the manuscript. Tongzheng Liu, Jiaxu Chen, Lei Liang and Hiroshi Kurihara consulted and discussed the data. All authors read and approved the final manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Chida Y., Hamer M., Wardle J., Steptoe A. Do stress-related psychosocial factors contribute to cancer incidence and survival?. Nat Clin Pract Oncol. 2008;5:466–475. doi: 10.1038/ncponc1134. [DOI] [PubMed] [Google Scholar]
- 2.Stommel M., Given B.A., Given C.W. Depression and functional status as predictors of death among cancer patients. Canc. 2002;94:2719–2727. doi: 10.1002/cncr.10533. [DOI] [PubMed] [Google Scholar]
- 3.Buccheri G. Depressive reactions to lung cancer are common and often followed by a poor outcome. Eur Respir J. 1998;11:173–178. doi: 10.1183/09031936.98.11010173. [DOI] [PubMed] [Google Scholar]
- 4.Lillberg K., Verkasalo P.K., Kaprio J., Teppo L., Helenius H., Koskenvuo M. Stressful life events and risk of breast cancer in 10,808 women: a cohort study. Am J Epidemiol. 2003;157:415–423. doi: 10.1093/aje/kwg002. [DOI] [PubMed] [Google Scholar]
- 5.Obradović M.M.S., Hamelin B., Manevski N., Couto J.P., Sethi A., Coissieux M.M., et al. Glucocorticoids promote breast cancer metastasis. Nature. 2019;567:540–544. doi: 10.1038/s41586-019-1019-4. [DOI] [PubMed] [Google Scholar]
- 6.Cui B., Luo Y., Tian P., Peng F., Lu J., Yang Y., et al. Stress-induced epinephrine enhances lactate dehydrogenase A and promotes breast cancer stem-like cells. J Clin Invest. 2019;129:1030–1046. doi: 10.1172/JCI121685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Q., Guo N., Zhou Y., Chen J., Wei Q., Han M. The role of tumor-associated macrophages (TAMs) in tumor progression and relevant advance in targeted therapy. Acta Pharm Sin B. 2020;10:2156–2170. doi: 10.1016/j.apsb.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y., Yu X., Li J. Manipulation of immune‒vascular crosstalk: new strategies towards cancer treatment. Acta Pharm Sin B. 2020;10:2018–2036. doi: 10.1016/j.apsb.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herwald H., Egesten A. Macrophages: past, present and future. J Innate Immun. 2013;5:657–658. doi: 10.1159/000356293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler M.O., Hirano N. Human cell-based artificial antigen-presenting cells for cancer immunotherapy. Immunol Rev. 2014;257:191–209. doi: 10.1111/imr.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eggermont L.J., Paulis L.E., Tel J., Figdor C.G. Towards efficient cancer immunotherapy: advances in developing artificial antigen-presenting cells. Trends Biotechnol. 2014;32:456–465. doi: 10.1016/j.tibtech.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauber K., Ernst A., Orth M., Herrmann M., Belka C. Dying cell clearance and its impact on the outcome of tumor radiotherapy. Front Oncol. 2012;2:116. doi: 10.3389/fonc.2012.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan W.J., Li Y.F., Liu F.L., Deng J., Wu Y.P., Yuan W.L., et al. A SIRT3/AMPK/autophagy network orchestrates the protective effects of trans-resveratrol in stressed peritoneal macrophages and RAW 264.7 macrophages. Free Radic Biol Med. 2016;95:230–242. doi: 10.1016/j.freeradbiomed.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 14.Izgüt-Uysal V.N., Tan R., Bülbül M., Derin N. Effect of stress-induced lipid peroxidation on functions of rat peritoneal macrophages. Cell Biol Int. 2004;28:517–521. doi: 10.1016/j.cellbi.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Tymen S.D., Rojas I.G., Zhou X., Fang Z.J., Zhao Y., Marucha P.T. Restraint stress alters neutrophil and macrophage phenotypes during wound healing. Brain Behav Immun. 2013;28:207–217. doi: 10.1016/j.bbi.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willingham S.B., Volkmer J.P., Gentles A.J., Sahoo D., Dalerba P., Mitra S.S., et al. The CD47–signal regulatory protein alpha (SIRPα) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A. 2012;109:6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaiswal S., Jamieson C.H., Pang W.W., Park C.Y., Chao M.P., Majeti R., et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138:271–285. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garg A.D., Romano E., Rufo N., Agostinis P. Immunogenic versus tolerogenic phagocytosis during anticancer therapy: mechanisms and clinical translation. Cell Death Differ. 2016;23:938–951. doi: 10.1038/cdd.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raghavan M., Wijeyesakere S.J., Peters L.R., Del Cid N. Calreticulin in the immune system: ins and outs. Trends Immunol. 2013;34:13–21. doi: 10.1016/j.it.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao M.P., Jaiswal S., Weissman-Tsukamoto R., Alizadeh A.A., Gentles A.J., Volkmer J., et al. Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Sci Transl Med. 2010;2:63ra94. doi: 10.1126/scitranslmed.3001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aderem A., Underhill D.M. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 22.Underhill D.M., Ozinsky A. Phagocytosis of microbes: complexity in action. Annu Rev Immunol. 2002;20:825–852. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X., Goncalves R., Mosser D.M. The isolation and characterization of murine macrophages. Curr Protoc Im. 2008;83:1–14. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyurin V.A., Balasubramanian K., Winnica D., Tyurina Y.Y., Vikulina A.S., He R.R., et al. Oxidatively modified phosphatidylserines on the surface of apoptotic cells are essential phagocytic ‘eat-me’ signals: cleavage and inhibition of phagocytosis by Lp-PLA2. Cell Death Differ. 2014;21:825–835. doi: 10.1038/cdd.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aaes T.L., Kaczmarek A., Delvaeye T., De Craene B., De Koker S., Heyndrickx L., et al. Vaccination with necroptotic cancer cells induces efficient anti-tumor immunity. Cell Rep. 2016;15:274–287. doi: 10.1016/j.celrep.2016.03.037. [DOI] [PubMed] [Google Scholar]
- 26.Liu W., Xiang L., Zheng T., Jin J., Zhang G. TranslatomeDB: a comprehensive database and cloud-based analysis platform for translatome sequencing data. Nucleic Acids Res. 2018;46:206–212. doi: 10.1093/nar/gkx1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obermoeller-McCormick L.M., Li Y., Osaka H., FitzGerald D.J., Schwartz A.L., Bu G. Dissection of receptor folding and ligand-binding property with functional minireceptors of LDL receptor-related protein. J Cell Sci. 2001;114:899–908. doi: 10.1242/jcs.114.5.899. [DOI] [PubMed] [Google Scholar]
- 28.Obermoeller L.M., Chen Z., Schwartz A.L., Bu G. Ca2+ and receptor-associated protein are independently required for proper folding and disulfide bond formation of the low density lipoprotein receptor-related protein. J Biol Chem. 1998;273:22374–22381. doi: 10.1074/jbc.273.35.22374. [DOI] [PubMed] [Google Scholar]
- 29.Bu G., Rennke S. Receptor-associated protein is a folding chaperone for low density lipoprotein receptor-related protein. J Biol Chem. 1996;271:22218–22224. doi: 10.1074/jbc.271.36.22218. [DOI] [PubMed] [Google Scholar]
- 30.Gyorffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 31.Antoni M.H., Lutgendorf S.K., Cole S.W., Dhabhar F.S., Sephton S.E., McDonald P.G., et al. The influence of bio-behavioural factors on tumour biology: pathways and mechanisms. Nat Rev Cancer. 2006;6:240–248. doi: 10.1038/nrc1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thaker P.H., Han L.Y., Kamat A.A., Arevalo J.M., Takahashi R., Lu C., et al. Chronic stress promotes tumor growth and angiogenesis in a mouse model of ovarian carcinoma. Nat Med. 2006;12:939–944. doi: 10.1038/nm1447. [DOI] [PubMed] [Google Scholar]
- 33.Justice A. Review of the effects of stress on cancer in laboratory animals: importance of time of stress application and type of tumor. Psychol Bull. 1985;98:108–138. [PubMed] [Google Scholar]
- 34.Ben-Eliyahu S. The promotion of tumor metastasis by surgery and stress: immunological basis and implications for psychoneuroimmunology. Brain Behav Immun. 2003;17(Suppl 1):S27–S36. doi: 10.1016/s0889-1591(02)00063-6. [DOI] [PubMed] [Google Scholar]
- 35.Oakley R.H., Cidlowski J.A. The biology of the glucocorticoid receptor: new signaling mechanisms in health and disease. J Allergy Clin Immunol. 2013;132:1033–1044. doi: 10.1016/j.jaci.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surjit M., Ganti K.P., Mukherji A., Ye T., Hua G., Metzger D., et al. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–241. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 37.Slezak-Prochazka I., Durmus S., Kroesen B.J., van den Berg A. MicroRNAs, macrocontrol: regulation of miRNA processing. RNA. 2010;16:1087–1095. doi: 10.1261/rna.1804410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 39.Mantuano E., Brifault C., Lam M.S., Azmoon P., Gilder A.S., Gonias S.L. LDL receptor-related protein-1 regulates NFκB and microRNA-155 in macrophages to control the inflammatory response. Proc Natl Acad Sci U S A. 2016;113:1369–1374. doi: 10.1073/pnas.1515480113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu D., Pan C., Li L., Bian Z., Lv Z., Shi L., et al. MicroRNA-17/20a/106a modulate macrophage inflammatory responses through targeting signal-regulatory protein alpha. J Allergy Clin Immunol. 2013;132:426–436. doi: 10.1016/j.jaci.2013.02.005. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen W., Li X., Wang J., Song N., Zhu A., Jia L. miR-378a modulates macrophage phagocytosis and differentiation through targeting CD47–SIRPalpha axis in atherosclerosis. Scand J Immunol. 2019;90 doi: 10.1111/sji.12766. [DOI] [PubMed] [Google Scholar]
- 42.Luo Z., Liu L.F., Jiang Y.N., Tang L.P., Li W., Ouyang S.H., et al. Novel insights into stress-induced susceptibility to influenza: corticosterone impacts interferon-β responses by Mfn2-mediated ubiquitin degradation of MAVS. Signal Transduct Target Ther. 2020;5:202. doi: 10.1038/s41392-020-00238-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaiswal S., Chao M.P., Majeti R., Weissman I.L. Macrophages as mediators of tumor immunosurveillance. Trends Immunol. 2010;31:212–219. doi: 10.1016/j.it.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dehne N., Mora J., Namgaladze D., Weigert A., Brüne B. Cancer cell and macrophage cross-talk in the tumor microenvironment. Curr Opin Pharmacol. 2017;35:12–19. doi: 10.1016/j.coph.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 45.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cassetta L., Pollard J.W. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov. 2018;17:887–904. doi: 10.1038/nrd.2018.169. [DOI] [PubMed] [Google Scholar]
- 47.Chao M.P., Alizadeh A.A., Tang C., Myklebust J.H., Varghese B., Gill S., et al. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell. 2010;142:699–713. doi: 10.1016/j.cell.2010.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan C., Luo Z., Li W., Li X., Dallmann R., Kurihara H., et al. Disturbed Yin–Yang balance: stress increases the susceptibility to primary and recurrent infections of herpes simplex virus type 1. Acta Pharm Sin B. 2020;10:383–398. doi: 10.1016/j.apsb.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y.C., He F., Feng F., Liu X.W., Dong G.Y., Qin H.Y., et al. Notch signaling determines the M1 versus M2 polarization of macrophages in antitumor immune responses. Cancer Res. 2010;70:4840–4849. doi: 10.1158/0008-5472.CAN-10-0269. [DOI] [PubMed] [Google Scholar]
- 50.Meng H., Zhang X., Lee S.J., Strickland D.K., Lawrence D.A., Wang M.M. Low density lipoprotein receptor-related protein-1 (LRP1) regulates thrombospondin-2 (TSP2) enhancement of Notch3 signaling. J Biol Chem. 2010;285:23047–23055. doi: 10.1074/jbc.M110.144634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Y., Zhao J.L., Zheng Q.J., Jiang X., Tian J., Liang S.Q., et al. Notch signaling modulates macrophage polarization andphagocytosis through direct suppression of signal regulatory protein alpha expression. Front Immunol. 2018;9:1744. doi: 10.3389/fimmu.2018.01744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.