Summary
Single-cell RNA sequencing (scRNA-seq) approach can broadly and specifically evaluate the individual cells with minimum detection bias. To explore the individual compositional and transcriptional alteration of intestinal leukocytes in the Dual Specificity Phosphatase six knockout (D6KO) mice, we performed a scRNA-seq followed by the cell type annotation based on ImmGen database. Composition assessments found that D6KO-derived intestinal leukocytes tend to stay inactivate or immature status. The enrichment analysis showed that D6KO-derived intestinal leukocytes are less sensitive to microbes. The mod PhEA phenotypic analysis showed that the D6KO leukocytes may link to not only immune-associated but also diverse previously non-immune-related diseases. Integrating our dataset with the published dataset GSE124880 generated a comprehensive dataset for exploring intestinal immunity. Down-regulation of Ccl17 gene was found in the D6KO-derived dendritic cells. Our results demonstrated the advantage of applying scRNA-seq for dissecting the individual alteration of intestinal leukocytes, particularly in the D6KO mice at a naive state.
Subject areas: Biological sciences, Immunology, Transcriptomics
Graphical abstract

Highlights
-
•
An scRNA-seq dataset includes CD45+ cells of epithelium and lamina propria from mice
-
•
The D6KO-derived intestinal leukocytes tend to stay inactivate or immature status
-
•
D6KO in leukocytes may link to certain previously non-immune-related diseases
-
•
Down-regulation of CCL17 gene was found in D6KO-derived dendritic cells
Biological sciences; Immunology; Transcriptomics
Introduction
The gut is constantly challenged by diverse environmental stimuli such as pathogenic/commensal microorganisms, dietary products, and inorganic materials and it has been considered as the largest immune organ in the human body. Besides a single layer of intestinal epithelial cells (IECs) providing a separate anatomical barrier protection, the gut is also equipped with an extremely complex, intrinsic immune system comprising intraepithelial lymphocytes (IELs), lymphocytes located immediately below the epithelial layer in the lamina propria (LPLs) and intestinal specific organized lymphoid structures, Peyer’s patches (PPs). Besides T and B lymphocytes providing adaptive immune responses, the innate immune cells (e.g., dendritic cells (DCs), monocytes/macrophages and the recently identified heterogeneous groups of innate lymphoid cells (ILCs) also contribute substantially to maintaining intestinal homeostasis.
The traditional antibody-based or gene-expression–based approaches in analyzing immune cell types in tissues such as intestines have some limitations due to certain shortcomings of the methodology. The histological analysis of immunohistochemistry or immunofluorescence staining and the breakthrough of three-dimensional imaging techniques have facilitated the understanding of the distribution of specific immune cell types located in diverse layers of intestinal tissues. However, the limited choice of antibodies and/or finite color channels of staining naturally restricts the number of target cells to be identified in one tissue section. These limitations also exist similarly in the flow cytometry approaches in which the cells are targeted by antigen-antibody reactions. Importantly, the antibody being pre-selected that might result in the misleading interpretation due to the bias of narrowed targets is indeed considered as the Achilles heel of these antibody-based approaches. While the quantitative reverse transcription PCR (RT-qPCR) is widely used for determining the mRNA expression of genes in tissue, this approach is also confined to certain pre-selected genes that are targeted by primers and lacks the information of each type of cells. Furthermore, transcriptome analysis based on bulk RNA-seq is not sufficient to assess the gene expression in one specific cell type, although it could provide an unbiased overall evaluation. Recently, the technological advances in single-cell mRNA sequencing (scRNA-seq) have provided exciting tools for single-cell level description of the cellular diversity and enabled identification of distinct cell types in various tissues (Papalexi and Satija, 2018). The gene expression signature, differentiation profiling, and response toward the gut microbiota therefore could be assessed within distinct cell types from intestinal tissue (Haber et al., 2017).
Dual-specificity phosphatase 6 (Dusp6), a negative feedback modulator of the mitogen-activated protein kinase (MAPK) pathway, has been found to be involved in the regulation of certain immune cells. For example, the CD4+T cells isolated from peripheral blood, spleen, lymph nodes and Peyer’s patch of Dusp6 knockout mice were shown to have increased proliferation, T helper one differentiation, IFN-g and IL-21 production, accompanied by decreased survival, regulatory T cell function and IL-17A secretion (Bertin et al., 2015; Hsu et al., 2018; Li et al., 2012). However, whether the deficiency of Dusp6 gene affected the other type of immune cells remains largely unclear. Moreover, the effects of Dusp6 knockout on the immune cells that resided within the intestinal epithelium and lamina propria are still unclear. While we have previously performed a gut transcriptome analysis by bulk RNA-seq and demonstrated that the immune system regulation in particularly lymphocytes regulation and T cell differentiation could be mediated by Dusp6 elimination under high-fat diet stimulation (Ruan et al., 2016), the effect of Dusp6 elimination on the intestinal immune cells at a naive state has still not been found potentially due to the limitations mentioned above regarding the bulk RNA-seq transcriptome analysis in whole tissue.
To assess whether Dusp6 deficiency exerts alteration to intestinal immune cells specifically, we further applied this state-of-art scRNA-seq with 10X Genomics platform in Dusp6-knockout (D6KO) mice and littermate wild-type (WT) control mice under chow diet. The intestines of wild-type and Dusp6-knockout mice (11 weeks old) were harvested and the live intestinal immune cells from the intraepithelial compartment and the lamina propria were collected with CD45+ marker by magnetic-activated cell sorting (MACS) Microbeads and fluorescence-activated cell sorting (FACS) for downstream 10X Genomics Chromium Single Cell 3′ Solution. A computational analysis based on the ImmGen database followed by the enrichment analysis of gene ontology, biological pathways, and phenotypes was carried out to elucidate the effects of Dusp6 elimination on diverse immune cells that lived in the small intestine.
Results
Dusp6 knockout induces the compositional changes of mucosal immune cells resided within the epithelium and lamina propria of the small intestine
To investigate the effects of Dusp6 knockout on intestinal immune homeostasis, the CD45+ leukocytes residing within the intraepithelial compartment and lamina propria of the small intestine in WT and D6KO mice were isolated. Since Peyer’s patches' immune functions could be significantly different from the intraepithelial compartment and lamina propria, they were not included for this scRNA-seq analysis and hence here intestinal immune cells refer to the intraepithelial compartment and lamina propria immune cells. After a single-cell RNA sequencing, a total of 27,998 genes were identified in 7,904 cells (3,996 cells in WT and 3,908 in D6KO) (Figure 1A). The visualization results displayed a segregated distribution of WT and D6KO cells, suggesting that the deviation existed between WT and D6KO in the original/unprocessed dataset (Figure S1A). To address the potential bias within the raw data, the generated gene-cell matrix was processed through serial steps including quality control (QC), filtering, normalization, and feature selection (Figures S1B–S1D).After data integration and dimensionality reduction, the visualization results showed a nicely overlapping of the distribution of WT and D6KO cells (Figure S1E), indicating that the gene-cell matrix was thoroughly cleaned and normalized. A subsequent graph-based clustering was performed (Figure S1F), and approximately 14 clusters were identified. Several clusters showed the significant difference between WT and D6KO cells (Figures S1G and S1H and Table S1); however, the biological importance of these differences is unknown due to the lack of information on the cell type or a possible error resulting from inappropriate classification.
Figure 1.
The scRNA-seq analysis with ImmGen-based annotation reveals the compositional rearrangement of intestinal CD45+ cells in Dusp6 knockout mice
(A) The workflow of scRNA-seq analysis for analyzing intestinal CD45+ leukocytes. The intestinal CD45+ cells from wild-type and Dusp6 knockout mice were isolated using MACS followed by FACS, and further analyzed with scRNA-seq according to the guideline of 10X Genomics. The generating matrix of gene counts versus cells were processed through serial steps including quality control and filtering, feature selection, library integration, clustering and cell-type annotation. A subsequent differential expression analysis identified the up- and down-regulated differential expressed genes in single-type of cell, which were further annotated by Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis and model organism phenotype enrichment analysis (modPhEA).
(B) The t-distributed stochastic neighbor embedding (t-SNE) plots of cells were annotated by ImmGen database.
(C) Composition of each type of intestinal CD45+ cells in wild-type or Dusp6 knockout mice.
(D) Statistical power of test by comparing the composition of each type of immune cells between wild-type and Dusp6 knockout. The dot lines indicate the p = 0.05, Fisher exact test.
See also Figures S1–S6, Tables 1 and S1.
To classify each cell type, we further performed a SingleR (single-cell recognition) computational analysis based on the ImmGen (Immunological Genome Project) reference set (Jojic et al., 2013; Robinette et al., 2015). Among the 14 clusters, 17 main cell types including Tgd (6,308 cells), CD8+T cells (232 cells), macrophages (195 cells), DC (194 cells), B cells (178 cells), NKT (82 cells), NK cells (64 cells), CD4+T cells (55 cells), Treg (40 cells), ILC1 (33 cells), ILC2 (14 cells), monocytes (13 cells), ILC3 (5 cells), other T cells (2 cells), neutrophils (2 cells), mast cells (2 cells) and basophils (1 cell) were assigned (Figure 1B, S2A and S2C). The distribution of each cell type was further verified by specific markers according to the PanglaoDB (Franzen et al., 2019) (Figure S3). However, diverse cell types were found within the same cluster in the 2-dimensional plot. For example, the macrophage and DC were co-distributed within Cluster 10, and NKT, CD4+T cells, Treg, and ILC1 were co-localized within Cluster 8. A subsequent permutational multivariate ANOVA (PERMANOVA) test demonstrated that these cells were mostly separable and distinguishable in each Cluster (Figures S4 and S5). In addition, the heatmap of marker gene expression revealed that the gene expression pattern was different within the phagocytes (Figure S6). These evidences indicated that the cell annotation is successful and suitable for the following analysis.
Because of the low isolation rate and/or identification limitation, it was insufficient to evaluate the alteration of Basophils, Mast cells, Neutrophils, and other T cells underDusp6 knockout (Figures 1C, 1D and Table 1). We found that the composition of DC was significantly decreased in D6KO mice as compared with that in WT mice while no notable change of the composition of Treg and Monocytes were found between WT and D6KO mice. Other immunocytes such as CD8+T cells, ILC1, ILC2, ILC3, B cells, and NK cells also showed a trend of decreasing in their composition in D6KO mice compared with that in WT mice. In contrast, the proportion of NKT cells was significantly increased in D6KO mice. The Tgd, macrophages and CD4+T cells also showed a trend of increased composition in D6KO mice as compared with that in WT mice. These results suggest that Dusp6 deficiency induces the compositional rearrangement of intestinal immune cells.
Table 1.
Main types of CD45+immunecells resided in intestinal epithelium and lamina propria of WT and D6KO mice
| Celltype | WT (3760 cells) | D6K (3660 cells) | WT (%) | D6KO (%) | Deviation | Odds ratio | pValue | Assessmenta |
|---|---|---|---|---|---|---|---|---|
| Tgd | 3176 | 3132 | 84.47% | 85.57% | 1.11% | 0.92 | 0.1825 | ↑ |
| CD8+T cell | 124 | 108 | 3.30% | 2.95% | −0.35% | 1.12 | 0.4235 | ↓ |
| Macrophage | 93 | 102 | 2.47% | 2.79% | 0.31% | 0.88 | 0.4249 | ↑ |
| DC | 121 | 73 | 3.22% | 1.99% | −1.22% | 1.63 | 0.0010 | ↓↓ |
| B cell | 96 | 82 | 2.55% | 2.24% | −0.31% | 1.14 | 0.4042 | ↓ |
| NKT | 20 | 62 | 0.53% | 1.69% | 1.16% | 0.31 | 0.0000 | ↑↑ |
| NK cell | 40 | 24 | 1.06% | 0.66% | −0.41% | 1.63 | 0.0603 | ↓ |
| CD4+T cell | 24 | 31 | 0.64% | 0.85% | 0.21% | 0.75 | 0.3438 | ↑ |
| Treg | 19 | 21 | 0.51% | 0.57% | 0.07% | 0.88 | 0.7522 | = |
| ILC1 | 20 | 13 | 0.53% | 0.36% | −0.18% | 1.50 | 0.2966 | ↓ |
| ILC2 | 10 | 4 | 0.27% | 0.11% | −0.16% | 2.44 | 0.1797 | ↓ |
| Monocyte | 6 | 7 | 0.16% | 0.19% | 0.03% | 0.83 | 0.7878 | = |
| ILC3 | 5 | 0 | 0.13% | 0.00% | −0.13% | INF | 0.0625 | ↓ |
| Other T cell | 2 | 0 | 0.05% | 0.00% | −0.05% | INF | 0.5000 | ND |
| Neutrophil | 2 | 0 | 0.05% | 0.00% | −0.05% | INF | 0.5000 | ND |
| Mast cell | 1 | 1 | 0.03% | 0.03% | 0.00% | 0.97 | 1.0000 | ND |
| Basophil | 1 | 0 | 0.03% | 0.00% | −0.03% | INF | 1.0000 | ND |
D6KO, Dusp6 knockout; INF, infinite; ND, not determined; WT, wild-type.
Assessment rules: ND, not determined due to insufficiency of sample size (total counts <5) that could be resulted from low isolation rate and/or identification limitation; =, no difference reflected by a value of Deviation between −0.1 and 0.1%; ↑, a trend of increase with no statistical significance; ↓, a trend of decrease with no statistical significance; ↑↑, significant increase; ↓↓, significant decrease.
Dusp6 knockout alters the activation status of mucosal immune cells in the small intestine
The Tgd cells, a dominant type of immunocytes residing in the intestinal epithelium in mice (Olivares-Villagomez and Van Kaer, 2018), were largely isolated and identified. However, only a slight difference of the abundance of Tgd cells was found between WT and D6KO mice. We further questioned whether the compositional change affected by Dusp6 knockout could be observed at the subtype levels. According to the ImmGen database, we found that the Tgd.VG5- (immature) cells were significantly increased in D6KO mice compared with WT mice (Figure 2; and Table 2). In contrast, the Tgd.VG5+ (mature) and Tgd.VG5+.ACT (mature and active) cells were decreased in D6KO mice. It has been known that the TCR gene Vg5 are expressed upon Tgd maturation in secondary lymphoid tissue (Xiong et al., 2008), and therefore the impoverishment of Vg5+ cells and enrichment of Vg5-cells that observed in D6KO mice indicate an attenuated maturation level of Tgd in the intestine of D6KO mice.
Figure 2.
Subpopulation of intestinal CD45+ cells are altered in Dusp6 knockout mice
Donut plots show the relative abundance of subtyped cells in the main type of intestinal CD45+ leukocytes isolated from wild-type and Dusp6 knockout mice. D6KO, Dusp6 gene knockout; WT, wild-type. See also Table 2.
Table 2.
Subtypes of CD45+ immune cells resided in intestinal epithelium and lamina propria of WT and D6KO mice
| Subtype | Cell counts |
% of CD45+ cell |
% of main type |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | D6KO | WT (%) | D6KO (%) | Deviation (%) | Assessmenta | Odds ratio | p-value | WT (%) | D6KO (%)< | Deviation (%) | Assessmenta | Odds ratio | p-value | |
| Tgd (3176 cells in WT; and 3132 cells in D6KO) | ||||||||||||||
| Tgd.VG5- | 291 | 551 | 7.74 | 15.05 | 7.32 | ↑↑ | 2.11 | 0.0000 | 9.16 | 17.59 | 8.43 | ↑↑ | 2.12 | 0.0000 |
| Tgd.VG5+ | 1530 | 1327 | 40.69 | 36.26 | -4.43 | ↓↓ | 0.83 | 0.0001 | 48.17 | 42.37 | -5.80 | ↓↓ | 0.79 | 0.0000 |
| Tgd.VG5+.ACT | 1334 | 1230 | 35.48 | 33.61 | -1.87 | ↓ | 0.92 | 0.0921 | 42.00 | 39.27 | -2.73 | ↓↓ | 0.89 | 0.0275 |
| Tgd.VG2+ | 16 | 22 | 0.43 | 0.60 | 0.18 | ↑ | 1.42 | 0.3306 | 0.50 | 0.70 | 0.20 | ↑ | 1.40 | 0.3324 |
| Tgd.mat.VG2+ | 4 | 1 | 0.11 | 0.03 | -0.08 | = | 0.26 | 0.3751 | 0.13 | 0.03 | -0.09 | = | 0.25 | 0.3749 |
| Tgd.imm.VG1+ | 0 | 1 | 0.00 | 0.03 | 0.03 | ND | INF | 0.4933 | 0.00 | 0.03 | 0.03 | ND | INF | 0.4965 |
| Tgd.mat.vg3 | 1 | 0 | 0.03 | 0.00 | -0.03 | ND | 0.00 | 1.0000 | 0.03 | 0.00 | -0.03 | ND | 0.00 | 1.0000 |
| CD8+T cell (124 cells in WT; and 108 cells in D6KO) | ||||||||||||||
| T.8NVE.OT1 | 1 | 0 | 0.03 | 0.00 | -0.03 | ND | 0.00 | 1.0000 | 0.81 | 0.00 | -0.81 | ND | 0.00 | 1.0000 |
| T.8EFF.OT1.12HR.LISOVA | 62 | 34 | 1.65 | 0.93 | -0.72 | ↓↓ | 0.56 | 0.0073 | 50.00 | 31.48 | -18.52 | ↓↓ | 0.46 | 0.0050 |
| T.8EFF.OT1.24HR.LISOVA | 12 | 9 | 0.32 | 0.25 | -0.07 | = | 0.77 | 0.6638 | 9.68 | 8.33 | -1.34 | ↓ | 0.85 | 0.8203 |
| T.8EFF.OT1.48HR.LISOVA | 4 | 2 | 0.11 | 0.05 | -0.05 | = | 0.51 | 0.6875 | 3.23 | 1.85 | -1.37 | ↓ | 0.57 | 0.6880 |
| T.8EFF.OT1.D10LIS | 7 | 17 | 0.19 | 0.46 | 0.28 | ↑↑ | 2.50 | 0.0406 | 5.65 | 15.74 | 10.10 | ↑↑ | 3.12 | 0.0163 |
| T.8MEM.OT1.D45.LISOVA | 28 | 34 | 0.74 | 0.93 | 0.18 | ↑ | 1.25 | 0.4445 | 22.58 | 31.48 | 8.90 | ↑ | 1.58 | 0.1388 |
| T.8EFF.TBET-.OT1LISOVA | 0 | 1 | 0.00 | 0.03 | 0.03 | ND | INF | 0.4933 | 0.00 | 0.93 | 0.93 | ND | INF | 0.4655 |
| T.8Mem | 0 | 1 | 0.00 | 0.03 | 0.03 | ND | INF | 0.4933 | 0.00 | 0.93 | 0.93 | ND | INF | 0.4655 |
| T.8NVE | 1 | 1 | 0.03 | 0.03 | 0.00 | ND | 1.03 | 1.0000 | 0.81 | 0.93 | 0.12 | ND | 1.15 | 1.0000 |
| T.CD8.1H | 9 | 8 | 0.24 | 0.22 | -0.02 | = | 0.91 | 1.0000 | 7.26 | 7.41 | 0.15 | ↑ | 1.02 | 1.0000 |
| T.CD8.5H | 0 | 1 | 0.00 | 0.03 | 0.03 | ND | INF | 0.4933 | 0.00 | 0.93 | 0.93 | ND | INF | 0.4655 |
| Macrophage (93 cells in WT; and 102 cells in D6KO) | ||||||||||||||
| MF.103-11B+ | 26 | 51 | 0.69 | 1.39 | 0.70 | ↑↑ | 2.03 | 0.0028 | 27.96 | 50.00 | 22.04 | ↑↑ | 2.58 | 0.0020 |
| MF.103-11B+.SALM3 | 18 | 10 | 0.48 | 0.27 | -0.21 | ↓ | 0.57 | 0.1852 | 19.35 | 9.80 | -9.55 | ↓ | 0.45 | 0.0673 |
| MF.103-11B+24- | 1 | 0 | 0.03 | 0.00 | -0.03 | ND | 0.00 | 1.0000 | 1.08 | 0.00 | -1.08 | ND | 0.00 | 0.4769 |
| MF.103CLOSER | 6 | 5 | 0.16 | 0.14 | -0.02 | = | 0.86 | 1.0000 | 6.45 | 4.90 | -1.55 | ↓ | 0.75 | 0.7598 |
| MF.11CLOSER | 17 | 14 | 0.45 | 0.38 | -0.07 | = | 0.85 | 0.7203 | 18.28 | 13.73 | -4.55 | ↓ | 0.71 | 0.4359 |
| MF.11CLOSER.SALM3 | 22 | 17 | 0.59 | 0.46 | -0.12 | ↓ | 0.79 | 0.5228 | 23.66 | 16.67 | -6.99 | ↓ | 0.65 | 0.2824 |
| MF.169+11CHI | 2 | 3 | 0.05 | 0.08 | 0.03 | = | 1.54 | 0.6833 | 2.15 | 2.94 | 0.79 | ↑ | 1.38 | 1.0000 |
| MF.II+480LO | 0 | 1 | 0.00 | 0.03 | 0.03 | ND | INF | 0.4933 | 0.00 | 0.98 | 0.98 | ND | INF | 1.0000 |
| MF.SBCAPS | 1 | 1 | 0.03 | 0.03 | 0.00 | ND | 1.03 | 1.0000 | 1.08 | 0.98 | -0.09 | ND | 0.91 | 1.0000 |
| DC (121 cells in WT; and 73 cells in D6KO) | ||||||||||||||
| DC | 10 | 5 | 0.27 | 0.14 | -0.13 | ↓ | 0.51 | 0.3019 | 8.26 | 6.85 | -1.42 | ↓ | 0.82 | 0.7888 |
| DC.103+11B- | 5 | 4 | 0.13 | 0.11 | -0.02 | = | 0.82 | 1.0000 | 4.13 | 5.48 | 1.35 | ↑ | 1.34 | 0.7310 |
| DC.103-11B+24+ | 16 | 6 | 0.43 | 0.16 | -0.26 | ↓ | 0.38 | 0.0526 | 13.22 | 8.22 | -5.00 | ↓ | 0.59 | 0.3543 |
| DC.8+ | 6 | 3 | 0.16 | 0.08 | -0.08 | = | 0.51 | 0.5079 | 4.96 | 4.11 | -0.85 | ↓ | 0.82 | 1.0000 |
| DC.8- | 1 | 0 | 0.03 | 0.00 | -0.03 | ND | 0.00 | 1.0000 | 0.83 | 0.00 | -0.83 | ND | 0.00 | 1.0000 |
| DC.8-4-11B+ | 6 | 0 | 0.16 | 0.00 | -0.16 | ↓↓ | 0.00 | 0.0313 | 4.96 | 0.00 | -4.96 | ↓↓ | 0.00 | 0.0851 |
| DC.8-4-11B- | 12 | 3 | 0.32 | 0.08 | -0.24 | ↓↓ | 0.26 | 0.0352 | 9.92 | 4.11 | -5.81 | ↓↓ | 0.39 | 0.1734 |
| DC.LC | 2 | 0 | 0.05 | 0.00 | -0.05 | ND | 0.00 | 0.5000 | 1.65 | 0.00 | -1.65 | ND | 0.00 | 0.5282 |
| DC.PDC.8+ | 50 | 39 | 1.33 | 1.07 | -0.26 | ↓ | 0.80 | 0.3373 | 41.32 | 53.42 | 12.10 | ↑ | 1.63 | 0.1052 |
| DC.PDC.8- | 13 | 13 | 0.35 | 0.36 | 0.01 | = | 1.03 | 1.0000 | 10.74 | 17.81 | 7.06 | ↑ | 1.80 | 0.1933 |
| B cell (96 cells in WT; and 82 cells in D6KO) | ||||||||||||||
| B.CD19CONTROL | 6 | 8 | 0.16 | 0.22 | 0.06 | = | 1.37 | 0.6024 | 6.25 | 9.76 | 3.51 | ↑ | 1.62 | 0.4157 |
| B.FrE | 1 | 0 | 0.03 | 0.00 | -0.03 | ND | 0.00 | 1.0000 | 1.04 | 0.00 | -1.04 | ND | 0.00 | 1.0000 |
| B.T3 | 0 | 1 | 0.00 | 0.03 | 0.03 | ND | INF | 0.4933 | 0.00 | 1.22 | 1.22 | ND | INF | 0.4607 |
| B.FO | 0 | 7 | 0.00 | 0.19 | 0.19 | ↑ | INF | 0.0071 | 0.00 | 8.54 | 8.54 | ↑ | INF | 0.0038 |
| B.GC | 3 | 1 | 0.08 | 0.03 | -0.05 | ND | 0.34 | 0.6250 | 3.13 | 1.22 | -1.91 | ND | 0.38 | 0.6255 |
| B.FrF | 1 | 0 | 0.03 | 0.00 | -0.03 | ND | 0.00 | 1.0000 | 1.04 | 0.00 | -1.04 | ND | 0.00 | 1.0000 |
| B.MEM | 0 | 1 | 0.00 | 0.03 | 0.03 | ND | INF | 0.4933 | 0.00 | 1.22 | 1.22 | ND | INF | 0.4607 |
| B.MZ | 1 | 0 | 0.03 | 0.00 | -0.03 | ND | 0.00 | 1.0000 | 1.04 | 0.00 | -1.04 | ND | 0.00 | 1.0000 |
| B1A | 84 | 64 | 2.23 | 1.75 | -0.49 | ↓ | 0.78 | 0.1361 | 87.50 | 78.05 | -9.45 | ↓ | 0.51 | 0.1099 |
| NKT (20 cells in WT; 62 cells in D6KO) | ||||||||||||||
| NKT.4+ | 11 | 27 | 0.29 | 0.74 | 0.45 | ↑↑ | 2.53 | 0.0086 | 55.00 | 43.55 | -11.45 | ↓ | 0.63 | 0.4436 |
| NKT.4- | 4 | 20 | 0.11 | 0.55 | 0.44 | ↑↑ | 5.16 | 0.0008 | 20.00 | 32.26 | 12.26 | ↑ | 1.90 | 0.4005 |
| NKT.44+NK1.1+ | 5 | 14 | 0.13 | 0.38 | 0.25 | ↑↑ | 2.88 | 0.0388 | 25.00 | 22.58 | -2.42 | ↓ | 0.88 | 1.0000 |
| NKT.44+NK1.1- | 0 | 1 | 0.00 | 0.03 | 0.03 | ND | INF | 0.4933 | 0.00 | 1.61 | 1.61 | ND | INF | 1.0000 |
| NK cell (40 cells in WT; 24 cells in D6KO) | ||||||||||||||
| LPL.NCR+CNK | 15 | 13 | 0.40 | 0.36 | -0.04 | = | 0.89 | 0.8507 | 37.50 | 54.17 | 16.67 | ↑ | 1.97 | 0.2073 |
| NK.DAP10- | 25 | 11 | 0.66 | 0.30 | -0.36 | ↓↓ | 0.45 | 0.0289 | 62.50 | 45.83 | -16.67 | ↓↓ | 0.51 | 0.2073 |
| CD4+T cell (24 cells in WT; and 31 cells in D6KO) | ||||||||||||||
| T.4EFF49D+11A+.D8.LCMV | 0 | 2 | 0.00 | 0.05 | 0.05 | ND | INF | 0.2433 | 0.00 | 6.45 | 6.45 | ND | INF | 0.4990 |
| T.4MEM44H62L | 4 | 7 | 0.11 | 0.19 | 0.08 | = | 1.80 | 0.3813 | 16.67 | 22.58 | 5.91 | ↑ | 1.46 | 0.7385 |
| T.4Mem | 0 | 1 | 0.00 | 0.03 | 0.03 | ND | INF | 0.4933 | 0.00 | 3.23 | 3.23 | ND | INF | 1.0000 |
| T.CD4.1H | 1 | 0 | 0.03 | 0.00 | -0.03 | ND | 0.00 | 1.0000 | 4.17 | 0.00 | -4.17 | ND | 0.00 | 0.4364 |
| T.CD4.5H | 6 | 5 | 0.16 | 0.14 | -0.02 | = | 0.86 | 1.0000 | 25.00 | 16.13 | -8.87 | ↓ | 0.58 | 0.5049 |
| T.CD4.CTR | 1 | 0 | 0.03 | 0.00 | -0.03 | ND | 0.00 | 1.0000 | 4.17 | 0.00 | -4.17 | ND | 0.00 | 0.4364 |
| T.CD4TESTCJ | 12 | 16 | 0.32 | 0.44 | 0.12 | ↑ | 1.37 | 0.4524 | 50.00 | 51.61 | 1.61 | ↑ | 1.07 | 1.0000 |
| Treg (19 cells in WT; and 21 cells in D6KO) | ||||||||||||||
| T.4FP3+25+ | 1 | 1 | 0.03 | 0.03 | 0.00 | ND | 1.03 | 1.0000 | 5.26 | 4.76 | -0.50 | ND | 0.90 | 1.0000 |
| T.Tregs | 18 | 20 | 0.48 | 0.55 | 0.07 | = | 1.14 | 0.7462 | 94.74 | 95.24 | 0.50 | ↑ | 1.11 | 1.0000 |
| ILC1 (20 cells in WT; and 13 cells in D6KO) | ||||||||||||||
| LIV.ILC1.DX5- | 4 | 1 | 0.11 | 0.03 | -0.08 | = | 0.26 | 0.3751 | 20.00 | 7.69 | -12.31 | ↓ | 0.33 | 0.6253 |
| LPL.NCR+ILC1 | 16 | 12 | 0.43 | 0.33 | -0.10 | ↓ | 0.77 | 0.5718 | 80.00 | 92.31 | 12.31 | ↑ | 3.00 | 0.6253 |
| ILC2 (10 cells in WT; and 4 cells in D6KO) | ||||||||||||||
| ILC2 | 10 | 4 | 0.27 | 0.11 | -0.16 | ↓ | 0.41 | 0.1797 | 100.00 | 100.00 | 0.00 | ND | ND | 1.0000 |
| Monocyte (6 cells in WT; and 7 cells in D6KO) | ||||||||||||||
| MO.6C+II- | 6 | 6 | 0.16 | 0.16 | 0.00 | = | 1.03 | 1.0000 | 100.00 | 85.71 | -14.29 | ↓ | 0.00 | 1.0000 |
| MO.6C-II+ | 0 | 1 | 0.00 | 0.03 | 0.03 | ND | INF | 0.4933 | 0.00 | 14.29 | 14.29 | ND | INF | 1.0000 |
| ILC3 (5 cells in WT; and 0 cell in D6KO) | ||||||||||||||
| ILC3.LTI.4+ | 1 | 0 | 0.03 | 0.00 | -0.03 | ND | 0.00 | 1.0000 | 20.00 | ND | ND | ND | ND | 1.0000 |
| ILC3.LTI.CD4- | 4 | 0 | 0.11 | 0.00 | -0.11 | ND | 0.00 | 0.1250 | 80.00 | ND | ND | ND | ND | 1.0000 |
| Other T cells (2 cells in WT; and 0 cell in D6KO) | ||||||||||||||
| T.ETP | 2 | 0 | 0.05 | 0.00 | -0.05 | ND | 0.00 | 0.5000 | 100.00 | ND | ND | ND | ND | 1.0000 |
| Neutrophil (2 cells in WT; and 0 cell in D6KO) | ||||||||||||||
| GN.ARTH | 2 | 0 | 0.05 | 0.00 | -0.05% | ND | 0.00 | 0.5000 | 100.00 | ND | ND | ND | ND | 1.0000 |
| Mast cell (1 cell in WT; and 1 cell in D6KO) | ||||||||||||||
| MC.ES | 1 | 0 | 0.03 | 0.00 | -0.03 | ND | 0.00 | 1.0000 | 100.00 | 0.00 | -100.00 | ND | 0.00 | 1.0000 |
| MC.TR | 0 | 1 | 0.00 | 0.03 | 0.03 | ND | INF | 0.4933 | 0.00 | 100.00 | 100.00 | ND | INF | 1.0000 |
| Basophils (1 cell in WT; and 0 cell in D6KO) | ||||||||||||||
| BA | 1 | 0 | 0.03 | 0.00 | -0.03 | ND | 0.00 | 1.0000 | 100.00 | ND | ND | ND | ND | 1.0000 |
D6KO, Dusp6 knockout; INF, infinite; ND, not determined; WT, wild-type.
See also Figure 2.
Assessment rules: ND, not determined due to insufficiency of sample size (total counts <5) that could be resulted from low isolation rate and/or identification limitation; =, no difference reflected by a value of Deviation between −0.1 and 0.1%; ↑, a trend of increase with no statistical significance; ↓, a trend of decrease with no statistical significance; ↑↑, significant increase; ↓↓, significant decrease.
The population of macrophages was slightly increased in the D6KO mice as compared with WT mice. By analyzing the composition at the subtype level, we found this increase was mainly due to the enrichment of MF.103-11B+ cells, a relatively non-infected/stimulated macrophage in the ImmGen dataset(Figure 2; and Table 2). In contrast, the MF.103-11B+.SLAM3 cells, another MF.103-110+ subset that had been stimulated by a 3-day infection of Salmonella typhimurium, was decreased in D6KO mice. These changes led to a higher inactivate-to-activate ratio of MF.103-110+ cells in D6KO mice (ratio = 5.10) as compared with that in WT mice (ratio = 1.44), and suggests that D6KO have a higher abundance of inactivated macrophages in the lamina propria of the small intestine.
While the Dusp6 knockout showed a slight increase of CD4+T cells at the main type level, no notable change was found at the subtype level. Interestingly, the significant decrease of main type CD8+T cells by Dusp6 knockout could be attributed to the decrease of subtype T.8EFF.OT1.12HR.LISOVA, the effector CD8+T cells that were activated by Listeria for 12 h. In contrast, T.8EFF.OT1.D10LIS, the subtype CD8+T cells that were activated by Listeria for 10 days, significantly increased in D6KO mice. These results reveal that the Dusp6 knockout alters both short-term and long-term immune responses of intestinal CD8+ cells in clearance of the invasive pathogens.
The decrease in main type B cells of D6KO mice can be attributed to the decrease in subtype B1A cells, a type of cells that can be linked to autoimmune diseases (Deng et al., 2016; Duan and Morel, 2006). Surprisingly, the subtype B.FO cells were dramatically increased by Dusp6 deficiency. The decrease in main type DC was mainly contributed by the decreases in subtype DC.103-11B+24+, DC.8-4-11B+ and DC.8-4-11B- cells. In contrast, the proportion of subtype DC.103 + 11B-, DC.PDC.8+ and DC.PDC.8- cells among main type DC were relatively increased in D6KO mice as compared with WT mice. Besides this, the number of all subtypes of NKT cells in D6KO was increased as compared with WT mice, leading to the increases in their percentage of isolated CD45+ cells. However, the increase in the percentage of total NKT cells was only observed in the subtype NKT.4- of D6KO mice compared with WT mice. The proportion of subtype NKT.4+ and NKT.44 + NK1.1+ were relatively reduced in NKT cells underDusp6 deficiency. The decrease in the main type NK cells was primarily due to the decrease in the subtype NK.DAP10- cells, but relatively, the proportion of LPL.NCR + CNK cells within total NK cells was increased in D6KO mice as compared with WT mice. Together, these results suggest that Dusp6 deficiency alters the proportion of intestinal immune cells, particularly the ratio of active and inactive subtype cells, which may affect the progress of certain immune-associated diseases.
Dusp6 knockout alters the gene expression patterns of intestinal lymphocytes and phagocytes
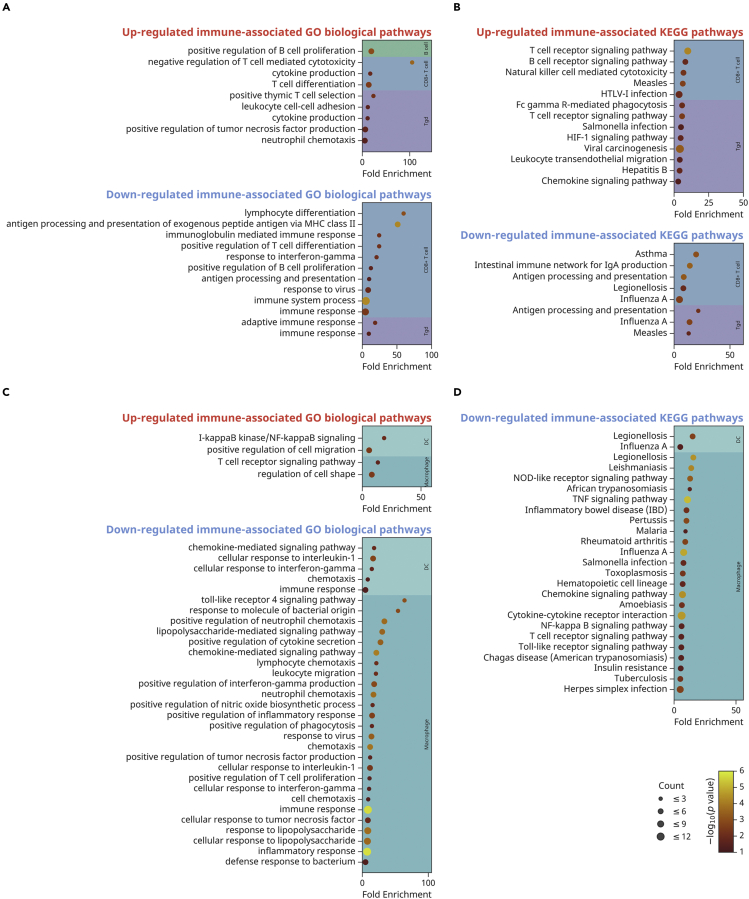
To understand the gene expression alteration effect of Dusp6 deficiency on the diverse intestinal immune cells, we then analyzed the differentially expressed genes (DE-Gs) of each type of immune cells (Figure 3; and Table S2) by Gene Ontology (GO) analysis (Figure 4; and Table S3). We found that the elimination of Dusp6 in mice affected the differentiation-associated biological pathways in intestinal B cells and CD8+T cells (Figure 4A; and Table S3). The Dusp6 deficient CD8+T cells also showed attenuated immune pathways, reflected by up-regulated negative regulation of T cell mediated cytotoxicity, and down-regulated immune system process, immune response, response to virus and response to interferon-gamma. Similarly, the D6KO-derived intestinal Tgd cells showed down-regulated pathways including adaptive immune response and immune response. Interestingly, the up-regulation of pathway of cytokine production was observed in both CD8+T cells and Tgd cells underDusp6 deficiency. An additional Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed that the D6KO-derived intestinal CD8+T cells exhibited the gene expression patterns of down-regulated influenza A, asthma and Legionellosis pathways, and up-regulated HTLV-I infection and Measles pathways (Figure 4B; and Table S4). The D6KO-derived intestinal Tgd cells also showed the down-regulation of KEGG pathway of influenza A like in D6KO-derived CD8+T cells, despite the measles pathway being down-regulated in D6KO-derived intestinal Tgd cells. The T cell receptor signaling pathway was up-regulated in both CD8+T cells and Tgd cells underDusp6 deficiency, and both the Chemokine signaling pathway and HIF-1 signaling pathway were found to be up-regulated in Dusp6 deficient Tgd cells. Of note, the hepatitis B and viral carcinogenesis pathways were also up-regulated in Dusp6 knockout Tgd cells. Together, Dusp6 knockout interfered with the intestinal lymphocytes, likely through the alteration of signaling, differentiation and immune pathways, and could intervene in the response for infectious diseases.
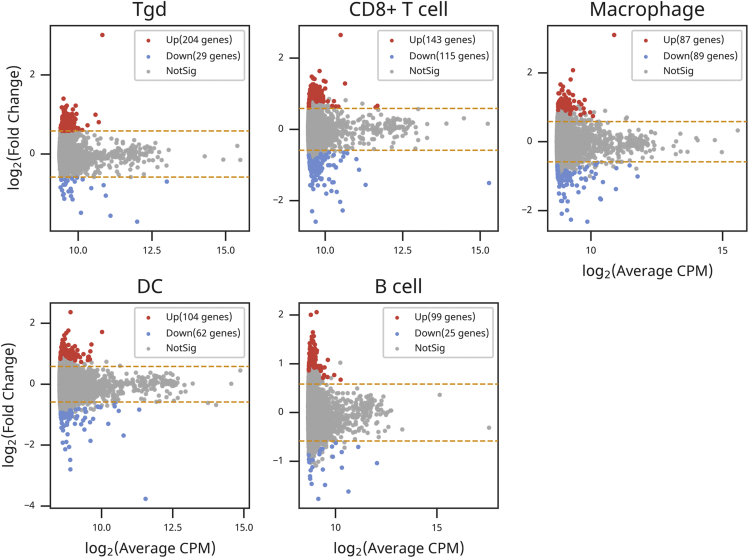
Figure 3.
Differential expression genes are found in the intestinal immune cells of Dusp6 knockout mice as compared with wild-type mice
Smear plot of all differential expression genes (DE-Gs) with count per million reads (CPM) from Tgd, CD8+T cell, macrophage, DC and B cell. Up-regulated DE-Gs are shown in red and down-regulated DE-Gs are shown in blue. Genes with False Discovery Rate (FDR) ≥ 0.05 or log2 fold change ≤±1.5 are defined as not significant (NotSig) and shown in grey. See also Table S2.
Figure 4.
GO and KEGG pathways reveal the alteration of immune-associated pathways in the intestinal CD45+ cells from Dusp6 knockout mice
(A and C)GO biological pathways and (B and D) KEGG pathways analyses for up-regulated or down-regulated DE-Gs identified from intestinal lymphocytes (B cell, CD8+T cell, and Tgd) and phagocytes (DC and macrophage) in Dusp6 knockout mice. Each GO or KEGG term is represented by a single circle, that color indicates the-log10 p-values of the term, and the size is proportional to the gene counts. GO, Gene Ontology. KEGG, Kyoto Encyclopedia of Genes and Genomes. See also Tables S3 and S4.
The GO analysis also showed that the intestinal phagocytes including DC and macrophages attenuated their immune response underDusp6 deficiency (Figure 4C; and Table S3). The D6KO-derived DC showed down-regulated pathways including cellular response to interleukin-1, chemokine-mediated signaling pathway, cellular response to interferon-gamma and chemotaxis, and these changes were also consistently observed in D6KO-derived macrophages. Moreover, the pathways that could be linked to the leukocyte chemotaxis and migration function were also down-regulated in D6KO-derived macrophages, accompanied with the down-regulation of several cytokine pathways including positive regulation of cytokine secretion, positive regulation of interferon-gramma production, cellular response to tumor necrosis factor, and positive regulation of tumor necrosis factor production. Of note, the D6KO-derived intestinal macrophages showed down-regulated pathways such as response to lipopolysaccharide, cellular response to lipopolysaccharide, lipopolysaccharide-mediated signaling pathway, defense response to bacterium, toll-like receptor four signaling pathway, response to molecule of bacterial origin, and positive regulation of phagocytosis. The KEGG analysis further showed that the pathways associated with bacterial, viral and parasitic infections were down-regulated in D6KO-derived macrophages (Figure 4D; and Table S4). Thus, the intestinal macrophages in the D6KO mice may have lower sensitivity in response to microbes. In contrast, pathways of certain inflammatory diseases or syndromes including rheumatoid arthritis, inflammatory bowel disease (IBD), and insulin resistance were also down-regulated in D6KO-derived macrophages. A lower sensitivity of immune response in D6KO macrophages may also provide the benefits of modulating certain inflammatory diseases.
Dusp6 knockout in intestinal immune cells links to phenotypes of mice
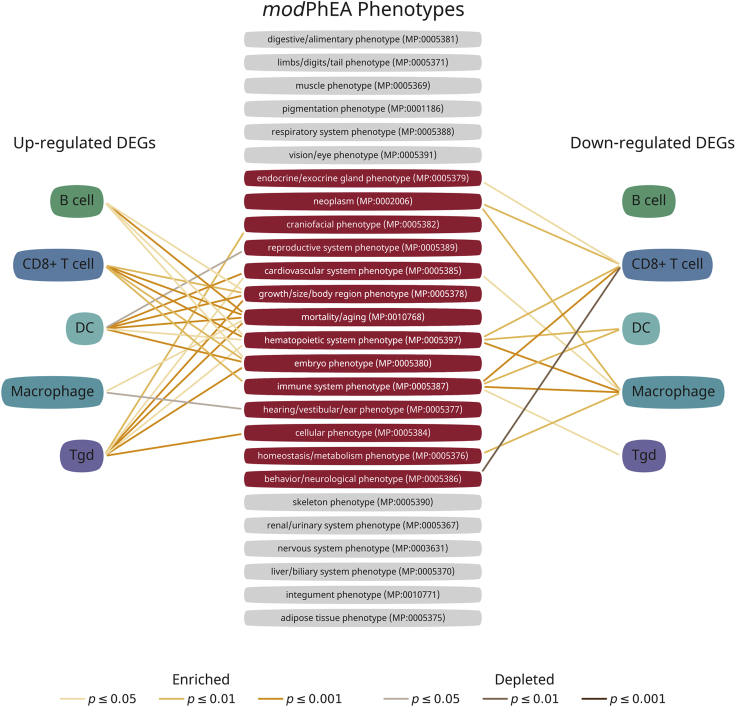
By using the single-cell RNA sequencing approach, we successfully evaluated the effects of Dusp6 knockout on intestinal immune cells that we were previously not able to distinguish the difference due to the technical limitation for whole-tissue lysate. This gave us a chance to further evaluate the possible links of Dusp6 deficient immune cells toward the diseases or phenotypes in the body. We therefore applied a model organism phenotype enrichment analysis (modPhEA) to both up- and down-regulated DE-Gs of D6KO-derived B cells, CD8 T cells, Tgd cells, macrophages and DC. As a result, the DE-Gs hit numerous phenotypes of modPhEA, which can be further classified into 26 main phenotypes (Figure 5; and Table S5).Among these, 14 main phenotypes showed significant links toward the DE-Gs from one cell type. Consistent with the results from the GO and KEGG analyses, most of the D6KO cells were predicted to have the hematopoietic system phenotype (MP:0005397) and immune system phenotype (MP:0005387). This result indicates that modPhEA is reliable and suitable for phenotypic annotation, and also evidences that the Dusp6 gene knockout affects the differentiation of each type of immune cells. Interestingly, we found that all the cell types showed the similar phenotype that their up-regulated DE-Gs implied the possible effects on the mortality/aging (MP:0010768) and embryo phenotype (MP:0005380). Furthermore, most of the D6KO-derived cells represented the up-regulated DE-Gs toward the growth/size/body region phenotype (MP:0005378). These results reveal that the genetic effects of Dusp6 gene knockout on differentiation exist in diverse intestinal immune cells.
Figure 5.
The modPhEA phenotypic analysis reveals the possible links between Dusp6 deficient immune cells and diverse phenotypes and diseases
A total of 26 main modPhEA phenotypes are found to be linked with up-regulated or down-regulated DE-Gs of B cell, CD8+T cell, DC, macrophage and Tgd underDusp6 knockout. The phenotypes that significantly enriched or depleted by DE-Gs of immune cell are highlighted in red. The Fisher's exact test is calculated for p-values. See also Table S5.
In addition to the immune system, the down-regulated DE-Gs of D6KO-derived CD8+T cells showed the depleted feature of behavior/neurological phenotype (MP:0005386). The up-regulated DE-Gs of D6KO-derived DC and Tgd and down-regulated DE-Gs of D6KO-derived macrophages showed enrichment of the cardiovascular system phenotype (MP:0005385). Down-regulated DE-Gs of D6KO-derived macrophages showed an enriched homeostasis/metabolism phenotype (MP:0005397). Up-regulated DE-Gs of D6KO-derived DC showed the depleted reproductive system phenotype (MP:0005389). The down-regulated DE-Gs of D6KO-derived CD8 T cells and macrophages showed the enriched phenotype of neoplasm (MP:0002006). Together, these results suggest that Dusp6 deficiency in immune cells can be linked to several pathophysiological phenotypes and play a role in the progress of multiple diseases.
A scRNA-seq dataset for comprehensively exploring intestinal immunity in mice
Histologically, the immune cells are mainly distributed within three tissue layers in the intestine: the epithelium, lamina propria, and Peyer’s patches. To increase the reads number and the resolution of the immune cells in the intestinal epithelium, we removed the Peyer’s patches and successfully observed the effects of Dusp6 knockout on mucosal immune cells at a naive state. A recent study of scRNA-seq by Xu et al. reported an atlas of intestinal immune cells from the lamina propria and Peyer’s patch for exploring intestinal inflammation (Xu et al., 2019). To generate a comprehensive database that covered entire intestinal immune cells, we integrated our dataset with this published dataset (GSE124880). The GSE124880 dataset that included 21,270 lamina propria cells and 36,797 Peyer’s patch cells was first processed through serial steps including QC, filtering, normalization, feature selection, and data integration according to the procedures described above, and a processed dataset including 19,062 genes and 55,184 cells were obtained. Finally, that processed data was merged with our processed data, and a matrix of 19,150 genes versus 62,604 cells was generated. The t-SNE plot revealed a nicely overlapping of not only the WT and D6KO cells but also the cells grouped by healthy, inflammation, lamina propria and Peyer’s patch (Figure S7).The plot based on individual cell type showed that the epithelial Tgd cells from WT and D6KO mice successfully filled the vacancy of Tgd cells in the GSE124880 dataset, in particular the region between the Gamma delta T cell (Xcl1+) and Gamma delta T cell (Gzma+) (Figure S8). In contrast, our B cells were not observed in the region of the germinal center B cells which is most abundant in the Peyer’s patch. In addition, the distribution of our CD4+T cells was closer to the activated CD4+T cells rather than the resting CD4+T cells that also mainly existed in the Peyer’s patch. Importantly, our macrophages and DC showed a nicely co-distribution with that from the GSE124880 dataset. These results reveal that these two datasets were successfully integrated and could complement each other.
By analyzing the DE-Gs of DC, four genes that were significantly regulated by both inflammation and Dsup6 deficiency were pulled out (Figure 6A). Among these, only the Ccl17 gene showed the down-regulation in DCs derived from both healthy mice and D6KO mice (Figure 6B), suggesting a potential link between inflammation, Dusp6 existence, and Ccl17 expression. Furthermore, the immunofluorescence staining of intestinal tissue revealed that the WT-derived CD209(+) cells expressed the chemokine CCL17 at basal state (Figure 6C), but this was not observed in the D6KO-derived CD209(+) cells. Collectively, the Dusp6 deficiency in mice could diminish the intestinal DCs in both quantitative and quality manner.
Figure 6.
Identification of Dusp6 deficiency- and healthy-associated genes in intestinal DCs
(A) Intersection of significant DE-Gs (p<0.05) of DC calculated by healthy mice vs. inflammation mice and D6KO mice vs. WT mice shows the enriched genes that could be modulated by Dusp6 deficiency and inflammation in DC. Values of DEG are log2 fold change. The up-regulations are indicated in red, and the down-regulations are indicated in blue.
(B) Expression means of intersecting genes are obtained from 500 bootstraps, followed by Levene’s test for p-value. Data are represented as mean ±SD. ∗∗∗p< 0.001.
(C) Immunofluorescence staining of the DC marker CD209 (green) and chemokine CCL17 (red) in the intestinal tissue from the wild-type (WT) and Dusp6 knockout (D6KO) mice. Scale bar = 25 μm.
See also Figures S7 and S8.
Discussion
In this study, we have successfully uncovered the previously unclear effects of Dusp6 gene knockout on diverse intestinal immune cells using the scRNA-seq approach. We found that Dusp6 deficiency induces a rearrangement of the subpopulation and alters their functional status of intestinal immune cells, including the depletions of DC, CD8+T cells, ILC1, ILC2, ILC3, B cells, and NK cells, and enrichments of NKT, Tgd, macrophages and CD4+T cells. The analysis at the subtype level further revealed that the intestinal immune cells in D6KO mice are likely more oriented to inactivated or immature status. A further analysis by GO and KEGG demonstrated that the D6KO-derived immune cells have lower sensitivity for detecting microbes, which might decrease the response in particular infectious or inflammatory diseases. The phenotype enrichment analysis by modPhEA has demonstrated that Dusp6 deficiency might affect the differentiation pathway of immune cell lineages and it may not only link to the immune-associated diseases but also includes neuropsychiatric, cardiovascular, reproductive, metabolic and oncologic diseases. These results suggest that the Dusp6 gene is critical for leukocyte differentiation and therefore involved in the progression of certain inflammatory diseases.
Our phenotypic analysis of D6KO-derived intestinal leukocytes is significantly consistent with numerous previously reported characteristics of Dusp6 deficient mice. For example, it has been reported that Dusp6 null mice had larger hearts and that could be associated with the increased proliferation and the decreased apoptosis rate of cardiomyocytes (Maillet et al., 2008). We here found that Dusp6 gene knockout could confer the cardiovascular system phenotype (MP:0005385) through up-regulation of genes in DC and Tgd or down-regulation of genes in the macrophage. The Dusp6 gene mutation also causes skeletal dwarfism, coronal craniosynostosis and hearing loss (Li et al., 2007), whereas the DE-Gs from D6KO-derived intestinal leukocytes can target the skeleton phenotype (MP:0005390), craniofacial phenotype (MP:0005382), and hearing/vestibular/ear phenotype (MP:0005377). The embryonic lethality was also observed in the mice with Dusp6 knockout (Li et al., 2007), and the DE-Gs identified from our D6KO intestinal leukocytes showed the possible effects on embryo phenotype (MP:0005380), growth/size/body region phenotype (MP:0005378), mortality/aging (MP:0010768), and reproductive system phenotype (MP:0005389). It has been reported that Dusp6 is significantly upregulated in the liver of obese and diabetic mice and promotes glucose output in both cultured liver cells and mouse livers (Feng et al., 2014a). Our recent study also showed that Dusp6 deficiency could decrease blood glucose levels, improve insulin sensitivity and confer the resistance to diet-induced obesity (Feng et al., 2014b). In the present study, we found that Dusp6 elimination in the intestinal leukocytes was predicted to be linked with metabolic organs, reflected by the adipose tissue phenotype (MP:0005375), digestive/alimentary phenotype (MP:0005381), liver/biliary system phenotype (MP:0005370) and muscle phenotype (MP:0005369). In addition, one functional Leu114Val polymorphism of Dusp6 gene in human was reported to be positively associated with bipolar disorder, and our phenotypic analysis showed that Dusp6 gene deficiency is associated with behavior/neurological phenotype (MP:0005386). Collectively, these findings raise the possibility that Dusp6 deficiency in immune cells and even in the intestinal resident leukocytes may mediate the diseases that have taken place in diverse organs/tissues of the body. These clues might have previously been hidden in the whole-body knockout model, in which disease progression is usually first thought to be caused by the intrinsic factors of non-immune cells in the target tissues or organs. Our findings have suggested that the neglected role of Dusp6 in the immune cells should be also taken into consideration for counteracting unsolved diseases. Hence, a subsequent study is required to validate the causative role of Dusp6-deficient leukocytes in the diseases.
The role of Dusp6 in immune cells is still largely unknown, despite It having been reported that Dsup6 deficiency can enhance the development of CD4+T cell lineage (Bertin et al., 2015; Hsu et al., 2018; Li et al., 2012). Here we also reported that Dusp6 deficiency enriched the CD4+T cells in the small intestine of mice. However, only a small amount of CD4+T cells were isolated and identified, which leads to the lower statistical power in identifying the difference between WT and D6KO mice. The insufficiency of cell count might also contribute to the decreased resolution of downstream subtyping analysis. Nevertheless, these results represented a real composition of leukocytes in the intestine of D6KO mice, and it is plausible to observe the trend of the rearrangement of certain immune cell types. For example, we found that the composition of intestinal CD8−CD4− DCs in WT mice (4.96% of DC.8-4-11B+ and 9.92% of DC.8-4-11B- = 14.88% of total DCs) was reduced in D6KO mice (0.00% of DC.8-4-11B+ and +4.11% 9.92% of DC.8-4-11B- = 4.11% of total DCs). Of note, a previous report showed a similar composition of the splenic CD8−CD4− DCs in control mice (15% of total DCs) (Flohe et al., 2006), but these DCs were relatively enriched in mice with polymicrobial sepsis induced by cecal ligation and puncture for 8 h (47% of total DCs). Interestingly it has been known that the DCs play not only protective but sometimes also pathogenic roles in the intestine. The DCs depletion was evidenced to resist dextran-sulfate sodium (DSS)-induced colitis (Berndt et al., 2007). In patients with IBD, the intestinal lamina propria DCs are found to be activated via up-regulation of microbial recognition receptors and certain pathologically relevant cytokines (Hart et al., 2005). Our GO biological pathway analysis also reveals that Dusp6 gene knockout may affect DCs on the migration, NF-kB signaling, and response to cytokines. Therefore the depletion of DCs by Dusp6 elimination is likely beneficial for the resistance to colitis. Interestingly, a subsequent DE-Gs analysis in both our and GSE124880 datasets indicates that the Ccl17 gene expression is down-regulated in intestinal DCs underDusp6 deficiency and/or a healthy/non-inflammation state. It has been reported that CCL17 promotes intestinal inflammation, and induces the IL12 expression in DCs (Heiseke et al., 2012). Of note, our data showed that the Il12b gene was significantly down-regulated in the Dusp6-derived DCs. Whether the Dusp6-CCL17-IL12 axis plays a role in IBD is interesting and a further evaluation is required.
In summary, we have demonstrated that the Dusp6 gene knockout alters the gene expression profile within intestinal leukocytes, leading to the change of their biological functions particularly deaffrication, microbial sensing, and immune responses. The phenotypic analysis has further demonstrated that these changes may have protective or pathogenic effects on various disease progressions. Importantly, although we have only evidenced that Dusp6 gene knockout affects the leukocytes residing in the small intestine, the resident immune cells in other tissues and organs may also be affected by Dusp6. The overall findings of our study will lay a solid foundation for a better design (e.g., a chemical compound to modulate intestinal lymphocyte-mediated immunity) in the future of more specific agents or strategies for the prevention or treatment of diverse inflammatory and/or immune-associated diseases.
Limitations of the study
In this study, we employed the scRNA-seq technique and ImmGen database to broadly assess the compositional and functional alterations of intestinal CD45+ cells underDusp6 gene knockout. However, certain type of immune cells might not be evaluated due to the limitation of resolution. A higher number of cell count for increasing the sequencing depth of scRNA-seq would be required to explore the rare immune cells resided in the tissue. In addition, due to the high-cost nature of scRNA-seq, the evaluation by biological repeats is lacking in the present work. A subsequent study for validating the effect of Dusp6 knockout on the immune cells particularly the DC is required.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE anti-mouse CD45 antibody | BioLegend | Cat#103105; RRID:AB_312970 |
| Rabb anti-CD209 antibody | LifeSpan BioSciences | Cat#LS-C782089; RRID: N/A |
| Goat anti-CCL17 antibody | R&D systems | Cat#AF529; RRID:AB_355416 |
| Alexa Fluor 488 Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody | ThermoFisher Scientific | Cat#A11034; RRID:AB_2576217 |
| Alexa Fluor 594 Donkey anti-Goat IgG (H + L) Cross-Adsorbed Secondary Antibody | ThermoFisher Scientific | Cat#A11058; RRID:AB_2534105 |
| Chemicals, peptides, and recombinant proteins | ||
| Hank’s Balanced Salt Solution without calcium and magnesium salts (HBSS w/o) | Biological Industries | Cat#02-018-1A |
| fetal bovine serum | ThermoFisher Scientific | Cat#10437-28 |
| phosphate-buffered saline | ThermoFisher Scientific | Cat#70011-044 |
| bovine serum albumin | Biological Industries | Cat#03-101-1B |
| EDTA | Sigma-Aldrich | Cat#03690 |
| DTT | Sigma-Aldrich | Cat#43816 |
| frozen sectionmedia | Leica | Cat#14020108926 |
| acetone | Sigma-Aldrich | Cat#32201 |
| Fluoroshield mounting medium | Abcam | Cat#ab104139 |
| Critical commercial assays | ||
| Lamina Propria Dissociation Kit | Miltenyi Biotec | Cat#130-097-410 |
| Dead Cell Removal Kit | Miltenyi Biotec | Cat#130-090-101 |
| CD45 MicroBeads | Miltenyi Biotec | Cat#130-052-301 |
| Live/Dead fluorescent reactive dye | Invitrogen | Cat#L34957 |
| Deposited data | ||
| Raw sequencing data | This paper | BioProject ID: PRJNA734865 (SRA: SRP322434) Gene Expression Omnibus (GEO) accession number: GSE176087 |
| Code | This paper | https://github.com/wenhsuanyuu/Int-CD45-scRNAseq |
| Experimental models: Organisms/strains | ||
| Mouse: B6.129S6-Dusp6tm1Jmol N10 | The Jackson Laboratory | JAX:009069 |
| Software and algorithms | ||
| Cell Ranger (version 2.0.1) | 10x Genomics | https://support.10xgenomics.com/single-cell-vdj/software/pipelines/2.0/what-is-cell-ranger |
| scater (version 1.14.6) | (McCarthy et al., 2017) | http://bioconductor.org/packages/scater/ |
| Seurat (version 3.1.5) | (Stuart et al., 2019) | https://satijalab.org/seurat/ |
| SingleR (version 1.4.1) | (Aran et al., 2019) | https://github.com/LTLA/SingleR |
| edgeR (version 3.32.1) | (McCarthy et al., 2012; Robinson et al., 2010) | http://bioinf.wehi.edu.au/edgeR |
| R | R Core Team | https://www.r-project.org/ |
| Python | Python Software Foundation | https://www.python.org/ |
| D.A.V.I.D. (DAVID 6.8 Oct. 2016) | (Huang da et al., 2009) | https://david.ncifcrf.gov/ |
| modPhEA (2021/01/31 updated) | (Weng and Liao, 2017) | https://evol.nhri.org.tw/phenome2/ |
| Other | ||
| gentleMACS Dissociator | Miltenyi Biotec | Cat#130-093-235 |
| 100 μm strainer | Miltenyi Biotec | Cat#130-098-463 |
| 70 μm strainer | FALCON | Cat#352350 |
| cell sorter | BD Biosciences | BD Influx |
| cell counter | Nexcelom Bioscience | Cellometer Auto T4 |
| confocal microscopy | Leica | TCS SP5 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Cheng-Yuan Kao (chengyuankao@nhri.edu.tw).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals
The Dusp6 gene knockout (D6KO) mice obtained from the Jackson Laboratory were transferred onto a C57BL/6J genetic background by backcrossing more than 10 generations. The D6KO mice and their littermate wild-type (WT) mice were weaned at 3–4 weeks after birth, and caged separately after weaning (3 mice/cage). The male mice at 11 weeks of age were euthanized by CO2 inhalation for the following experiments. The female mice and the mice of different ages were not examined in the present study, and the results were only associated with the adult male mice. Animal protocols were approved by the Institutional Animal Care and Use Committee of the National Health Research Institutes, Taiwan. All mice were housed in a 12-hours light/dark cycle, fed with autoclaved chow diet, and maintained under semi-specific pathogen-free conditions.
Method details
Isolation of intestinal CD45 + immune cells
The cells living in the epithelial layer and lamina propria of the small intestine were isolated via a Lamina Propria Dissociation Kit (130-097-410, Miltenyi Biotec) and the procedures were according to the manufacturer's instructions. Three male D6KO mice and three littermate male WT mice at 11 weeks of age were euthanized by CO2 inhalation, and perfused with 4°C Hank’s Balanced Salt Solution without calcium and magnesium salts (HBSS w/o) containing 5% fetal bovine serum (FBS) to remove the blood leukocytes. Approximately 20 cm of intestine was harvested, and the luminal content was removed by flushing with HBSS w/o. Peyer’s patches were also removed to exclude the leukocytes of secondary gut-associated lymphoid tissue. The intestine was longitudinally slit, washed by HBSS w/o, and laterally cut into pieces of around 0.5 cm in length. The intestinal pieces were incubated with predigestion solution (HBSS w/o containing 5 mM EDTA, 5% FBS, and 1 mM DTT) for 20minsat 37°C on a shaker at 220 rpm, and the solution was passed through the 100 μm strainer. The flow-through fraction was placed on ice for 10 mins, and the supernatant containing the suspended intraepithelial leukocytes was collected. The intestinal pieces were conducted with the above procedures thrice to obtain the cells from the epithelial layer and to remove the entire epithelial layer. For the isolation of cells from lamina propria, the remaining pieces were further incubated with enzymes for 20mins at 37°C on a shaker at 220 rpm, churned by gentleMACS Dissociator, and passed through the 100 μm strainer. All fractions were centrifugated at 300 rcf for 5minsat 4°C, and the pellet was resuspended in 4°C PB buffer (Phosphate-buffered saline, PBS, containing 0.5% bovine serum albumin). The cells isolated from the epithelial layer and lamina propria were pooled together, passed through the 70 μm strainer, and washed with 4°C PB buffer once. The live cells were isolated using a Dead Cell Removal Kit (130-090-101, Miltenyi Biotec). The CD45 + cells were selected through a MACS column with CD45 MicroBeads (130-052-301, Miltenyi Biotec). To increase the purity of live CD45 + intestinal cells, the cells were further stained with Live/Dead fluorescent reactive dye (L34957, Invitrogen) and PE anti-mouse CD45 antibody (103105, BioLegend) and sorted by BD Influx™ Cell Sorter (BD Biosciences) in 100% FBS. The sorted cells were centrifugated at 300 rcf for 5 mins at 4°C, and finally resuspended in 4°C PBS. Due to low sorting efficacy, one D6KO sample was abandoned. Ultimately, three WT samples and two D6KO samples were chosen for following single-cell RNA sequencing.
Single-cell RNA sequencing
The isolated intestinal CD45 + cells were counted by Cellometer Auto T4 (Nexcelom Bioscience). Three WT samples and two D6KO samples were pooled separately. Subsequently, WT and D6KO samples were processed by BIOTOOLS Co., Ltd through the GemCode Single Cell Platform using the GemCode Gel Bead, Chip and 3′-Library v2 Kits (10X Genomics, Pleasanton) according to the manufacturer’s instructions. In brief, cell suspensions were loaded onto a Chromium Single-Cell Chip along with the reverse transcription master mix and single cell 3′ gel beads, aiming for 5,000 cells per channel. Following generation of single-cell gel bead-in-emulsions, reverse transcription was carried out in a Mastercycler ep gradient S thermocycler (Eppendorf). The amplified cDNA was subsequently purified using SPRIselect beads and sequenced on an Illumina HiSeq X Ten PE150 platform. The raw data contained the following read regions: 26 bp Read 1 (16 bp single cell barcode, 10x barcode; 10 bp Unique molecular identifier, UMI), 98 bp Read 2 (transcript insert), 8 bp i7 Index (sample index). The Cell Ranger software pipeline (version 2.0.1, 10X Genomics) was used to demultiplex cellular barcodes, map reads to the genome and transcriptome using the STAR aligner, and down-sample reads as required to generate normalized aggregate data across samples, producing a matrix of gene counts (27,998 genes) versus cells (3,996 cells in WT and 3,908 cells in D6KO).
Quality control and filtering of single-cell RNA sequencing data
The scater (version 1.14.6) package (McCarthy et al., 2017) provides methods to compute relevant quality control (QC) metrics for an SCESet object. The genes that were not expressed in any cell were first excluded. The cell-level QC was performed via evaluating the detected genes, library size, and percentage of mitochondrial genes, in which the cells that out of range of 3 median absolute deviation (MAD) were identified as the outliers for removal. For gene filtering, only the gene with at least 2 cells and contained more than 1 transcript was kept. Finally, a total of 7,420 cells (3,760 cells in WT and 3,660 cells in D6KO) and 27,153 genes (13,509 genes in WT and 13,644 genes in D6KO) were chosen for downstream analysis.
Feature selection, data integration and cell-type annotation
The gene-cell matrices of WT and D6KO libraries were subsequently processed by Seurat (version 3.1.5) (Stuart et al., 2019) for normalization, feature selection, data integration, dimension reduction, graph-based clustering, and visualization. In brief, library-size normalization to each cell was performed by the NormalizeData function. The Seurat v3 using canonical correlation analysis (CCA) and mutual nearest neighbors (MNNs) was conducted to integrate multiple experimental datasets. A feature selection process was performed via the FindVariableFeatures function, and the top 2,000 highly variable genes (HVGs) in either WT or D6KO library were identified. From the union of two HVG lists, 2,000 genes were selected as the input genes for CCA. The cells were then aligned with the FindIntegrationAnchors function using 30 CC dimensions. Subsequently, the WT and D6KO libraries were combined together by the IntegrateData function. The integrated expression data were scaled and centered for each gene by the ScaleData function before principal component analysis (PCA). The PCA for dimensionality reduction was performed on the scaled integrated expression matrix with the first 50 principal components (PCs) using the RunPCA function. We used the top 18 PCs as input for the downstream analysis, such as clustering and 2D visualization. The FindClusters function was used to cluster cells using the first 18 PCs at a resolution of 0.5 (total 14 clusters). The clustering results were visualized with t-distributed stochastic neighbor embedding (t-SNE) dimensionality reduction using the RunTSNE function. Finally, a single-cell recognition (SingleR, version 1.4.1) package (Aran et al., 2019) with its built-in Immunological Genome Project (ImmGen) reference dataset (Jojic et al., 2013; Robinette et al., 2015) were used for achieving cell-type annotation at main cell type and subtype levels. The SingleR function returns the best main cell type and subtype annotation for each cell in the integrated data. The annotation cells were verified based on the Celltype gene expression markers selected from PanglaoDB (2020/03/27 updated).
Differentially expressed genes analysis
Differentially expressed genes (DEGs) were calculated using a gene-wise quasi-likelihood negative binomial generalized log-linear model (glmQLFit function) from the edgeR (version 3.32.1) package (McCarthy et al., 2012; Robinson et al., 2010). In brief, binomial dispersion was estimated using the estimateDisp function, followed by model fitting using the glmQLFit function, and finally, likelihood ratios for differential expression were calculated using the glmQLFTest function. Subsequent Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed using D.A.V.I.D. (DAVID 6.8 Oct. 2016) (Huang da et al., 2009). The phenotype analyses were performed by the model organism phenotype enrichment analysis (modPhEA; 2021/01/31 updated) (Weng and Liao, 2017).
Immunofluorescence staining
Intestinal tissue was embedded in frozen section media (Leica, Richmond, IL, USA) and snap-frozen in liquid nitrogen. The 5 μm-thick sections were fixed in ice-cold acetone (Sigma-Aldrich). The sections were immuno-stained with anti-CD209 antibody (1:100; LS-C782089, LifeSpan BioSciences) and anti-CCL17 antibody (1:50; AF529, R&D systems) followed by Alexa Fluor 488 Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody (1:400; A11034, ThermoFisher Scientific) and Alexa Fluor 594 Donkey anti-Goat IgG (H + L) Cross-Adsorbed Secondary Antibody (1:400; A11058, ThermoFisher Scientific). The sections were washed with PBS and mounted in Fluoroshield mounting medium (ab104139, Abcam). Immunofluorescent images of intestinal tissue were obtained by confocal microscopy (TCS SP5, Leica).
Quantification and statistical analysis
Statistical analysis
The two-tailed Fisher’s exact test was used to determine statistical significance. The differences and associations are considered to be statistically significant at p< 0.05.
Acknowledgments
We are grateful to Nir Hacohen at Broad Institute of MIT and Harvard for the suggestions on the experimental design of scRNA-seq. We thank Chao-Yang Hsiao at the Core Instrument Center in National Health Research Institutes (NHRI) for assistance of FACS experiment and the technical support for scRNA-seq platform by BIOTOOLS Co., Ltd. This work was supported by the following grants: IM-108-PP-04 from NHRI, Taiwan; CYC109003 from National Cheng Kung University, Taiwan and Ditmanson Medical Foundation Chia-Yi Christian Hospital, Taiwan; and 106-2628-B-400-001-MY3, 106-2923-B-400-001-MY3, 108-2321-B-400-011-, 109-2221-E-010-012-MY3, 109-2327-B-400-001-, 109-2320-B-400-008-MY3, 109-2221-E-002-161-MY3, 109-2221-E-010-012-MY3 from MOST, Taiwan.
Author contributions
C.-S.C., W.-H.Y., C.-C.S., J.-W. R., H.-C.H., H.-F.J., and C.-Y.K. designed the study and interpreted the data. C.-S.C., J.-W.R., C.-T.H., C.-M.L., Y.-T.T., and C.-Y.L. performed animal and isolation experiments. W.-H.Y., I.-J.L., H.-C.H., and H.-F.J. performed computational analysis. T.-H.T. provided D6KO mice. C.-S.C. and W.-H.Y. wrote the manuscript. J.-W.R., C.-C.S., T.-H.C., W.-J.L., H.-C.H., H.-F.J., and C.-Y.K. edited the manuscript. C.-Y.K. performed overall supervision of the project.
Declaration of interests
The authors declare no competing interests.
Published: February 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103738.
Supplemental information
. Clustering of intestinal CD45+ cells from WT mice and D6KO mice, related to Figures 1 and S1–S6 and Table 1
. DEGs of intestinal CD45+ cells from WT mice and D6KO mice, related to Figure 3
. GO analysis of intestinal CD45+ cells from WT mice and D6KO mice, related to Figure 4 and Table S4
. KEGG analysis of intestinal CD45+ cells from WT mice and D6KO mice, related to Figure 4 and Table S3
. modPhEA analysis of intestinal CD45+ cells from WT mice and D6KO mice, related to Figure 5
Data and code availability
-
•
Raw sequencing data files have been uploaded to NCBI underGene Expression Omnibus (GEO) accession number GSE176087 of BioProject PRJNA734865 (SRA: SRP322434).
-
•
The codes that are used in this study could be retrieved from https://github.com/wenhsuanyuu/Int-CD45-scRNAseq.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R., et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt B.E., Zhang M., Chen G.H., Huffnagle G.B., Kao J.Y. The role of dendritic cells in the development of acute dextran sulfate sodium colitis. J. Immunol. 2007;179:6255–6262. doi: 10.4049/jimmunol.179.9.6255. [DOI] [PubMed] [Google Scholar]
- Bertin S., Lozano-Ruiz B., Bachiller V., Garcia-Martinez I., Herdman S., Zapater P., Frances R., Such J., Lee J., Raz E., Gonzalez-Navajas J.M. Dual-specificity phosphatase 6 regulates CD4+ T-cell functions and restrains spontaneous colitis in IL-10-deficient mice. Mucosal Immunol. 2015;8:505–515. doi: 10.1038/mi.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Wang X., Chen Q., Sun X., Xiao F., Ko K.H., Zhang M., Lu L. B1a cells play a pathogenic role in the development of autoimmune arthritis. Oncotarget. 2016;7:19299–19311. doi: 10.18632/oncotarget.8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan B., Morel L. Role of B-1a cells in autoimmunity. Autoimmun Rev. 2006;5:403–408. doi: 10.1016/j.autrev.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Feng B., He Q., Xu H. FOXO1-dependent up-regulation of MAP kinase phosphatase 3 (MKP-3) mediates glucocorticoid-induced hepatic lipid accumulation in mice. Mol. CellEndocrinol. 2014;393:46–55. doi: 10.1016/j.mce.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B., Jiao P., Helou Y., Li Y., He Q., Walters M.S., Salomon A., Xu H. Mitogen-activated protein kinase phosphatase 3 (MKP-3)-deficient mice are resistant to diet-induced obesity. Diabetes. 2014;63:2924–2934. doi: 10.2337/db14-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flohe S.B., Agrawal H., Schmitz D., Gertz M., Flohe S., Schade F.U. Dendritic cells during polymicrobial sepsis rapidly mature but fail to initiate a protective Th1-type immune response. J. Leukoc Biol. 2006;79:473–481. doi: 10.1189/jlb.0705413. [DOI] [PubMed] [Google Scholar]
- Franzen O., Gan L.M., Bjorkegren J.L.M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database (Oxford) 2019;2019:baz046. doi: 10.1093/database/baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y., et al. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart A.L., Al-Hassi H.O., Rigby R.J., Bell S.J., Emmanuel A.V., Knight S.C., Kamm M.A., Stagg A.J. Characteristics of intestinal dendritic cells in inflammatory bowel diseases. Gastroenterology. 2005;129:50–65. doi: 10.1053/j.gastro.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Heiseke A.F., Faul A.C., Lehr H.A., Forster I., Schmid R.M., Krug A.B., Reindl W. CCL17 promotes intestinal inflammation in mice and counteracts regulatory T cell-mediated protection from colitis. Gastroenterology. 2012;142:335–345. doi: 10.1053/j.gastro.2011.10.027. [DOI] [PubMed] [Google Scholar]
- Hsu W.C., Chen M.Y., Hsu S.C., Huang L.R., Kao C.Y., Cheng W.H., Pan C.H., Wu M.S., Yu G.Y., Hung M.S., et al. DUSP6 mediates T cell receptor-engaged glycolysis and restrains TFH cell differentiation. Proc Natl. Acad. Sci.U S A. 2018;115:E8027–E8036. doi: 10.1073/pnas.1800076115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jojic V., Shay T., Sylvia K., Zuk O., Sun X., Kang J., Regev A., Koller D., Best A.J., Knell J., et al. Identification of transcriptional regulators in the mouse immune system. Nat. Immunol. 2013;14:633–643. doi: 10.1038/ni.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Scott D.A., Hatch E., Tian X., Mansour S.L. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development. 2007;134:167–176. doi: 10.1242/dev.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Yu M., Lee W.W., Tsang M., Krishnan E., Weyand C.M., Goronzy J.J. Decline in miR-181a expression with age impairs T cell receptor sensitivity by increasing DUSP6 activity. Nat.Med. 2012;18:1518–1524. doi: 10.1038/nm.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet M., Purcell N.H., Sargent M.A., York A.J., Bueno O.F., Molkentin J.D. DUSP6 (MKP3) null mice show enhanced ERK1/2 phosphorylation at baseline and increased myocyte proliferation in the heart affecting disease susceptibility. J. Biol. Chem. 2008;283:31246–31255. doi: 10.1074/jbc.M806085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D.J., Campbell K.R., Lun A.T., Wills Q.F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. NucleicAcids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares-Villagomez D., Van Kaer L. Intestinal intraepithelial lymphocytes: sentinels of the mucosal barrier. Trends Immunol. 2018;39:264–275. doi: 10.1016/j.it.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalexi E., Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat.Rev. Immunol. 2018;18:35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- Robinette M.L., Fuchs A., Cortez V.S., Lee J.S., Wang Y., Durum S.K., Gilfillan S., Colonna M. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J.W., Statt S., Huang C.T., Tsai Y.T., Kuo C.C., Chan H.L., Liao Y.C., Tan T.H., Kao C.Y. Dual-specificity phosphatase 6 deficiency regulates gut microbiome and transcriptome response against diet-induced obesity in mice. Nat. Microbiol. 2016;2:16220. doi: 10.1038/nmicrobiol.2016.220. [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M.P., Liao B.Y. modPhEA: model organism Phenotype Enrichment Analysis of eukaryotic gene sets. Bioinformatics. 2017;33:3505–3507. doi: 10.1093/bioinformatics/btx426. [DOI] [PubMed] [Google Scholar]
- Xiong N., Zhang L., Kang C., Raulet D.H. Gene placement and competition control T cell receptor gamma variable region gene rearrangement. J. Exp. Med. 2008;205:929–938. doi: 10.1084/jem.20071275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Ding J., Porter C.B.M., Wallrapp A., Tabaka M., Ma S., Fu S., Guo X., Riesenfeld S.J., Su C., et al. Transcriptional atlas of intestinal immune cells reveals that neuropeptide alpha-CGRP modulates group 2 innate lymphoid cell responses. Immunity. 2019;51:696–708.e9. doi: 10.1016/j.immuni.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
. Clustering of intestinal CD45+ cells from WT mice and D6KO mice, related to Figures 1 and S1–S6 and Table 1
. DEGs of intestinal CD45+ cells from WT mice and D6KO mice, related to Figure 3
. GO analysis of intestinal CD45+ cells from WT mice and D6KO mice, related to Figure 4 and Table S4
. KEGG analysis of intestinal CD45+ cells from WT mice and D6KO mice, related to Figure 4 and Table S3
. modPhEA analysis of intestinal CD45+ cells from WT mice and D6KO mice, related to Figure 5
Data Availability Statement
-
•
Raw sequencing data files have been uploaded to NCBI underGene Expression Omnibus (GEO) accession number GSE176087 of BioProject PRJNA734865 (SRA: SRP322434).
-
•
The codes that are used in this study could be retrieved from https://github.com/wenhsuanyuu/Int-CD45-scRNAseq.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.